Abstract
Shwachman-Diamond syndrome (SDS) is an autosomal recessively inherited disorder characterized by exocrine pancreatic insufficiency and bone marrow failure. The gene for this syndrome, SBDS, encodes a highly conserved novel protein. We characterized Shwachman-Bodian-Diamond syndrome (SBDS) protein expression and intracellular localization in 7 patients with SDS and healthy controls. As predicted by gene mutation, 4 patients with SDS exhibited no detectable full-length SBDS protein. Patient DF277, who was homozygous for the IVS2 + 2 T>C splice donor mutation, expressed scant levels of SBDS protein. Patient SD101 expressed low levels of SBDS protein harboring an R169C missense mutation. Patient DF269, who carried no detectable gene mutations, expressed wild-type levels of SBDS protein to add further support to the growing body of evidence for additional gene(s) that might contribute to the pathogenesis of the disease phenotype. The SBDS protein was detected in both the nucleus and the cytoplasm of normal control fibroblasts, but was particularly concentrated within the nucleolus. SBDS localization was cell-cycle dependent, with nucleolar localization during G1 and G2 and diffuse nuclear localization during S phase. SBDS nucleolar localization was intact in SD101 and DF269. The intranucleolar localization of SBDS provides further supportive evidence for its postulated role in rRNA processing.
Introduction
Shwachman-Diamond syndrome (SDS)1,2 is an autosomal recessive disease3 characterized by exocrine pancreatic insufficiency, bone marrow failure, and leukemia predisposition (reviewed in Dror and Freedman4 and Smith5 ). Additional features commonly associated with this syndrome include skeletal malformations,6 liver abnormalities, and short stature.4,5 Neutropenia is the most common manifestation of bone marrow failure in patients with SDS, although a subset of patients develops aplastic anemia. Lymphocyte defects7 and bone marrow stromal-cell dysfunction8 have also been described in patients with SDS. Thus, the disease phenotype is not restricted to myeloid cells but affects multiple hematopoietic lineages and multiple organ systems.
The Shwachman-Diamond gene, SBDS, was recently cloned.9 SBDS is located on chromosome 7 near the centromere.10,11 An adjacent pseudogene, SBDSP, shares a high degree of homology with SBDS but contains deletions and nucleotide changes that prevent the generation of a functional protein.9 The majority of patients (nearly 75%) with Shwachman-Diamond syndrome were found to harbor SBDS mutations arising from a gene conversion event with this pseudogene.9
The SBDS gene is predicted to encode a novel 250–amino acid protein with no homology to other proteins of known function. The SBDS gene is highly conserved throughout evolution, and orthologues can be found in organisms ranging from plants, Archaea, and yeast to vertebrate animals. The SBDS mRNA is widely expressed throughout human tissues9 ; however, little is yet known about SBDS protein expression patterns. Ubiquitous SBDS mRNA tissue expression, together with multiorgan involvement in patients with SDS, suggests that the SBDS protein likely functions in a global cellular process.
To further investigate the molecular pathways affected in SDS, we examined the SBDS protein in 7 patients with Shwachman-Diamond. The SBDS gene was sequenced for all 7 patients. Using an antibody generated against the carboxyl terminus of the SBDS protein, we assessed SBDS protein expression in cell lines derived from these 7 patients. We also examined the intracellular localization of the SBDS protein in wild-type and SDS patient-derived cells.
Materials and methods
Cell culture
Dana-Farber Cancer Institute (DFCI) Institutional Review Board–approved informed consent was obtained from participating patients.
Peripheral blood mononuclear cells from patients with Shwachman-Diamond syndrome followed at Children's Hospital or from healthy volunteers were isolated by centrifugation over a Histopaque 1077 (Sigma, St Louis, MO) cushion for 30 minutes at 250g. Samples were stripped of identifying information and assigned a code number to maintain patient confidentiality. Epstein-Barr virus–immortalized lymphoblast lines were established and maintained in RPMI 1640 containing 15% heat-inactivated fetal bovine serum (FBS; Sigma), 2 mM glutamine, 1% penicillin and streptomycin. Cell cultures were maintained in a humidified incubator containing 5% carbon dioxide at 37°C.
Primary fibroblast cultures were established from bone marrow aspirates and grown in Chang Medium D (Irvine Scientific, Santa Ana, CA).
SBDS gene sequencing
SBDS sequencing was performed on patient samples by GeneDx (GeneDx, Gaithersburg, MD).
SBDS antibody
A polyclonal antibody was generated against the terminal 18 amino acids of the SBDS protein (Ac-SLEVLNLKDVEEGDEKFE-amide) (BioSource, Hopkinton, MA). The peptide was injected into rabbits using standard techniques. The antibody was affinity purified over a column bearing the 18–amino acid peptide.
Retroviral infection
Fibroblasts or lymphoblasts were infected with a pBabe-puro retrovirus containing the SBDS cDNA as previously described.12
Western blotting
The Western blot protocol was modified from a previous protocol.13 Cells from lymphoblasts or primary fibroblasts were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in lysis bufferA(50 mM Tris [tris(hydroxymethyl) aminomethane] pH 6.8, 2% sodium dodecyl sulfate, and 86 mM 2-mercaptoethanol) or buffer B (NETN [50 mM Tris pH 7.4, 150 mM NaCl, 5 mM EGTA (ethylene glycol-bis-(B-aminoethyl ether)-N-N-N′-N′-tetraacetic acid)], 1% NP-40 (Nonidet P-40, [Octylphenoxy]polyethylene glycol ether), 2 × protease inhibitor tablet (Roche, Indianapolis, IN), 1 μM pepstatin (Roche) for 15 minutes on ice. If lysed in buffer B, the lysate was centrifuged at full speed in an Eppendorf Microfuge centrifuge (Eppendorf, Westbury, NY) for 20 minutes at 4°C. Protein concentration was determined using a Bradford (BioRad, Hercules, CA) or Lowry (BioRad) assay with a bovine serum albumen standard curve. For Western blots, 20 to 30 μg protein were loaded per lane on a 10% sodium dodecyl sulfate–polyacrylamide gel with a 5% stacking gel. The proteins were transferred to a nitrocellulose membrane for 45 minutes at 0.8 amps, 4°C in Tris-glycine buffer (25 mM Tris base, 200 mM glycine, 20% methanol). The filter was washed in TBST (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Tween 20), blocked for 1 hour in 5% nonfat dry milk in TBST at room temperature, and then incubated in primary antibody for 1 hour at room temperature or overnight at 4°C. The anti-SBDS antibody was incubated at a 1:10 000 dilution in TBST for the anti–C-terminal SBDS antibody. The antitubulin antibody (Sigma) was incubated at a 1:800 dilution in TBST for 1 hour at room temperature. The filter was then washed 3 times (10 minutes per wash) with TBST. The secondary antibody (anti–rabbit-horseradish peroxidase; Amersham, Piscataway, NJ) was added at a 1:10 000 dilution in 1% milk/TBST for 1 hour. The filter was washed 3 times (10 minutes per wash) in TBST followed by chemiluminescence.
Immunofluorescence
Primary fibroblasts were plated onto sterile coverslips. The following day, the cells were washed twice in PBS and fixed in 4% paraformaldehyde in PBS for 10 minutes. Following another 3 washes with PBS, the cells were permeabilized for 10 minutes with 0.3% Triton X-100 in PBS. The cells were washed 3 times in PBS and incubated with blocking buffer (10% goat serum, 0.1% NP-40 in PBS) for 1 hour. The anti-SBDS antibody (1:400 dilution in blocking buffer) and/or the C-23 antinucleolin antibody (1:800; Santa Cruz Biotechnology, Santa Cruz, CA) was added for 1 to 2 hours at room temperature. The cells were washed 5 times with 0.1% NP-40 in PBS, 5 minutes per wash, then incubated for 1 hour with the secondary fluorescein isothiocyanate (FITC)–labeled antirabbit antibody (1:1000 in blocking buffer) and/or Texas-red antimouse antibody (1:400) (Jackson Immunoresearch, West Grove, PA) in the dark. After 5 washes in 0.1% NP-40/PBS, the coverslips were mounted onto slides with Vectashield (Vector Laboratories, Burlingame, CA). The cells were visualized with a Zeiss Axioplan 2 microscope (Thornwood, NY), and digital images were obtained using OpenLab software (Improvision, Lexington, MA).
Confocal microscopy was performed with a Zeiss L5M510META NLO in the MRRC (Mental Retardation Research Center) Imaging Core at Children's Hospital, Boston.
Results
Characterization of SBDS-patient mutations
Seven patients fulfilled the diagnostic criteria for Shwachman-Diamond syndrome as outlined in a recent consensus statement from an expert panel.14 All patients had evidence of exocrine pancreatic insufficiency by fulfilling one of the following criteria: (1) direct pancreatic enzyme measurement following pancreatic stimulation, (2) low serum trypsinogen or isoamylase levels, or (3) abnormal fecal fat balance study without intestinal pathology and a pancreatic imaging study showing a small fatty pancreas. The patients had peripheral blood cytopenias as noted in Table 1. All patients had neutropenia, 2 patients had concomitant anemia, and 1 patient underwent a bone marrow transplantation for aplastic anemia with an isochromosome 7 clonal marrow population in the absence of overt myelodysplastic syndrome or acute myeloid leukemia.
The SBDS genes were sequenced for each patient (Table 1). The broad distribution of these mutations within the gene is illustrated in Figure 1A. Five of the patients exhibited mutations predicted to lead to premature protein truncation. The intron 2 donor splice site mutation is the most common mutation associated with SDS9 and was seen in 6 of the patients in our cohort. Patient DF277 was homozygous for this mutation, whereas the other 5 patients harbored different mutations on the second allele. Of note, patient SD101 harbored a 505C>T missense mutation, leading to an amino acid substitution of a cysteine for an arginine at amino acid position 169. Patient DF269 was particularly interesting. Although this patient met the clinical diagnosis for SDS, he had no identified mutations in the SBDS gene. Some types of mutations, such as promotor mutations, gene inversions, or large gene deletion mutations, would not be detected by the exon-directed gene sequencing approach used here.
Patients with SDS exhibit variable levels of SBDS protein expression
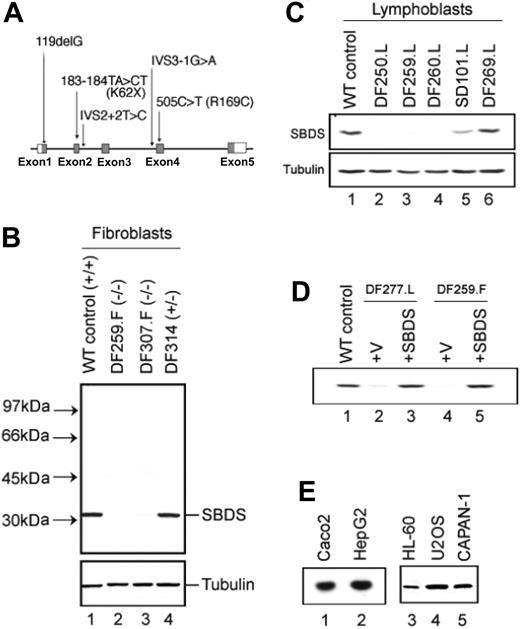
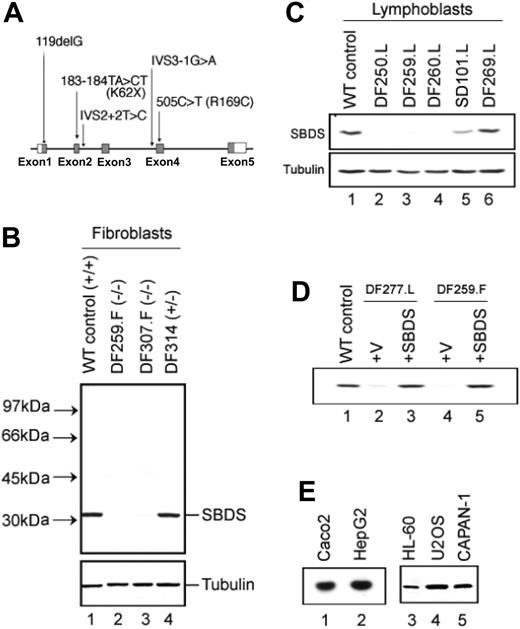
To assess the effects of these mutations on SBDS protein expression, a rabbit polyclonal antibody was generated against an SBDS C-terminal peptide. The ability of the antibody to recognize SBDS protein was first validated against epitope-tagged SBDS cDNA-encoded proteins as well as against glutathione-S-transferase (GST)–tagged recombinant SBDS expressed in bacteria (data not shown). We next used this antibody to assess endogenous SBDS protein expression. Lysates from normal control fibroblasts were analyzed by immunoblotting with an antibody directed against the C-terminus of SBDS. As shown in Figure 1B, lane 1, a protein of slighter more than 30 kDa was detected in normal control lysates. This size was close to the predicted size of 28.8 kDa based on the protein sequence and correlated with findings by Woloszynek et al.15 SBDS protein expression was noted in all cell types examined, including lymphocytes (Figure 1C, lane 1), myeloid cells (HL-60), pancreatic cells (CAPAN-1), osteosarcoma cells (U2OS), liver cells (HepG2), and intestinal epithelial cells (Caco2) (Figure 1E, lanes 1-5).
Patients with SDS express variable levels of SBDS protein. (A) Schematic diagram of SBDS mutations. The locations of the SBDS mutations identified in our patient series are diagramed (not drawn to scale). The SBDS gene is composed of 5 exons (depicted with boxes). The translated mRNA regions are shaded gray. (B) SBDS expression in fibroblasts. Bone marrow fibroblast protein lysates were analyzed by immunoblotting for SBDS protein (top). The SBDS genotypes of each cell line are indicated in parentheses. The blot was also probed for tubulin to ascertain equivalent protein loading (bottom). DF259 and DF307 harbored SBDS mutations predicted to lead to premature translation termination. The wild-type (WT) control (NMF-100) is a marrow fibroblast cell line derived from a healthy control patient. (C) SBDS expression in lymphoblasts. Lymphoblast protein lysates were analyzed by immunoblotting for SBDS protein (top) and tubulin (bottom). DF250, DF259, and DF260 carried SBDS mutations predicted to lead to premature translation termination. SD101 carried a missense (R169C) mutation on 1 SBDS allele. DF269 is derived from a patient with SDS who lacked any identified SBDS mutations. (D) Retroviral transduction of the SBDS cDNA restores SBDS protein expression. A lymphoblast (DF277.L) or fibroblast (DF259.F) SDS cell line was infected with a pBabe retrovirus containing a wild-type SBDS cDNA (lanes 3 and 5) or empty vector (lanes 2 and 4). Cell lysates were immunoblotted for SBDS protein. A normal lymphoblast control cell line (DF213) was run in lane 1 (WT control). (E) SBDS protein is expressed in a variety of tissues. Lysates from the indicated human cell lines were immunoblotted for SBDS protein expression. Caco2 indicates intestinal epithelial cell line; HepG2, hepatocellular carcinoma cell line; HL-60, myeloid leukemia cell line; U2OS, osteosarcoma cell line; CAPAN-1, pancreatic adenocarcinoma cell line.
Patients with SDS express variable levels of SBDS protein. (A) Schematic diagram of SBDS mutations. The locations of the SBDS mutations identified in our patient series are diagramed (not drawn to scale). The SBDS gene is composed of 5 exons (depicted with boxes). The translated mRNA regions are shaded gray. (B) SBDS expression in fibroblasts. Bone marrow fibroblast protein lysates were analyzed by immunoblotting for SBDS protein (top). The SBDS genotypes of each cell line are indicated in parentheses. The blot was also probed for tubulin to ascertain equivalent protein loading (bottom). DF259 and DF307 harbored SBDS mutations predicted to lead to premature translation termination. The wild-type (WT) control (NMF-100) is a marrow fibroblast cell line derived from a healthy control patient. (C) SBDS expression in lymphoblasts. Lymphoblast protein lysates were analyzed by immunoblotting for SBDS protein (top) and tubulin (bottom). DF250, DF259, and DF260 carried SBDS mutations predicted to lead to premature translation termination. SD101 carried a missense (R169C) mutation on 1 SBDS allele. DF269 is derived from a patient with SDS who lacked any identified SBDS mutations. (D) Retroviral transduction of the SBDS cDNA restores SBDS protein expression. A lymphoblast (DF277.L) or fibroblast (DF259.F) SDS cell line was infected with a pBabe retrovirus containing a wild-type SBDS cDNA (lanes 3 and 5) or empty vector (lanes 2 and 4). Cell lysates were immunoblotted for SBDS protein. A normal lymphoblast control cell line (DF213) was run in lane 1 (WT control). (E) SBDS protein is expressed in a variety of tissues. Lysates from the indicated human cell lines were immunoblotted for SBDS protein expression. Caco2 indicates intestinal epithelial cell line; HepG2, hepatocellular carcinoma cell line; HL-60, myeloid leukemia cell line; U2OS, osteosarcoma cell line; CAPAN-1, pancreatic adenocarcinoma cell line.
Fibroblast cell lines derived from patients with SDS were then analyzed for SBDS protein expression. As expected, SBDS protein was not readily discernible in DF259 and DF307, who harbored biallelic SBDS gene mutations predicted to lead to protein truncation (Figure 1B, lanes 2 and 3). Equivalent protein loading was ascertained by both Ponceau stain and by probing the same blot with an antibody against tubulin as an internal standard (Figure 1B, bottom). Since the antibody is directed against the carboxyl terminus of SBDS, the production of smaller truncated SBDS mutant proteins cannot be ruled out. Western blots of lymphocytes from patients DF250, DF259, and DF 260 also lacked full-length SBDS protein (Figure 1C, lanes 2-4). A small quantity of SBDS protein expression was detectable in lysates from DF277 lymphoblasts (Figure 1D, lane 2). This patient was homozygous for mutations at the IVS2 + 2 T>C intron 2 splice donor site. A trace amount of SBDS protein could sometimes be discerned in some SDS cell lines wherein one affected allele bore the IVS2 + 2T>C splice site mutation. This mutation could potentially produce a small quantity of alternatively spliced SBDS mRNA encoding a functional protein, and the additive production from 2 alleles in DF277 likely facilitated the detection of this scant SBDS protein product. A recent study of the yeast SBDS orthologue, YLR022C, suggested that pathogenic SBDS mutations were likely to represent the effects of hypomorphic alleles and that complete loss of SBDS function was likely to be lethal.16 This observation provides a possible rationale for the prevalence of this splice site mutation in patients with SDS. In further support of the antibody specificity, the introduction of a wild-type SBDS cDNA into the DF259 fibroblast cell line or the DF277 lymphocyte cell line restored SBDS protein expression (Figure 1D). DF314 is a cell line derived from a 6-year-old patient who had exocrine pancreatic insufficiency but no significant marrow failure and was heterozygous for SBDS mutations (IVS2 + 2 T>C on one allele, no mutations detected on the other allele). DF314 expressed wild-type levels of SBDS protein (compare Figure 1B, lanes 1 and 4).
Patient SD101 harbored the arginine to cysteine point mutation in the SBDS gene, and this patient expressed reduced levels of SBDS protein (Figure 1C, lane 5). Interestingly, Patient DF269, for whom no SBDS gene mutation was detected by sequencing, expressed SBDS protein at levels comparable to that observed in wild-type control cells (compare Figure 1C, lanes 1 and 6). The presence of normal levels of full-length protein ruled out the presence of other undetected gene mutations that might affect protein expression. It was possible that this patient harbors a defect in a different gene contributing to the SDS molecular pathway. Another possibility was that there might be a separate second molecular pathway that culminates in a phenotype similar to SDS.
SBDS protein is located in both the nucleus and the cytoplasm but particularly concentrated within the nucleolus
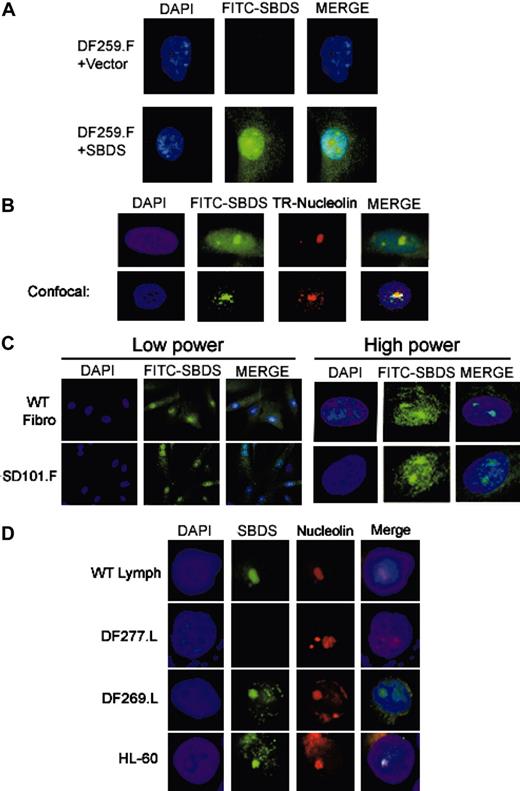
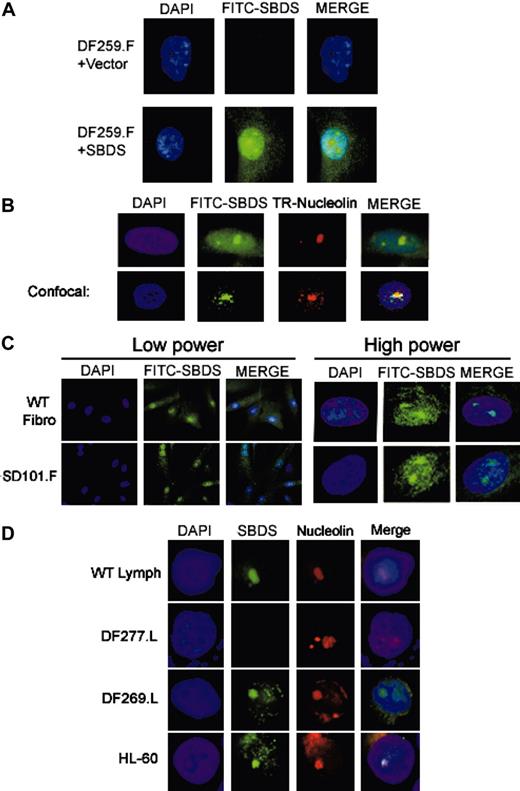
We next investigated the intracellular localization of SBDS protein by immunofluorescence. We first validated the antibody specificity in this assay by probing DF259 cells, which lack detectable SBDS protein expression by Western blot. No SBDS protein was detected by immunofluorescence in either DF259.F fibroblasts infected with empty vector retrovirus (Figure 2A, top) or uninfected DF259.F cells (data not shown). SBDS expression was restored following stable infection with a retrovirus containing a wild-type SBDS cDNA (Figure 2A, bottom). SBDS protein was present throughout the cell in both the cytoplasm and the nucleus, but it was most prominent within subregions of the nucleus corresponding to the nucleolus by DAPI stain in the majority of cells.
To confirm whether endogenous SBDS protein exhibited a similar localization pattern, we probed normal control fibroblasts (Figure 2C, top) or HeLa cells (Figure 2B, top) with the anti-SBDS antibody. Again, endogenous SBDS was localized throughout the cell but was concentrated within the nucleolus in the majority of cells. To confirm the nucleolar localization of SBDS, HeLa cells were simultaneously probed with an antibody against nucleolin (reviewed in Ginisty et al17 and Srivastava and Pollard18 ) (Figure 2B, top). Cells were also analyzed by confocal microscopy (Figure 2B, bottom) to ascertain the colocalization of SBDS and nucleolin. Wild-type lymphocyte controls also exhibited nucleolar SBDS localization (Figure 2D). This pattern of localization was not restricted to fibroblasts and lymphocytes since a similar pattern of SBDS localization was also observed in a myeloid cell line (Figure 2D).
SBDS localizes to the nucleolus. (A) Retroviral SBDS transduction restores SBDS expression by immunofluorescence. DF259.F fibroblasts infected with retrovirus containing empty vector (top) or SBDS cDNA (bottom) were fixed and probed with the anti-SBDS antibody followed by a FITC-labeled secondary antibody. The cells were counterstained with DAPI (4′, 6-diamino-2-phenylindole) to visualize the nuclei. SBDS was located throughout the cell but was particularly prominent in the region corresponding to the nucleolus. (B) Endogenous SBDS localizes to the nucleolus. HeLa cells were fixed and probed with both an anti-SBDS polyclonal antibody and an antinucleolin monoclonal antibody. SBDS was detected with an antirabbit FITC-labeled secondary antibody, and nucleolin was detected with an antimouse Texas-Red (TR) secondary antibody. Cells were visualized by standard fluorescence microscopy (top) or by confocal microscopy (bottom). (C) SBDS nucleolar localization is intact in SD101 fibroblasts. WT control fibroblasts (NMF-100) or SD101.F fibroblasts were analyzed by immunofluorescence for SBDS. Low power, × 63 magnification; high power, × 100 magnification. (D) SBDS localizes to the nucleolus in DF269 lymphoblasts and in myeloid cells. The indicated lymphoblast cell lines or the myeloid leukemia HL-60 cell lines were subjected to immunofluorescent staining for SBDS protein and nucleolin. Nuclei were counterstained with DAPI.
SBDS localizes to the nucleolus. (A) Retroviral SBDS transduction restores SBDS expression by immunofluorescence. DF259.F fibroblasts infected with retrovirus containing empty vector (top) or SBDS cDNA (bottom) were fixed and probed with the anti-SBDS antibody followed by a FITC-labeled secondary antibody. The cells were counterstained with DAPI (4′, 6-diamino-2-phenylindole) to visualize the nuclei. SBDS was located throughout the cell but was particularly prominent in the region corresponding to the nucleolus. (B) Endogenous SBDS localizes to the nucleolus. HeLa cells were fixed and probed with both an anti-SBDS polyclonal antibody and an antinucleolin monoclonal antibody. SBDS was detected with an antirabbit FITC-labeled secondary antibody, and nucleolin was detected with an antimouse Texas-Red (TR) secondary antibody. Cells were visualized by standard fluorescence microscopy (top) or by confocal microscopy (bottom). (C) SBDS nucleolar localization is intact in SD101 fibroblasts. WT control fibroblasts (NMF-100) or SD101.F fibroblasts were analyzed by immunofluorescence for SBDS. Low power, × 63 magnification; high power, × 100 magnification. (D) SBDS localizes to the nucleolus in DF269 lymphoblasts and in myeloid cells. The indicated lymphoblast cell lines or the myeloid leukemia HL-60 cell lines were subjected to immunofluorescent staining for SBDS protein and nucleolin. Nuclei were counterstained with DAPI.
We next assessed SBDS localization in patient SD101, who carried a R169C SBDS missense mutation. By immunofluorescence, the R169C SBDS mutant protein was still able to localize to the nucleolus (Figure 2C, bottom). Quantitation of the percentage of cells with SBDS nucleolar localization failed to show any significant difference between SD101 and a wild-type control cell line (data not shown). We also examined DF269 cells, which were derived from a patient with SDS lacking SBDS mutations. It was possible that the defect in this patient might involve pathways affecting SBDS localization; however, SBDS was observed in the nucleolus in DF269 cells (Figure 2D). A slightly more prominent cytoplasmic localization was generally noted in the DF269 cells; however, quantitation of the percentage of DF269 cells exhibiting nucleolar SBDS localization did not show any significant difference from that observed in the wild-type lymphocyte controls (data not shown).
SBDS nucleolar localization is cell-cycle dependent
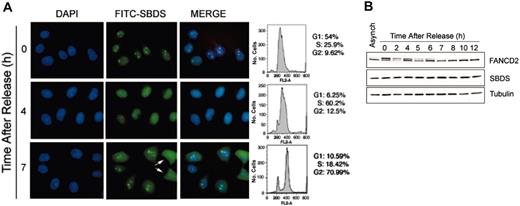
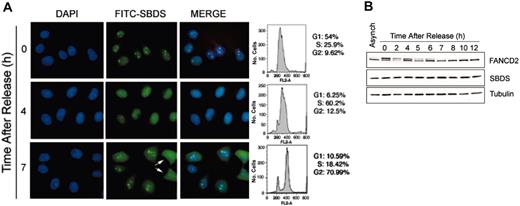
The observation that SBDS was localized to the nucleolus in only a subset of cells (approximately 70%-80%) raised the intriguing possibility that SBDS intracellular localization was dynamic, as is the case for many nucleolar proteins.19 To investigate the regulation of SBDS localization, we visualized SBDS in synchronized HeLa cells following release from a double thymidine block. The cell-cycle profile was analyzed by flow cytometry (Figure 3A). SBDS protein was localized to the nucleolus when cells were predominantly in the G1 or G2 phases of the cell cycle (Figure 3A, 0 hour and 7 hours). SBDS became diffusely localized throughout the nucleus when cells were predominantly in S phase (Figure 3A, 4 hours). The nucleolus is disassembled during mitosis,20,21 and, accordingly, SBDS nucleolar localization was also lost during that time. A pair of cells in telophase can be seen on the right-hand side of Figure 3A at 7 hours (arrows). SBDS is diffusely dispersed throughout the mitotic cells, although some reaccumulation within the nucleolus could be detected. The same cell-cycle–dependent pattern of SBDS intracellular localization was observed following synchronization with mimosine (data not shown).
SBDS nucleolar localization is cell-cycle dependent. (A) HeLa cells were synchronized with a double thymidine block, then released for the times indicated. Cells were fixed and stained for SBDS. An aliquot of cells was also fixed for flow cytometry (right). Cells in late telophase are indicated with arrows. (B) SBDS protein levels remain constant throughout the cell cycle. HeLa cells synchronized with a double thymidine block were released for the indicated times. Cell lysates were immunoblotted for Fanconi anemia, complementation group D2 (FANCD2), SBDS, and tubulin.
SBDS nucleolar localization is cell-cycle dependent. (A) HeLa cells were synchronized with a double thymidine block, then released for the times indicated. Cells were fixed and stained for SBDS. An aliquot of cells was also fixed for flow cytometry (right). Cells in late telophase are indicated with arrows. (B) SBDS protein levels remain constant throughout the cell cycle. HeLa cells synchronized with a double thymidine block were released for the indicated times. Cell lysates were immunoblotted for Fanconi anemia, complementation group D2 (FANCD2), SBDS, and tubulin.
We also assessed SBDS protein levels by Western blot at different phases of the cell cycle (Figure 3B). The blot was also probed for tubulin to confirm equivalent protein loading. FANCD2 was simultaneously immunoblotted as a marker for progression through the cell cycle.22 FANCD2 is predominantly monoubiquitinated following a double thymidine block and is gradually deubiquitinated as the cells are released to progress through the cell cycle.22 As shown by Western blot analysis, SBDS protein expression remained constant throughout the cell cycle (Figure 3B).
Discussion
We characterized a rabbit polyclonal antibody raised against the carboxyl terminus of SBDS. Using this antibody, we have identified wild-type SBDS as an approximately 31-kDa protein located throughout the cell but primarily in the nucleus with accumulation in the nucleolus in a cell-cycle–dependent pattern. Protein expression levels varied between patients with SDS in a manner corresponding with that predicted from the associated SBDS gene mutations.
The data from patient DF269 was provoking. This patient fulfilled the current diagnostic criteria for SDS, yet carried no SBDS gene mutations and expressed normal levels of SBDS protein. Furthermore, SBDS nucleolar localization is intact in this patient. It is possible that DF269 represents a different disease process with phenotypic features that are similar to SDS. Another possibility is that this patient could harbor a mutation in a second gene involved in the pathogenesis of Shwachman-Diamond syndrome. The identification of a cellular phenotype or biochemical pathway for Shwachman-Diamond syndrome would facilitate further investigation into this question. Additional patients with SDS lacking identifiable SBDS gene mutations have been previously described,9,15 and SBDS protein expression has been documented in some of these patients.15
Immunofluorescence studies showed that SBDS was localized to both the nucleus and the cytoplasm but was particularly concentrated within the nucleolus. The nucleolus is best known as a site of ribosome biogenesis. RNA processing factors are localized in the nucleolus, which is intriguing because the SBDS protein has been postulated to play a role in RNA processing based on data from SBDS orthologues.9 The yeast orthologue of SBDS clusters with RNA processing genes in microarray expression profiles.23 Yeast microarray studies of noncoding RNAs revealed a potential defect in rRNA processing in the yeast mutant strain lacking the SBDS ortholog YLR022C.24 The Archaeal orthologue resides within conserved operons encoding RNA processing genes.25 The Arabidopsis orthologue has an extended C-terminus that contains an RNA-binding motif.9 More direct evidence is provided by a recent study in yeast, where the SBDS orthologue YLR022C protein was shown to be associated with other proteins involved in ribosome biosynthesis.26 Genetic interactions between the yeast nonessential gene YHR087W, which shares structural similarities to YLR022C, and other rRNA processing genes, have also been reported.26 Our studies describing the intranucleolar accumulation of SBDS in human cells provide further supportive evidence for a role for SBDS in RNA processing.
The shuttling of SBDS protein in and out of the nucleolus raises the intriguing possibility that its localization may play an important role in SBDS protein function. To date, neither of the 2 patients with SDS who produce SBDS protein manifested any gross defect in SBDS nucleolar localization. Whether the regulation of SBDS nucleolar localization is disrupted in these patients is currently under investigation. Further systematic mutational analysis of the SBDS protein will help clarify this question. The dynamic state of the nucleolus was demonstrated in a recent study of nucleolar proteomic analysis following different stressors.19 Recent studies have implicated the nucleolus in cellular stress responses27 and cell-cycle regulation.28 An intriguing discovery from proteomic studies of the nucleolus29-31 was that approximately 30% of nucleolar proteins constituted either novel or uncharacterized proteins. Such data raise the possibility that the nucleolus may harbor hitherto unappreciated molecular functions. The question of whether SBDS might contribute to cellular processes in addition to rRNA processing remains to be explored.
The nucleolus and RNA processing have been implicated in other inherited marrow failure syndromes. A defect in the RNA component of RNAse MRP is responsible for cartilage-hair hypoplasia,32 which shares clinical features of SDS, including metaphyseal dysostosis, marrow failure, and short stature. Another inherited marrow failure syndrome, dyskeratosis congenita,33 is associated with mutations in DKC1 that encodes the nucleolar protein, dyskerin.34 Dyskerin shares some homology with RNA pseudouridine synthases, which play a role in the isomerization of uridine bases in ribosomal RNAs. The role of dyskerin in rRNA processing is not clear, because dyskerin also associates with telomerase RNA and affects telomerase function.35 Interestingly, telomerase has also been observed in the nucleolus,36 and the telomeres in patients with SDS are short.37 It is unknown whether telomere shortening contributes to disease pathogenesis or merely represents a secondary effect of marrow failure because shortened telomeres are a common feature of inherited and acquired marrow failure.38-42 One of the genes responsible for the inherited marrow failure syndrome, Diamond-Blackfan anemia (DBA), is the gene encoding the ribosomal protein RSP19. RPS19 has recently been shown to be localized to the nucleolus of normal cells, but it can be mislocalized when it carries pathogenic mutations.43 Further investigation of SBDS protein function may provide insights into the emerging molecular themes underlying general pathogenic mechanisms of the inherited marrow failure syndromes.
Prepublished online as Blood First Edition Paper, April 28, 2005; DOI 10.1182/blood-2005-02-0807.
Supported by the National Institutes of Health (grants 1 K23 HL68632-01 and 1 R01 HL079582-01) (A.S.) and the Gracie Fund.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank David Nathan for critical discussions and advice throughout this project. We thank Shannon MacKeigan for assistance with patient samples and Matthew Salanga for performing the confocal microscopy in the Children's Hospital Imaging Core. We thank Jim Huang for helpful discussions. We thank Dr Richard Grand and Dr Robert Montgomery for the Caco2 and HepG2 cells. We are especially grateful for the support of the Gracie Fund.