Abstract
Interferon regulatory factor-8 (IRF-8)/interferon consensus sequence–binding protein (ICSBP) is a transcription factor that controls myeloid-cell development. Microarray gene expression analysis of Irf-8-/- myeloid progenitor cells expressing an IRF-8/estrogen receptor chimera (which differentiate into macrophages after addition of estradiol) was used to identify 69 genes altered by IRF-8 during early differentiation (62 up-regulated and 7 down-regulated). Among them, 4 lysosomal/endosomal enzyme-related genes (cystatin C, cathepsin C, lysozyme, and prosaposin) did not require de novo protein synthesis for induction, suggesting that they were direct targets of IRF-8. We developed a reporter assay system employing a self-inactivating retrovirus and analyzed the cystatin C and cathepsin C promoters. We found that a unique cis element mediates IRF-8–induced activation of both promoters. Similar elements were also found in other IRF-8 target genes with a consensus sequence (GAAANN[N]GGAA) comprising a core IRF-binding motif and an Ets-binding motif; this sequence is similar but distinct from the previously reported Ets/IRF composite element. Chromatin immunoprecipitation assays demonstrated that IRF-8 and the PU.1 Ets transcription factor bind to this element in vivo. Collectively, these data indicate that IRF-8 stimulates transcription of target genes through a novel cis element to specify macrophage differentiation.
Introduction
Transcription factors play a major role in orchestrating the gene expression programs governing hematopoietic-cell differentiation.1 Dysregulation of such programs can result in hematopoietic disorders such as leukemia.2
Interferon regulatory factor (IRF)-8/interferon consensus sequence–binding protein (ICSBP) is a hematopoietic transcription factor belonging to the IRF family.3 A series of studies performed with Irf-8-/- mice have indicated that this factor plays critical roles in the development and function of immune cells such as macrophages, dendritic cells, and B cells.4-8 Consistent with its developmental role, IRF-8 expression is initiated in hematopoietic progenitors and increases during the development of the above-mentioned immune cells.7,9
In myeloid progenitor cells, IRF-8 controls lineage selection by stimulating macrophage differentiation while inhibiting the growth and differentiation of granulocytes that are another myeloid-progeny cell type.9,10 This finding partially explains why Irf-8-/- mice develop a chronic myelogenous leukemia (CML)–like syndrome with expansion of granulocytes and impaired macrophage development.4,9,11 In addition, IRF-8 was shown to inhibit the activity of breakpoint cluster region/abelson murine leukemia (Bcr/Abl), which is the causal oncoprotein of CML, both in vitro and in vivo.12-14 Expression of IRF-8 is severely impaired in human patients with CML and acute myelogenous leukemia (AML), providing additional evidence that IRF-8 is a critical antileukemogenic factor in myeloid cells.15,16 We previously showed that the c-Myc repressors, B-lymphocyte–induced maturation protein-1 (Blimp-1) and mitogenic Ets transcriptional suppressor (METS), and the cyclin-dependent kinase inhibitor, cyclin-dependent kinase inhibitor 2B (CDKN2B/INK4B), are direct target genes of IRF-8 in differentiating myeloid progenitors.13,17 However, to our knowledge, there is no report on a genome-wide gene expression study to identify IRF-8–regulated genes in differentiating myeloid cells.
IRF-8 acts either as an activator or a repressor, depending on interacting factors and target DNA elements.3 For example, IRF-8 interacts with IRF-1 or IRF-2 to repress transcription of genes such as interferon (IFN)–stimulated gene-15 and 2′5′-oligoadenylate synthetase, which both contain an IFN-stimulated response element (ISRE; A/BNGAAANNGAAACT).18-20 It has been suggested that in immune cells, a subset of ISREs called the Ets/IRF response element (EIRE; GGAAANNGAAA), is activated by IRF-8 in cooperation with the IRF-8–interacting Ets transcriptional activator, PU.1.21 Another DNA element activated by IRF-8 and PU.1 is the Ets/IRF composite element (EICE; GGAANNGAAA), which is found in various genes such as those encoding immunoglobulin (Ig) κ and λ, glycoprotein (gp) 91phox, scavenger receptor, toll-like receptor-4, and interleukin-1β.22-25 Since PU.1 is essential for macrophage development,26 IRF-8 may exert its activity through the EICE and EIRE to stimulate macrophage differentiation. However, it has been unclear whether IRF-8 regulates endogenous genes through these elements during macrophage differentiation. Indeed, the molecular mechanism by which IRF-8 regulates myeloid development is still poorly understood.
In this study, we performed complementary DNA (cDNA) microarray analysis using an in vitro differentiation system in which Irf-8-/- myeloid progenitor cells undergo macrophage differentiation upon introduction of IRF-8. We identified 69 genes regulated by IRF-8 in an early phase of differentiation, including 4 novel direct target genes. Furthermore, we identified a unique target DNA element (GAAANN[N]GGAA) that mediates IRF-8 induction of target genes during macrophage differentiation. This element consists of a core IRF binding site and an Ets binding site, with the polarity opposite of that in the EICE and EIRE sequences. Finally, we showed that IRF-8 binds to this element in vivo, concomitant with an increased binding of PU.1.
Materials and methods
Cells and retroviral transduction
The establishment and culture conditions of cell line Tot2, a granulocyte macrophage–colony-stimulating factor (GM-CSF)–dependent myeloid progenitor-cell line derived from an Irf-8-/- mouse were described previously.10 Murine stem cell virus (MSCV)–CD8t vectors carrying IRF-8, IRF-8/estrogen receptor (ER), and ER were constructed by inserting the relevant cDNAs into the EcoRI site of MSCV-CD8t.27 Other MSCV retrovirus vectors and transduction procedures for Tot2 cells and bone marrow lineage–negative (lin-) cells were as described previously.9,10,13 Peritoneal macrophages were harvested from mice that were intraperitoneally injected with 3 mL of 3% thioglycollate (Difco, Detroit, MI) 4 days earlier. All animal work conformed to the guidelines of the animal care and use committee of the National Institute of Child Health and Human Development. Morphologic differentiation was monitored by Wright-Giemsa staining of cytospin preparations using Eclipse E400 (Nikon, Melville, NY).
Microarray analysis
Tot2 cells transduced with MSCV-ER-puro or MSCV-IRF-8/ER-puro were treated with β-estradiol for 4 hours, and total RNA was purified using the Trizol reagent (Invitrogen, Carlsbad, CA). The NIA 15K mouse cDNAs microarray, probe preparation, and hybridization were as previously described.28 Briefly, the cDNA probe was prepared from 300 μg total RNA by reverse transcription in the presence of α-[33P] deoxycytidine triphosphate (dCTP). Hybridization with the purified probe was carried out overnight at 65°C with prehybridized nylon membranes spotted with 15 247 mouse developmental cDNA clones. After washing, membranes were exposed to a phosphoscreen, and the screen was scanned by a phosphorimager (Storm860; Amersham Pharmacia, Piscataway, NJ). Images were analyzed with the IMAGEQUANT 5.0 software package (Amersham Pharmacia). The expression levels of each gene were expressed in arbitrary units after subtraction of the background. Two independent experiments were used to identify differentially expressed genes with a statistical significance of P less than .05 by t test.
Semiquantitative and quantitative polymerase chain reactions with reverse transcription
Reverse transcription (RT) and semiquantitative and quantitative polymerase chain reactions (PCRs) were performed as previously described.13 The amount of cDNA appropriate for each reaction was determined prior to actual experiments. cDNA derived from 100 ng of total RNA was used for amplification of the scavenger receptor (SR), cDNA from 25 ng for c-fms and protein kinase C (PKC) δ, and cDNA from 2 ng for the other genes. The following primers were used: cystatin C (sense 5′-gaggcagatgccaatgaggaag-3′ and antisense 5′-actgcaagaagagtggagccag-3′); lysozyme M (5′-cctgctttctgtcactgctcag-3′ and 5′-ctccgcagttccgaatatactg-3′); cathepsin C (5′-cccgaagcgacattaactgctc-3′and 5′-ggccacttctccttatcagatc-3′), prosaposin (5′-ggagatccttcattacctggag-3′ and 5′-tgacctcattacagaagccagc-3′), and PKCδ (5′-ctgtgctgtgaagatgaaggag-3′ and 5′-cctgcatttgtagccttgcttg-3′). The primers for the other tested genes were as described elsewhere.10 Real-time fluorescent quantitative PCR was performed in duplicate using the SYBR green PCR master kit (Applied Biosystems, Foster City, CA) and an ABI PRISM 7000 sequence detection system (Applied Biosystems). cDNAs derived from 2 ng of total RNA were used for PCR. The primer sets used were: cystatin C (5′-gcgttggacttcgctgtga-3′ and 5′-ggctgtggtacgcatcgtt-3′); cathepsin C (5′-ccgggcattttaccctcat-3′ and 5′-ttgaaaaacgcaaaccatttgt-3′); lysozyme M (5′-ttagctcagcacgagagcaatt-3′ and 5′-gctgcgcttcagctcgtt-3′); and c-fms (5′-gatgaggcctgtctcgacttct-3′and 5′-ctggcctctttgtccagatctt-3′). The primers for the other examined genes were as described.10 Each primer sets gave a unique product, and transcript levels were normalized against the expression levels of glyceraldehyde-3′-phosphate dehydroge-nase (GAPDH).
Reporter assay
For generation of reporter constructs, we first replaced the neomycin resistance gene in the self-inactivating retrovirus pSIR (Clontech, Palo Alto, CA) with a puromycin resistance gene. We then inserted the green fluorescent protein (GFP) cDNA along with a new multiple cloning site (MCS) containing XhoI and HindIII sites into the original MCS to generate retroviral reporter SIRV-GFP. The DNA fragments used for this construction were generated from the appropriate plasmids by restriction enzyme digestion or PCR using Pfu DNA polymerase (Stratagene, La Jolla, CA). The utilized GFP was a destabilized form known as d2EGFP (Clontech) that has a half-life of 2 hours. We used the QuikChange site-directed mutagenesis kit (Stratagene) to introduce a point mutation abolishing a HindIII site within the d2EGFP cDNA without altering the amino acid sequence, thus enabling us to use the HindIII site for the new MCS. The cystatin C and cathepsin C promoter regions were PCR amplified using Pfu polymerase and the genomic DNA of Tot2 cells, and the amplified fragments were inserted into SIRV-GFP. Mutants were generated by PCR using primers that introduced the desired mutations and relevant restriction enzyme sites for ligation. The nucleotide sequences of all constructs containing PCR-derived DNA fragments were confirmed by sequencing. For reporter assay, cells were transduced with SIRV-GFP reporter constructs, selected with puromycin, and transduced with MSCV-CD8t vectors. Cells were then either purified by immunomagnetic cell sorting, or directly stained with anti–human CD8 conjugated with Cy-Chrome (BD Pharmingen, San Diego, CA) and analyzed by FACSCalibur (BD Biosciences, Mountain View, CA) to acquire GFP signals in CD8+ cells. The data were analyzed with the CellQuest software (BD Biosciences).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to manufacturer's instructions with slight modifications. Briefly, 6.5 × 106 cells were treated with 1% formaldehyde for 30 minutes at room temperature to crosslink proteins and genomic DNA. After washing, cells were lysed in 650 μL sodium dodecyl sulfate (SDS) lysis buffer, and then sonicated (20 seconds each, 10 times at 15% of maximum power on wet ice using an XL2007 sonicator; Misonix, Farmingdale, NY) to shear the genomic DNA into 200- to 1000–base pair (bp) fragments. After centrifugation, 100 μL supernatant was diluted with 1 mL ChIP Dilution Buffer. Chromatin was precleared with 80 μL of protein A/G agarose–25% slurry (Santa Cruz Biotechnology, Santa Cruz, CA) supplemented with salmon sperm DNA (200 μg/mL; Invitrogen) for 30 minutes at 4°C. Precleared chromatin was incubated with 5 μg of normal goat immunoglobulin G (IgG), normal rabbit IgG, goat anti–IRF-8 antibody (C-19; Santa Cruz Biotechnology) or rabbit anti-PU.1 antibody (T-21; Santa Cruz Biotechnology) overnight at 4°C with rotation. Protein A/G–25% slurry with salmon sperm DNA (60 μL) was added and samples were rotated for 1 hour at 4°C to collect the antibody-chromatin complexes. Agarose-antibody-chromatin complexes were then washed twice with each of the following in order: low salt wash buffer, high salt wash buffer, LiCl wash buffer, and 10 mM tris(hydroxymethyl)aminomethane (Tris)–HCl (pH 8), 1 mM ethylenediamine-tetraacetic acid (EDTA). After elution of the chromatin complexes, the crosslink was reversed, and RNA and proteins were digested with RNase and proteinase K, respectively. DNA was then recovered by phenol/chloroform extraction followed by ethanol precipitation, and resuspended in 100 μL of 5 mM Tris, pH 8. Each sample (5 μL) was used for quantification of the specific region of genomic DNA (50-150 bp) by duplicate real-time PCR amplifications. Input DNA (0.1%) was used for normalization. The primers used for PCR were: cystatin C promoter (5′-gcaatgaccaacttctctggtg-3′ and 5′-cttaccagttcctcttctgtgc-3′); cathepsin C promoter (5′-ggagtcagaaatgcaggaaagtg-3′ and 5′-gggttgacaagagtcttaaagtctactg-3′); HPRT promoter (5′-gggcctaaatcttgaggaatcac-3′ and 5′-gtctcccagaggattcccagata-3′), and SIRV-IECS-Ld40 (5′-tgaagtcgatgccgcttttc-3′ and 5′-aaccagcgtccgcagatct-3′).
Results
cDNA microarray analysis of genes regulated by IRF-8
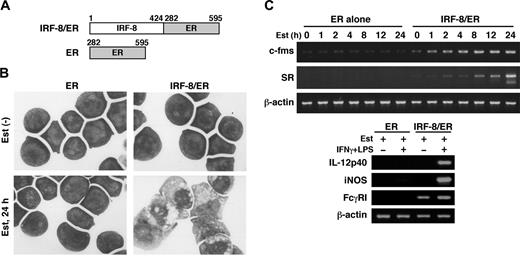
To begin understanding the mechanism by which IRF-8 controls myeloid-cell development, we sought to identify IRF-8–regulated genes. We had previously established an in vitro macrophage differentiation system in which introduction of IRF-8 drives Irf-8-/- myeloid progenitor Tot2 cells to fully differentiate into macrophages.10 To examine early changes caused by IRF-8, we employed an estrogen-inducible IRF-8/ER chimera (Figure 1A).13 In the absence of β-estradiol, IRF-8/ER–transduced Tot2 cells remained undifferentiated. However, after 24 hours of estradiol treatment, IRF-8/ER cells showed an intermediate stage of differentiation as judged by Wright-Giemsa staining (Figure 1B). In contrast, cells transduced with ER alone did not show any sign of differentiation in the absence or presence of estradiol. RT-PCR revealed that estradiol treatment rapidly induced expression of 2 macrophage-specific genes, c-fms (CSF-1/M-CSF receptor) and scavenger receptor (SR), only in IRF-8/ER–transduced cells (Figure 1C). These results confirmed that the estrogen-inducible form of IRF-8 induced rapid differentiation in these cells. The differentiated cells were growth arrested, and showed the property of functional macrophages as assessed by their ability to express IL-12p40 and inducible nitric oxide synthase (iNOS) mRNAs in response to IFNγ and lipopolysaccharide (LPS; Figure 1C). Fcγ receptor I (FcγRI) expression was detected in the differentiated cells and was further induced by the immune stimuli.
Macrophage differentiation by estradiol-inducible IRF-8/ER chimera in Irf-8-/- myeloid progenitor cells. (A) Diagram of the IRF-8/hormone-binding domain of the estrogen receptor (IRF-8/ER). (B) Wright-Giemsa–stained Irf-8-/- myeloid progenitor Tot2 cells transduced with a retrovirus carrying ER or IRF-8/ER (original magnification, × 1000). Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 24 hours. (C) Semiquantitative RT-PCR analysis for macrophage differentiation-related genes. RNA from cells treated with 1 μM β-estradiol for the indicated time was subjected to semiquantitative RT-PCR for c-fms and scavenger receptor (SR) expression. Cells treated with 1 μM β-estradiol for 3 days were also analyzed for expression of IL-12p40, inducible nitric oxide synthase (iNOS), and Fcγ receptor I (FcγRI) transcripts. β-actin was used as the loading control.
Macrophage differentiation by estradiol-inducible IRF-8/ER chimera in Irf-8-/- myeloid progenitor cells. (A) Diagram of the IRF-8/hormone-binding domain of the estrogen receptor (IRF-8/ER). (B) Wright-Giemsa–stained Irf-8-/- myeloid progenitor Tot2 cells transduced with a retrovirus carrying ER or IRF-8/ER (original magnification, × 1000). Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 24 hours. (C) Semiquantitative RT-PCR analysis for macrophage differentiation-related genes. RNA from cells treated with 1 μM β-estradiol for the indicated time was subjected to semiquantitative RT-PCR for c-fms and scavenger receptor (SR) expression. Cells treated with 1 μM β-estradiol for 3 days were also analyzed for expression of IL-12p40, inducible nitric oxide synthase (iNOS), and Fcγ receptor I (FcγRI) transcripts. β-actin was used as the loading control.
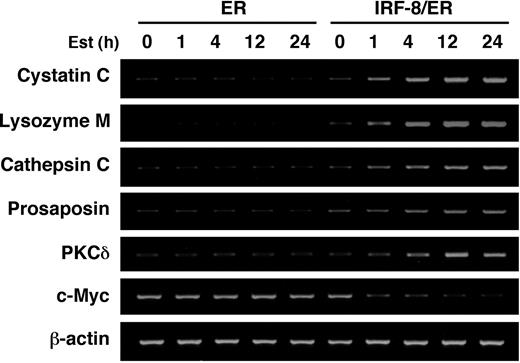
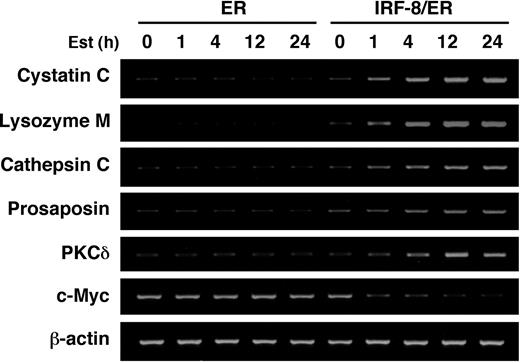
IRF-8/ER– and ER-transduced Tot2 cells treated with estradiol for 4 hours were then subjected to a genome-wide expression study using the NIA 15K mouse developmental cDNA microarray.28 We identified 69 genes (including 23 known/annotated genes) as displaying more than 2-fold differences between IRF-8/ER and ER cells (Tables 1, 2; also Supplemental Table S1, available online by clicking on the Supplemental Table link at the top of the article on the Blood website). Notably, the majority of the altered genes (62 genes) showed up-regulation, while only 7 genes were down-regulated by IRF-8/ER, suggesting that IRF-8 may act mainly as a transcriptional activator in this system. The microarray data were confirmed by semiquantitative RT-PCR analysis of 7 selected genes (Figure 2).
Somewhat unexpectedly, the genes showing the strongest induction at this early time point were those related to lysosomal/endosomal enzymes (Table 1). The cystatin C gene (induced ∼10-fold) encodes an endogenous inhibitor of the lysozomal cysteine proteases called cathepsins.29 We also observed significant induction of 2 cathepsins, cathepsin C/dipeptidyl-peptidase I and cathepsin L, as well as lysozyme M, a principal myeloid protease that has bactericidal activity,30,31 and prosaposin, which encodes a precursor of the sphingolipid activator proteins called saposins.32
Six of the more moderately up-regulated genes were related to signal transduction, including kinases and molecules involved in cyclic adenosine monophosphate (cAMP)/G-protein signaling. Notably, 1 of the up-regulated genes encodes PKCδ, which was previously shown to render myeloid progenitor 32Dcl.3 cells to respond to phorbol ester by differentiating into monocytic cells.33 There were 3 growth-related genes including c-Myc; this was strongly repressed by IRF-8, consistent with our previous finding.13 Other up-regulated genes included a major histocompatibility complex II gene central to the antigen presenting capability of macrophages, and a C-type lectin that acts as a pattern recognition receptor responsible for sensing foreign glycoproteins. The biologic significance of the noted changes in some of those genes, such as the cAMP/G-protein–related genes, is unclear at present and will require further investigation.
Identification of direct target genes of IRF-8
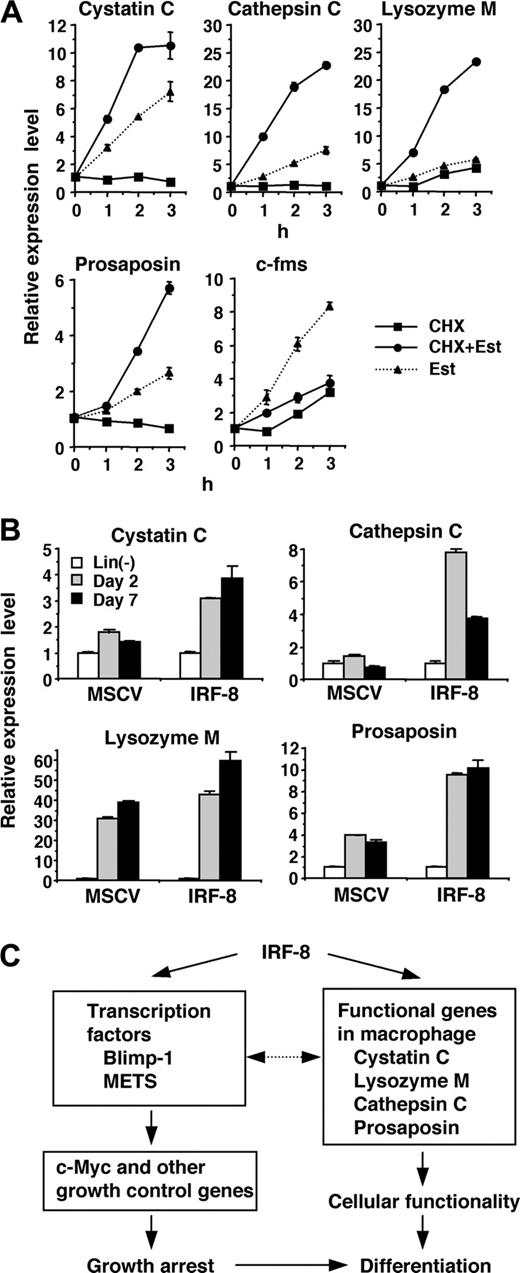
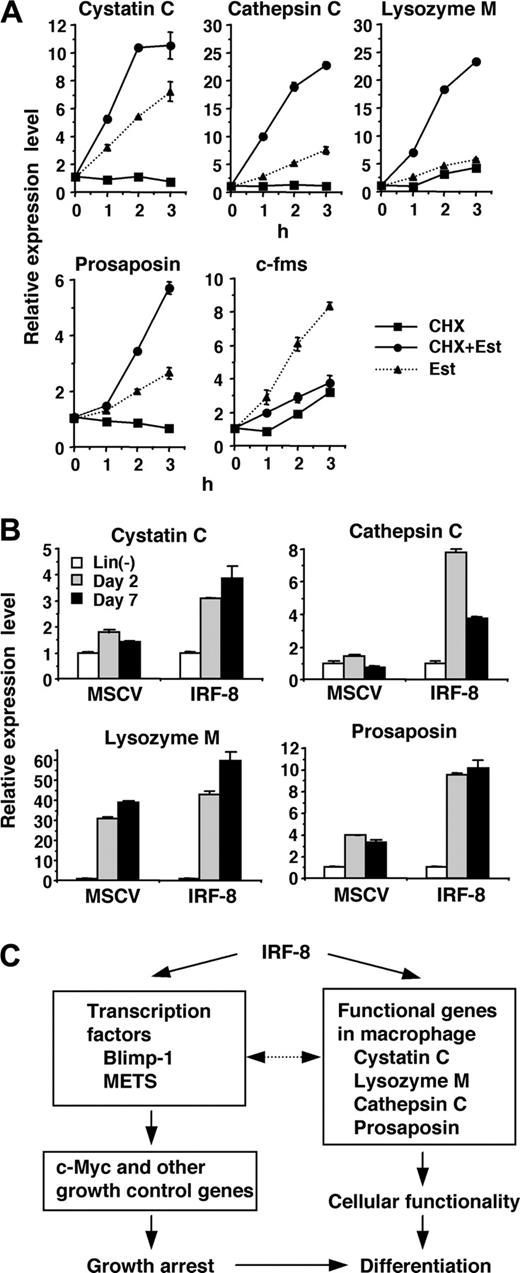
The rapid and strong induction of lysosomal/endosomal enzyme and related genes observed in our microarray and RT-PCR analyses led us to examine whether IRF-8 directly regulates the expression of these genes. To distinguish direct and indirect targets, we examined the effect of a protein synthesis inhibitor, cycloheximide (CHX), on the observed induction. If the gene induction occurs directly, estradiol should induce transcript expression in the presence of CHX, whereas CXH should abolish induction if the induction is indirect (ie, mediated by one or more proteins newly induced by IRF-8/ER). Accordingly, CHX was added 10 minutes prior to addition of estradiol. Real-time RT-PCR analysis demonstrated that the induction of the cystatin C, cathepsin C, lysozyme M, and prosaposin genes was not abrogated in the presence of CHX (Figure 3A), but rather increased over that treated with estradiol alone, indicating that these 4 genes are direct targets of IRF-8. In contrast, CHX inhibited the up-regulation of c-fms, indicating that this gene is an indirect target. We also observed that the suppression of c-Myc was inhibited by CHX (data not shown), in agreement with our previous findings that c-Myc is repressed through action of other transcription factors, Blimp-1 and METS.13
Consistent with the IRF-8–dependent induction of these genes, expression of cystatin C, cathepsin C, and prosaposin transcripts was lower in Irf-8-/- cells than that in wild-type cells (data not shown). To confirm that IRF-8 is responsible for the induction of the target genes in freshly isolated bone marrow cells, we transduced IRF-8 or control vector into Irf-8-/- lin- cells and cultured them in the presence of stem-cell factor (SCF) and M-CSF (Figure 3B). Consistent with the data with Tot2 cells, expression of the cystatin C, cathepsin C, and prosaposin genes was significantly induced in IRF-8–transduced bone marrow cells during the culture, whereas low or no up-regulation of these genes was observed in control vector–transduced cells. Expression of the lysozyme M gene was induced both in IRF-8–transduced and control cells. This is presumably because Irf-8-/- bone marrow progenitor cells differentiate into granulocytes that also express lysozyme M.
Together, IRF-8 directly induces transcription factors such as Blimp-1 and METS as well as macrophage proteases and related genes. These genes are in turn likely to affect subsequent gene expression patterns and cellular functions (Figure 3C).
Analysis of the cystatin C gene promoter
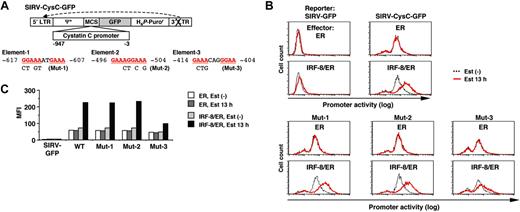
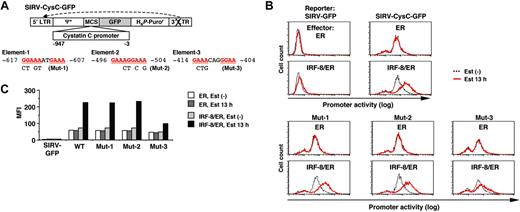
We next sought to identify the target DNA element through which IRF-8 activates transcription of the isolated target genes. We cloned the approximately 1 kilobase (kb) genomic sequence upstream of the cystatin C coding region, and analyzed the promoter activity using a novel reporter construct, SIRV-GFP (Figure 4A).
SIRV is a self-inactivating retrovirus containing a 176-bp deletion in the 3′ long terminal repeat (LTR) that removes the enhancer sequence. Following reverse transcription, the 3′ LTR is copied and replaces the 5′ LTR, resulting in inactivation of the 5′ LTR promoter. This enables us to use this vector for our reporter assay. We inserted a destabilized form of GFP (d2EGFP) into SIRV as a reporter, and placed the promoter sequences upstream of GFP. With this system the activity of chromatin-integrated promoters can be monitored in live cells as GFP signals via flow cytometry. In a preliminary experiment, introduction of SIRV-GFP containing the IFNγ activation site (GAS) along with the Ld40 minimal promoter allowed cells to express GFP in response to IFNγ, indicating that this promoter-reporter system functioned properly (data not shown).
Tot2 cells expressing ER alone or IRF-8/ER were transduced with empty SIRV-GFP or SIRV-GFP containing the cystatin C promoter (SIRV-CysC-GFP), and were then treated with estradiol for 13 hours. The GFP signals in empty SIRV-GFP–transduced ER and IRF-8/ER cells were very low (comparable to the autofluorescence of untransduced Tot2 cells) and were not altered by estradiol treatment, confirming that the LTRs were inactivated in this system (Figure 4B-C). In the absence of estradiol, SIRV-CysC-GFP cells showed modest levels of GFP signals, reflecting the basal promoter activity in undifferentiated Tot2 cells. Following estradiol treatment, GFP expression increased approximately 4-fold in IRF-8/ER cells but not in ER cells (Figure 4B-C), indicating that IRF-8 activated transcription of the cystatin C promoter–driven reporter.
The 1-kb cystatin C promoter did not contain the typical EICE sequence, but an EIRE was located at -617 (element-1; Figure 4A). We chose 2 additional DNA sequences (elements -2 and -3) as candidate target elements for IRF-8; these elements consisted of an IRF-binding motif (GAAA) followed by an Ets-binding motif (GGAA) without spacing nucleotides (element-2) or with 2 spacer nucleotides (element-3). We introduced mutations in all 3 elements and examined the ability of IRF-8 to activate the reporter gene. The mutation in element-3 (Mut-3) severely diminished IRF-8–induced activation of the promoter, whereas the mutations in element-1 and element-2 (Mut-1 and Mut-2) had no effect (Figure 4B-C), indicating that element-3 is important for the IRF-8 response.
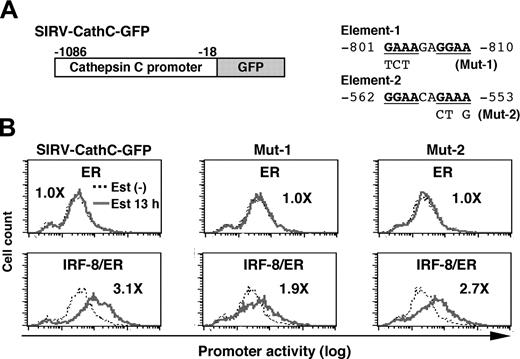
Analysis of the cathepsin C promoter
We next analyzed the cathepsin C promoter in the same manner. As shown in Figure 5, IRF-8/ER plus estradiol activated transcription through the 1-kb genomic sequence upstream of the open reading frame, whereas ER alone did not. We found that the cathepsin C promoter contained a DNA element (element-1) very similar to the important element-3 identified in the cystatin C promoter (Figure 5A). In addition, a typical EICE was found (element-2). In mutational experiments shown in Figure 5B, we found that a mutation in element-1 (Mut-1), but not that in element-2 (Mut-2) diminished IRF-8/ER–induced activation of the cathepsin C promoter, indicating that this element is required for induction of promoter activity by IRF-8. Thus, similar DNA elements mediate IRF-8–induced transcription from both cystatin C and cathepsin C promoters, indicating that these elements may represent a common target DNA sequence for IRF-8 during an early stage of macrophage differentiation. It should be noted, however, that the residual induction of the Mut-1 cathepsin C promoter (1.9-fold) suggests that there may be an additional element targeted by IRF-8.
RT-PCR confirmation of IRF-8–inducible genes identified by microarray analysis. Tot2 cells transduced with ER or IRF-8/ER were treated with β-estradiol for the indicated time and analyzed for gene expression by semiquantitative RT-PCR.
RT-PCR confirmation of IRF-8–inducible genes identified by microarray analysis. Tot2 cells transduced with ER or IRF-8/ER were treated with β-estradiol for the indicated time and analyzed for gene expression by semiquantitative RT-PCR.
Direct activation of lysosomal/endosomal enzyme-related genes by IRF-8. (A) Effect of cycloheximide (CHX) on IRF-8/ER–mediated induction of the indicated genes. IRF-8/ER–transduced Tot2 cells were treated with β-estradiol (Est) and/or CHX (10 μg/mL). CHX was added 10 minutes before addition of β-estradiol. Transcript levels were quantified in duplicate by real-time RT-PCR. Data were normalized by GAPDH levels and shown as values relative to those in untreated cells (mean ± standard deviation). (B) Expression of the indicated genes in IRF-8–transduced bone marrow lin- cells. Irf-8-/- bone marrow lin- cells were cultured in the presence of SCF, IL-6, and IL-3 for 1 day and were transduced with MSCV–IRF-8 or control MSCV retrovirus on the following 2 days. Next day, cells were washed and reinoculated in the presence of SCF and M-CSF. Cells were harvested 2 and 7 days after addition of M-CSF. Transcript levels were determined as in panel A. (C) A model for transcriptional pathways activated by IRF-8. IRF-8 activates transcription regulators and macrophage proteases. IRF-8–induced transcription factors regulate their target genes, including those that regulate cell growth. The proteases are critical for the functionality of macrophages, and might also influence transcription factor activity.
Direct activation of lysosomal/endosomal enzyme-related genes by IRF-8. (A) Effect of cycloheximide (CHX) on IRF-8/ER–mediated induction of the indicated genes. IRF-8/ER–transduced Tot2 cells were treated with β-estradiol (Est) and/or CHX (10 μg/mL). CHX was added 10 minutes before addition of β-estradiol. Transcript levels were quantified in duplicate by real-time RT-PCR. Data were normalized by GAPDH levels and shown as values relative to those in untreated cells (mean ± standard deviation). (B) Expression of the indicated genes in IRF-8–transduced bone marrow lin- cells. Irf-8-/- bone marrow lin- cells were cultured in the presence of SCF, IL-6, and IL-3 for 1 day and were transduced with MSCV–IRF-8 or control MSCV retrovirus on the following 2 days. Next day, cells were washed and reinoculated in the presence of SCF and M-CSF. Cells were harvested 2 and 7 days after addition of M-CSF. Transcript levels were determined as in panel A. (C) A model for transcriptional pathways activated by IRF-8. IRF-8 activates transcription regulators and macrophage proteases. IRF-8–induced transcription factors regulate their target genes, including those that regulate cell growth. The proteases are critical for the functionality of macrophages, and might also influence transcription factor activity.
We scanned the genomic sequence of 5 other genes directly activated by IRF-8 (lysosome M, prosaposin, Blimp-1, METS, and CDKN2B), looking for similar DNA elements. Indeed, we found that all of them except one gene (METS) contain a similar element in their promoter regions with the consensus sequence GAAANN(N)GGAA (Table 3). We designated this element the IRF-Ets composite sequence (IECS). In most cases, the element is located within approximately 1 kb upstream of the start codon. The IECS is distinct from the previously reported EICE and EIRE sequences, as the IRF-binding motif in the IECS is located 5′ of the Ets-binding, whereas the reverse is true in the latter 2 elements.
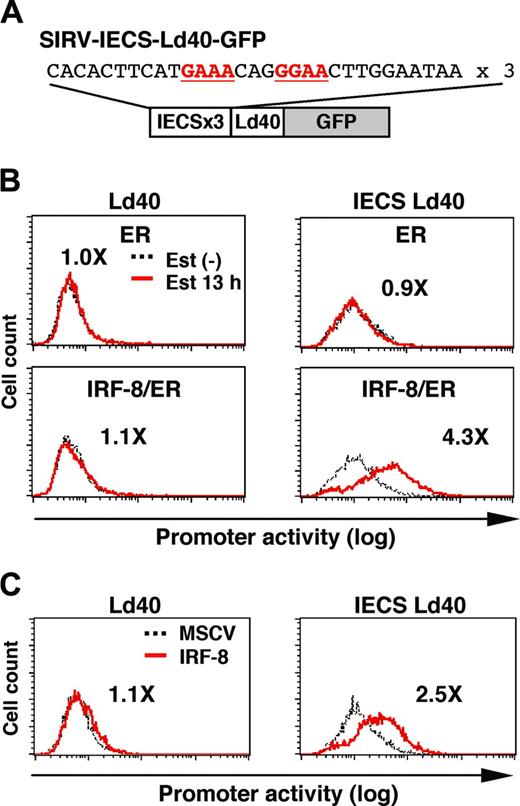
The IECS mediates IRF-8 activation of transcription
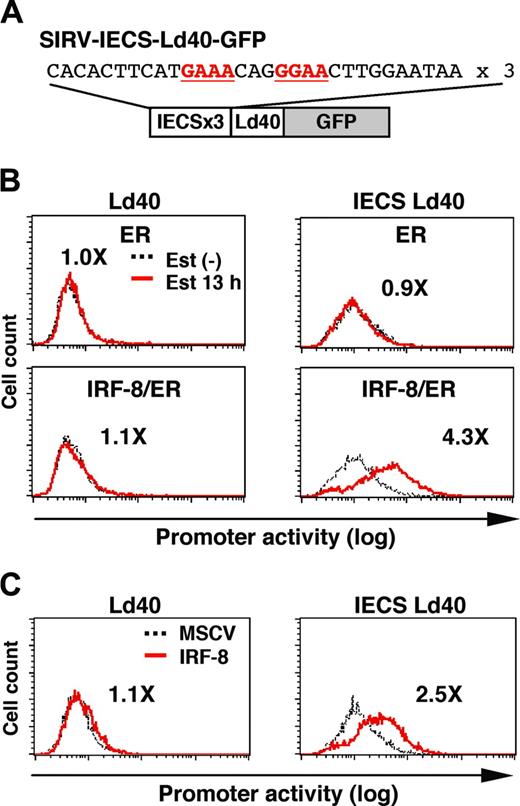
To examine whether the IECS is sufficient to confer IRF-8–induced activation of transcription, we constructed SIRV-IECS-Ld40-GFP, in which GFP transcription is driven by 3 copies of the IECS from the cystatin C gene followed by minimal promoter Ld40 (Figure 6A). SIRV-Ld40-GFP lacking IECS was used as a control. Ld40 alone had essentially no promoter activity in ER and IRF-8/ER cells before and after estradiol treatment (Figure 6B). IECS-Ld40 showed a modest basal transcriptional activity in untreated ER and IRF-8/ER cells. Importantly, transcription via the IECS was dramatically induced by estradiol treatment only in IRF-8/ER cells but not in ER cells.
We also examined whether wild-type (nonchimeric) IRF-8 can activate the IECS. Tot2 cells were transduced with SIRV-Ld40-GFP or SIRV-IECS-Ld40-GFP, and then with empty MSCV or MSCV-IRF-8. Promoter activity was measured on day 2 of transduction, when cells did not yet show any morphologic changes. IRF-8 clearly activated transcription through the IECS under these conditions (Figure 6C). Taken together, our results show that the IECS is sufficient to mediate IRF-8 activation of transcription in differentiating myeloid progenitor cells.
IRF-8 binds to target promoters and the IECS in vivo
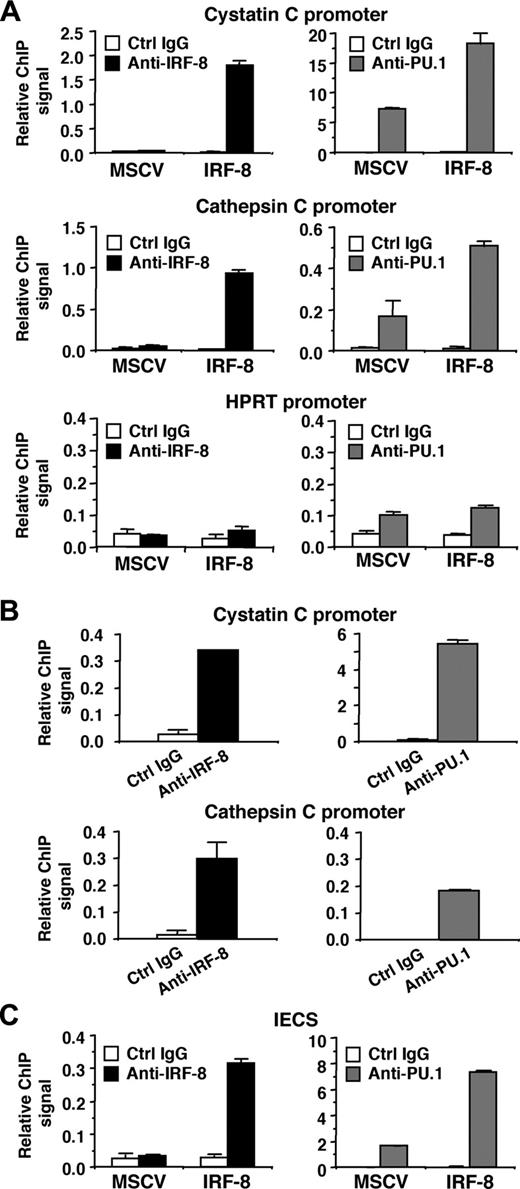
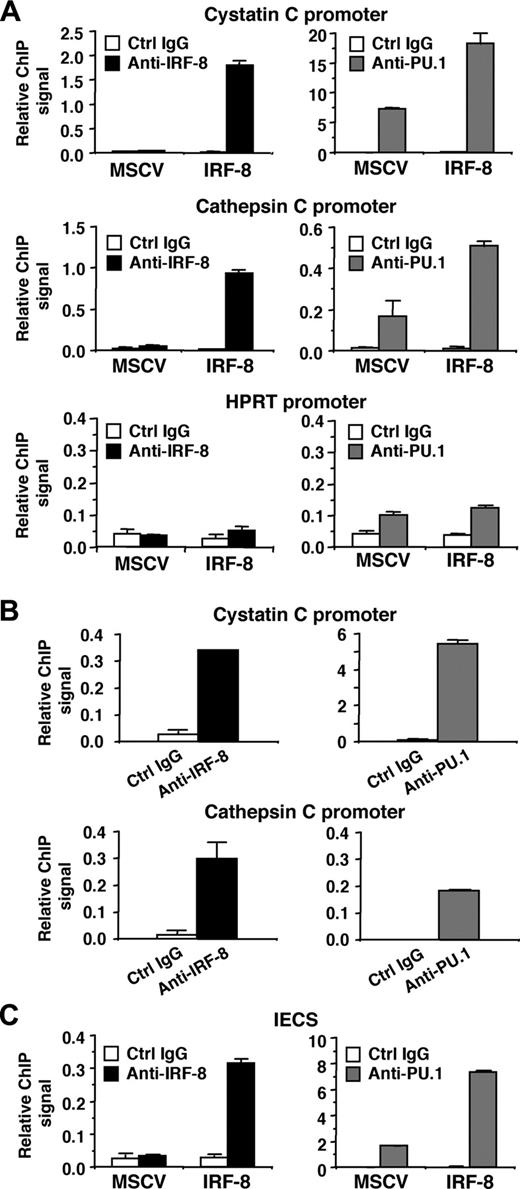
We next analyzed whether IRF-8 binds to endogenous target gene promoters containing the IECS. ChIP assays were performed in MSCV-IRF-8–transduced Tot2 cells, and specific PCR primers were used to amplify 50 to 150 nucleotides spanning the IECSs in the cystatin C and cathepsin C promoters. Real-time PCR analysis demonstrated that in IRF-8–transduced cells, both promoter fragments were significantly immunoprecipitated by anti–IRF-8 antibody, compared with the low levels of background signals generated by control IgG (Figure 7A, top 2 of the left panels). As expected, the ChIP signals generated by IRF-8 antibody in control MSCV-transduced cells were low, similar to background levels. We also used an anti-PU.1 antibody, which revealed that PU.1 bound to both promoters even in control MSCV-transduced cells. Interestingly, IRF-8 transduction increased PU.1 binding to the promoters approximately 3-fold (Figure 7A, top 2 of the right panels). Binding of IRF-8 and PU.1 to the irrelevant, but active hypoxanthine phosphoribosyltransferase (HPRT) promoter was also tested (Figure 7A, bottom panels). The ChIP signals generated by IRF-8 antibody were as low as background levels. PU.1 antibody showed only slightly elevated binding over control antibody in both control and IRF-8–transduced cells. These results suggest that IRF-8 specifically binds to endogenous promoters containing IECSs in vivo, and that IRF-8 binding is concomitant with enhanced binding of PU.1. We also confirmed the binding of endogenous IRF-8 protein to the promoter region of both cystatin C and cathepsin C genes in peritoneal macrophages (Figure 7B). As expected, PU.1 binding was also detected on both promoters.
Analysis of the cystatin C promoter. (A) Diagram of the self-inactivating retrovirus reporter (SIRV-GFP) carrying the cystatin C promoter. Both LTRs are inactivated and have no promoter activity. Ψ+, the extended viral packaging signal; MCS, multiple cloning site; H4P-Puror; the puromycin resistant gene driven by the histone H4 promoter. Sequences and mutations in element-1 to element-3 are shown. The numbers indicate the nucleotide positions relative to the start codon. (B) Cystatin C promoter activity in live cells, as monitored by flow cytometry of GFP signals. Tot2 cells were transduced with SIRV-GFP carrying wild-type (WT) or mutant (Mut-1 to Mut-3) promoters, and then with MSCV-ER or MSCV–IRF-8/ER. Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 13 hours. (C) Quantification of the promoter activity measured in panel B. The activity is shown as mean fluorescent intensity (MFI) of GFP signals.
Analysis of the cystatin C promoter. (A) Diagram of the self-inactivating retrovirus reporter (SIRV-GFP) carrying the cystatin C promoter. Both LTRs are inactivated and have no promoter activity. Ψ+, the extended viral packaging signal; MCS, multiple cloning site; H4P-Puror; the puromycin resistant gene driven by the histone H4 promoter. Sequences and mutations in element-1 to element-3 are shown. The numbers indicate the nucleotide positions relative to the start codon. (B) Cystatin C promoter activity in live cells, as monitored by flow cytometry of GFP signals. Tot2 cells were transduced with SIRV-GFP carrying wild-type (WT) or mutant (Mut-1 to Mut-3) promoters, and then with MSCV-ER or MSCV–IRF-8/ER. Cells were either left untreated or treated with 1 μM β-estradiol (Est) for 13 hours. (C) Quantification of the promoter activity measured in panel B. The activity is shown as mean fluorescent intensity (MFI) of GFP signals.
Finally, we examined whether IRF-8 and PU.1 bind to the IECS itself in vivo (Figure 7C). Tot2 cells were transduced with SIRV-IECS-Ld40-GFP, and then with MSCV–IRF-8. ChIP assays were performed using the same antibodies, and the presence of the IECS in immunoprecipitated chromatin was determined using PCR primers designed to specifically amplify the element. IRF-8 binding to the IECS was clearly detected in IRF-8–transduced Tot2 cells. PU.1 bound to the IECS even in the absence of IRF-8, but the binding was enhanced 4-fold in the presence of transduced IRF-8. Collectively, these results indicate that IRF-8 binds to the IECS concomitant with enhanced binding of PU.1, leading to stimulated transcription of IRF-8 target genes.
Analysis of the cathepsin C promoter. (A) Diagram of SIRV-GFP reporter carrying the cathepsin C promoter. Sequences and mutations of element-1 and element-2 are shown on right. The numbers indicate the nucleotide positions relative to the start codon. (B) Cathepsin C promoter activity in live cells monitored by flow cytometry. The promoter activity was analyzed as in Figure 4B. Values represent fold changes in MFI caused by estradiol treatment.
Analysis of the cathepsin C promoter. (A) Diagram of SIRV-GFP reporter carrying the cathepsin C promoter. Sequences and mutations of element-1 and element-2 are shown on right. The numbers indicate the nucleotide positions relative to the start codon. (B) Cathepsin C promoter activity in live cells monitored by flow cytometry. The promoter activity was analyzed as in Figure 4B. Values represent fold changes in MFI caused by estradiol treatment.
Discussion
Here, we used cDNA microarray analysis to identify genes that are rapidly induced or repressed by IRF-8 in differentiating myeloid progenitor cells. Most of these genes have not previously been described as IRF-8–regulated genes. Furthermore, we identified a unique target DNA element, the IECS, through which IRF-8 activates transcription.
We found that IRF-8 directly induces expression of genes encoding multiple lysosomal/endosomal enzymes (cathepsin C and L, lysozyme M, and prosaposin) as well as their inhibitor (cystatin C). This suggests that IRF-8 regulates not only “regulatory” genes such as those encoding transcription factors and signaling molecules mediating expression of other genes, but also “functional” genes that operate macrophage functions (Figure 3C). The latter enzymes might also have “regulatory” roles in addition to their involvement in end-stage protein breakdown. For example, cathepsin L was recently reported to be capable of cleaving a transcription factor, resulting in activation of transcription.34 Other cathepsins were demonstrated to degrade signal transduction molecules such as epidermal growth factor receptor and internalized insulin.35,36 Thus, it is possible that lysosomal/endosomal enzymes identified as IRF-8 targets in this study may also contribute to regulation of gene expression. As a related issue, Irf-8-/- macrophages are shown to terminate CSF-1/M-CSF signaling prematurely due to increased CSF-1 receptor ubiquitination, which may be caused by impaired expression of cathepsin B and accumulation of the ubiquitin ligase c-casitas B-lineage lymphoma (c-Cbl).37 However, it should be noted that regulation of CSF-1 signaling is not a sole mechanism of IRF-8–induced macrophage differentiation, since CSF-1 does not play major roles in Tot2-cell differentiation.10,13
IRF-8 activation of transcription through the IECS. (A) Diagram of the SIRV reporter driven by the IECS. (B) Activation of transcription through the IECS by IRF-8/ER. Tot2 cells were transduced with SIRVIECS-Ld40-GFP or SIRV-IECS-Ld40-GFP, and then with MSCV-ER or MSCV–IRF-8/ER. Cells were either left untreated or were treated with 1 μM β-estradiol (Est) for 13 hours. Values represent fold changes in MFI induced by estradiol treatment. (C) Activation of transcription through the IECS by wild-type IRF-8. Tot2 cells were transduced with SIRV-Ld40-GFP or SIRV-IECS-Ld40-GFP and then with empty MSCV or MSCV–IRF-8. Cells were analyzed on day 2 after transduction of MSCVs. Values represent fold changes in MFI induced by IRF-8.
IRF-8 activation of transcription through the IECS. (A) Diagram of the SIRV reporter driven by the IECS. (B) Activation of transcription through the IECS by IRF-8/ER. Tot2 cells were transduced with SIRVIECS-Ld40-GFP or SIRV-IECS-Ld40-GFP, and then with MSCV-ER or MSCV–IRF-8/ER. Cells were either left untreated or were treated with 1 μM β-estradiol (Est) for 13 hours. Values represent fold changes in MFI induced by estradiol treatment. (C) Activation of transcription through the IECS by wild-type IRF-8. Tot2 cells were transduced with SIRV-Ld40-GFP or SIRV-IECS-Ld40-GFP and then with empty MSCV or MSCV–IRF-8. Cells were analyzed on day 2 after transduction of MSCVs. Values represent fold changes in MFI induced by IRF-8.
We previously reported that the Blimp-1 and METS genes are direct targets of IRF-8.13 Blimp-1 and METS suppress transcription of growth-promoting genes such as c-Myc, and are also implicated in macrophage differentiation.38,39 Consistent with this, our microarray analysis detected c-Myc as a gene down-regulated by IRF-8. We also identified the c-fms gene as another “indirect” target but in this case up-regulated by IRF-8. These data suggest that regulatory genes directly induced by IRF-8 cause a number of subsequent changes in gene expression. Since Blimp-1 and METS are repressors, there may be 1 or more yet-unidentified transcriptional activators that mediate IRF-8 induction of indirect target genes. Such activators may be additional direct targets of IRF-8, and are likely to cooperate with other IRF-8 targets to orchestrate the molecular program for macrophage differentiation (Figure 3C).
In this work, we developed a reporter system (SIRV-GFP) using a self-inactivating retrovirus. Unlike the traditional reporter assay in which reporter constructs and effectors are transiently overexpressed, our system requires low copies of both reporters and effectors. Moreover, the reporter gene is integrated into the genome unlike transiently transfected plasmids, providing a more physiologic system of promoter analysis.
The SIRV-GFP system-based promoter analysis of the cystatin C and cathepsin C genes allowed us to identify a common DNA element that is responsible for IRF-8 activation of these 2 direct target genes. This element, the IECS, consists of an IRF-binding motif followed by an Ets-binding motif; the order of the 2 motifs is reversed relative to the previously reported EICE and EIRE sequences. The IECS was sufficient to mediate IRF-8 activation of transcription. Furthermore, IRF-8 bound to endogenous target promoters containing the IECS, and to the IECS itself in vivo. Notably, we observed that IRF-8 enhanced PU.1 binding to this element, whereas lower levels of PU.1 binding to the irrelevant HPRT promoter were not enhanced by IRF-8. Previous work showed that PU.1 transcript levels do not change during the early stages of differentiation in Tot2 cells.10 Therefore, the enhanced PU.1 binding may be due to stabilization of PU.1-DNA binding by physical interaction between IRF-8 and PU.1. It is also possible that IRF-8 may change the chromatin to a state more accessible to PU.1. Since PU.1 levels can affect lineage selection among macrophages, granulocytes, and B cells,40-42 IRF-8 enhancement of PU.1 binding may be a key event in IRF-8–mediated macrophage differentiation.
Binding of IRF-8 to target gene promoters and to the IECS in vivo. (A) Binding of IRF-8 and PU.1 to endogenous target gene promoters detected by chromatin immunoprecipitation (ChIP) assay. Tot2 cells were transduced with empty MSCV or MSCV–IRF-8, and were analyzed on day 3. Crosslinked, sheared chromatin was precipitated by goat anti–IRF-8, rabbit anti-PU.1 antibody, or control IgG (normal goat IgG for IRF-8 ChIP and normal rabbit IgG for PU.1 ChIP). Precipitated DNA was analyzed in duplicate by real-time PCR to detect genomic DNA from the cystatin C and cathepsin C promoter regions using primers that amplified the IECS region of each promoter. Levels of precipitated promoter DNA (ChIP signals) are shown as values relative to 0.1% input DNA (mean ± standard deviation). TheHPRT promoter was analyzed as a control irrelevant to IRF-8 and PU.1. (B) Binding of endogenous IRF-8 and PU.1 to the target gene promoters detected by ChIP assay. Peritoneal macrophages from wild-type mice were analyzed as in panel A. (C) ChIP assay for binding of IRF-8 and PU.1 to the IECS in vivo. Tot2 cells were transduced with SIRV-IECS-Ld40-GFP, and then with MSCV or MSCV–IRF-8. Cells were analyzed on day 3 after transduction of MSCVs. The IECS DNA was detected using primers designed to amplify the IECS sequence in SIRV.
Binding of IRF-8 to target gene promoters and to the IECS in vivo. (A) Binding of IRF-8 and PU.1 to endogenous target gene promoters detected by chromatin immunoprecipitation (ChIP) assay. Tot2 cells were transduced with empty MSCV or MSCV–IRF-8, and were analyzed on day 3. Crosslinked, sheared chromatin was precipitated by goat anti–IRF-8, rabbit anti-PU.1 antibody, or control IgG (normal goat IgG for IRF-8 ChIP and normal rabbit IgG for PU.1 ChIP). Precipitated DNA was analyzed in duplicate by real-time PCR to detect genomic DNA from the cystatin C and cathepsin C promoter regions using primers that amplified the IECS region of each promoter. Levels of precipitated promoter DNA (ChIP signals) are shown as values relative to 0.1% input DNA (mean ± standard deviation). TheHPRT promoter was analyzed as a control irrelevant to IRF-8 and PU.1. (B) Binding of endogenous IRF-8 and PU.1 to the target gene promoters detected by ChIP assay. Peritoneal macrophages from wild-type mice were analyzed as in panel A. (C) ChIP assay for binding of IRF-8 and PU.1 to the IECS in vivo. Tot2 cells were transduced with SIRV-IECS-Ld40-GFP, and then with MSCV or MSCV–IRF-8. Cells were analyzed on day 3 after transduction of MSCVs. The IECS DNA was detected using primers designed to amplify the IECS sequence in SIRV.
Similar elements were found in 6 out of 7 IRF-8 target gene promoters, further supporting the notion that the IECS is a common target DNA element for IRF-8. In contrast, the EIRE and EICE elements identified in the cystatin C and cathepsin C promoters, respectively, did not contribute to IRF-8 activation of these promoters. In addition, we found that 8 of 66 previously reported PU.1 binding sequences (those found in FcγRI, myeloperoxidase, p47phox, IL-7R, TAL1/SCL, Epstein-Barr virus LMP/TP2, lymphotropic papovovirus, and simian virus 40) are similar to the IECS.43,44 This implies that IRF-8 and PU.1 may cooperate to mediate transcription from these promoters, as well.
Despite clear IRF-8 binding to the IECS detected by our ChIP assay, we did not detect significant binding of IRF-8 to an IECS oligonucleotide by electrophoretic mobility shift assay (data not shown). The discrepancy between in vivo and in vitro binding assays suggests that IRF-8 may act on chromatinized templates, but not on naked DNA. Given that histones are known to play central roles in regulating transcription, we speculate that IRF-8 binding to the IECS might require an interaction between histones and the protein complex composed of IRF-8, PU.1, and possibly other proteins. Future work will be required to identify such an interaction.
In conclusion, our study describes identification of target genes activated/repressed by IRF-8, and shows a new target element by which IRF-8 regulates transcription in myeloid progenitor cells. These findings add to our broad understanding of the molecular mechanism by which specific transcription factors direct myeloid cell differentiation.
Prepublished online as Blood First Edition Paper, June 9, 2005; DOI 10.1182/blood-2005-01-0080.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Kevin Becker for microarray production, and Ben-Zion Levi, Hee Jeong Kong, Akira Nishiyama, and Tomonori Uno for valuable help.
Supplemental data
The table can be downloaded at http://ozatolab.nichd.nih.gov/tamura/TamuraRawData.xls. Expression levels of each gene in 2 independent experiments were placed in the NIA mouse 15K gene list table, downloaded from http://lgsun.grc.nia.nih.gov/download.html. Averages and variance were calculated to perform t test as indicated. Fold changes are also shown.