Abstract
An abundance of research has entrenched the view that the Ets domain containing transcription factor PU.1 is fundamental to the development and function of B lymphocytes. In this study, we have made use of a conditional PU.1 allele to test this notion. Complete deletion of PU.1 resulted in the loss of B cells and all other lineage-positive cells in the fetal liver and death between E18.5 and birth; however, specific deletion of PU.1 in the B lineage had no effect on B-cell development. Furthermore, deletion of PU.1 in B cells did not compromise their ability to establish and maintain an immune response. An increased level of apoptosis was observed in vitro upon B-cell receptor (BCR) cross-linking; however, this was partially rescued by interleukin-4 (IL-4). These findings suggest that PU.1 is not essential for the development of functional B lymphocytes beyond the pre-B stage. (Blood. 2005;106:2083-2090)
Introduction
As the sole source of immunoglobulin in the body, the development of functional B cells is essential in generating an adaptive immune system. B lymphocytes are produced in a stepwise process by the hematopoietic stem cell (HSC), first in the fetal liver and subsequently in the bone marrow. The self-renewing HSC is able to do this by generating a cascade of progenitors each of which is more restricted to a B-cell fate.1 The molecular mechanisms underpinning this process have come under intense scrutiny and, as a result, the transcription factor PU.1 has emerged as an essential component.
PU.1 and its close relative Spi-B are members of the Ets domain-containing transcription factor family that are expressed exclusively in the hematopoietic system. Mice carrying a targeted deletion of PU.1 lack all B cells; however, they also have no T cells or macrophages and die at E18.5.2 The importance of PU.1 in B-cell development has been confirmed by an alternative knock-out strategy which, despite having a less severe T-cell and survival phenotype, lacked immunoglobulin M-positive (IgM+) mature B cells. These mice did, however, have a population of B220+ cells that did not correspond to any known B-cell fraction and did not undergo IgH rearrangement.3 There remains some controversy regarding which of these deletion strategies best represents a PU.1 null phenotype.4
The loss of multiple lineages in PU.1-deficient mice has led to the hypothesis that it is able to influence commitment to both myeloid and lymphoid fates. Experiments in which PU.1-/- fetal liver progenitors (FLPs) were transduced with PU.1 retrovirus suggest this control is exerted by different doses of PU.1, with high levels directing myeloid differentiation and low levels supporting lymphoid commitment.5 This conclusion is, however, in stark contrast to our recent finding showing the level of PU.1 in the first definable myeloid and lymphoid progenitors is the same, implying that the level of PU.1 is not deterministic at the initial definition of myeloid versus lymphoid fate.6
The absence of normal B cells in PU.1-/- mice may be due to a failure to up-regulate the transcription factor early B-cell factor (EBF), which when knocked out blocks B-cell development prior to IgH gene rearrangement.7 PU.1 has been purported to directly activate Ebf1 expression, and transducing PU.1-/- FLPs with Ebf1 has been shown to rescue early-stage B-cell development, implying the lack of Ebf1 in PU.1-/- mice blocks B-cell development.8 In addition, PU.1 has also been shown to bind to regulatory sequences in the Il7r gene,9 a fundamental component of the receptor for the cytokines interleukin-7 (IL-7) and thymic stromal lymphopoietin (TSLP), both of which support lymphoid development.10-13 The loss of signals through the IL-7R in PU.1-/- mice may, however, block the generation of lymphoid progenitors rather than commitment to a B-cell fate, because Il7r is expressed from the earliest stages of lymphoid development.14
Several studies have linked PU.1 with the expression of B cell-specific genes including B220,8 IgH,15 Igκ,16 Igλ,17 J chain,18 and CD7219 ; however, the lethality of deleting PU.1 has obstructed studying the effect its absence would have on adult B-cell development and function. Mice lacking the related factor Spi-B do have B cells; however, they proliferate poorly in response to B-cell receptor (BCR) cross-linking and have reduced antigen-specific IgG in T-dependent immunization experiments, a phenotype attributed to increased apoptosis.20 When a PU.1+/- background is combined with Spib deficiency, the phenotype is enhanced21 as a result of a reduction in the level of the nuclear factor-κB (NF-κB) family member crel,22 thus implicating PU.1 in mature B-cell function and suggesting an overlap in the roles of PU.1 and Spi-B.23 This redundancy, however, is finite, because Spi-B is able to rescue myeloid but not lymphoid development in the absence of PU.1.24
To further investigate the role PU.1 plays in B-cell development and function, we have generated a conditional PU.1 allele, which has enabled the specific deletion of PU.1 in the B-cell lineage. Surprisingly, these mice have relatively normal numbers of B cells in all defined developmental stages and are able to respond, albeit with reduced efficiency, to T-dependent immunization. The reduced number of antigen-specific cells may be due to increased apoptosis resulting from BCR cross-linking.
Materials and methods
Mouse strains and deletion of PU.1
The PU.1 alleles PU.1FL/FL, PU.1Δ/ΔCD19, and PU.1Δ/Δdeleter and Ly5.1 congenic mouse lines were maintained on a C57BL/6 background.25 Nonconditional PU.1Δ/Δdeleter was created by breeding with Deleter-Cre26 and conditional deletion PU.1Δ/ΔCD19 with CD19-Cre.27 Deletion frequency was determined by polymerase chain reaction (PCR) amplification using the primers PU.1-D GCACACATGCGTGTTTGTGGATGCT, PU.1-C GTGCTTCCTTGGGAGTCTGGCGCT, and PU.1-B CTGTCTGCCACCACCTGCCTACATT and yielded a band of 732 bp for the targeted PU.1 allele compared with and a band of 866 bp when deleted. In all experiments equal amplification of both bands was ensured by including tail DNA of a heterozygote genotype.
Monoclonal antibodies (mAbs) and flow cytometry
Antigens were detected with fluorescent or biotinylated conjugated monoclonal antibodies (mAbs) as follows: Mac-1 (M1/70), Gr-1 (RA6-8C5), B220 (RA3-6B2), IgM (331.12), and CD19 (ID3) were purified from hybridoma supernatants and conjugated in the authors' laboratory. c-Kit (2B8), IL-7Rα (A7R34), CD21 (7G6), CD23 (B3B4), CD25 (7D4), Igκ (R8-140), and syndecan-1 (281-2) were from BD Biosciences (San Jose, CA). Sorting was performed on a FACSDiVa high-speed flow cytometer (BD Biosciences) and analysis on a FAScanII (BD Biosciences).
Western blotting
Protein extracts were prepared and Western blotting carried out as described by Rosenbauer et al.28 Antibodies used were rabbit anti-mouse PU.1 (T21) and goat anti-mouse β-actin (I-19) (Santa Cruz Biotechnology, Santa Cruz, CA).
Immunizations
Mice were immunized by intraperitoneal injection of 100 μg alum-precipitated nitrophenyl (NP) conjugated to keyhole limpet hemocyanin (NP-KLH) (ratio, 18:1), as described.29 Mice were boosted by a further intraperitoneal injection of 20 μg soluble NP-KLH per mouse on day 28.
ELISPOTs and ELISAs
The frequency of antibody-secreting cells (ASCs) was determined by enzyme-linked immunospot (ELISPOT) as reported previously.30 Briefly, 96-well filtration plates (Millipore, Bedford, MA) were coated with 100 μL of 15 μg/mL NP2BSA or NP20BSA in phosphate-buffered saline (PBS) for 12 hours at 4°C. Plates were washed with PBS and bone marrow cells or splenocytes added at 1 × 104, 1 × 105, or 1 × 106 cells per well in RPMI, 5% fetal calf serum (FCS), and 10-4 M β-mercaptoethanol and incubated for 18 hours. ELISPOTs were revealed using goat anti-mouse IgG1-horseradish peroxidase (IgG1-HRP) (Southern Biotech, Birmingham, AL) and color developed by the addition of 100 μL of 250 mg/mL 3-amino-9-ethylcarbazole (AEC; Sigma-Aldrich, St Louis, MO) in 0.05% sodium acetate buffer and 0.03% H2O2. Plates were counted using an AID ELISPOT reader (Autoimmun Diagnostika, Strassberg, Germany). Resting levels of immunoglobulin were detected by enzyme-linked immunosorbent assay (ELISA). Goat anti-mouse IgM, IgG1, IgG2a, IgG2b, IgG3, IgA, Igκ, and Igλ (Southern Biotech) were used as plate coats. Purified mouse monoclonal IgM (TEPC 183), IgG1κ (MOPC 21), IgG2a (UPC 10), IgG2b (MOPC 141), IgG3 (FLOPC 21), and IgA (MOPC 315) (Sigma-Aldrich) were used to quantify immunoglobulin concentration. Isotype-specific biotinylated secondary antibodies were IgM, IgG1, IgG2a, IgG2b, IgG3, IgA, Igκ, and Igλ (Southern Biotech). Streptavidin-HRP was used in combination with the color substrate diammonium 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (Sigma-Aldrich) and absorbance detected with a Vmax kinetic microplate reader (Molecular Devices, Sunnyvale, CA).
Immunohistochemistry
Tissue samples were embedded in optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA) and snap frozen in isopentane on dry ice. Six-micrometer cryostat sections were stained with a combination of purified GL7 (BD Pharmingen, San Diego, CA) and biotinylated IgD (Southern Biotech). An anti-rat HRP was used to detect GL7, with subsequent staining carried out with the addition of normal rat serum to block cross-reactivity. IgD-biotin was detected using streptavidin-alkaline phosphatase (Southern Biotech) and visualized using the Fast Blue kit (Vector Laboratories, Burlingame, CA). HRP was visualized using the 3-amino-9-ethylcarbazole substrate kit (Vector Laboratories). Slides were mounted under coverslips in Aqua polymount (Polysciences, Warrington, PA). Images were analyzed with an Axioplan 2 microscope, under a 100 × /1.3 NA objective and 10 × eyepiece (Carl Zeiss, North Ryde, NSW, Australia). Images were captured with an Axiocam digital camera (Zeiss) and processed with Axiovision software (Zeiss).
RT-PCR
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA) and reverse transcribed with Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT; Promega, Madison, WI) according to the manufacturer's instructions. Semiquantitative RT-PCR was performed on samples previously normalized by equilibrating the level of hypoxanthine guanine phosphoribosyltransferase (Hprt) expression. All the primer combinations, the sequences for which are available upon request, spanned introns.
Isolation and stimulation of B cells
Small resting B cells were isolated by macerating total spleens, treating with red cell lysis buffer, and spinning in a 80:65:50 gradient of Percoll.31 The small dense cells were removed from the 80:65 interface and the B cells positively sorted by magnetic-activated cell separation (MACS) using anti-B220 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). B cells were cultured in RPMI supplemented with 5% FCS and 10-4 M β-mercaptoethanol. Cells were stimulated with 10 μg/mL goat anti-mouse IgM F(ab′)2 fragments (Jackson ImmunoResearch Labs, West Grove, PA), 10 μg/mL anti-CD40 (FGK45) with IL-4 and IL-5 (produced as per Karasuyama and Melchers32 ), or 20 μg/mL lipopolysaccharide (LPS; Sigma-Aldrich). To determine proliferation, cells were pulsed with 1 μCi (37 Bq) [3H]thymidine (Amersham, Piscataway, NJ) for 6 hours. Cell viability was assessed by fluorescence-activated cell sorting (FACS) with the addition of a known number of microbeads and staining with annexin V (BD Biosciences) and propidium iodide.
Results
Conditional deletion of PU.1 in the B lineage
Of the 2 strategies previously used to delete PU.1, one resulted in the complete loss of B cells in the fetal liver2 and the other reported a population of aberrant B220+ cells in the spleen and bone marrow.3 We have generated a conditional allele, which allows for spatially or temporally controlled or complete deletion of PU.1 (Figure 1A). When PU.1 was inactivated throughout the mouse by crossing to the Deleter-Cre strain, no expression of either B220 or CD19 was observed in the fetal liver at E18.5 (Figure 1B). Furthermore, no Mac-1+Gr-1+ cells were present, and knock-out mice died between E18.5 and birth despite being present in normal mendelian frequency at E18.5 (data not shown). This phenotype most closely represents that previously reported by Scott et al.2
Conditional deletion of PU.1. (A) PU.1 conditional allele. The PU.1 targeting construct containing exon 5 of PU.1 surrounded by LoxP sites (▴) and a reporter cassette flanked by 2 Frt sites (•) containing both the internal ribosome entry site-green fluorescent protein (IRESGFP) and the phosphoglycerate kinase (PGK) promoter driving the selection marker Neomycin (Neo) was introduced into the PU.1 locus by homologous recombination to generate the PU.1gfp allele. When crossed to mice harboring flp recombinase, the reporter/selection cassette is lost, generating PU.1fl allele. Exon 5 of PU.1 is deleted by crossing with mice carrying Cre recombinase, creating the allele PU.1Δ. (B) Expression of lineage markers in E18.5 fetal livers. Following isolation, fetal livers were treated with red cell lysis buffer and analyzed by flow cytometry for the presence of B cells (CD19+B220+) and monocytes/granulocytes (Gr-1+Mac-1+). (C) Deletion efficiency of PU.1fl/fl alleles in the B-cell fractions of PU.1Δ/ΔCD19 mice. Isolated B-cell fractions from the fetal liver (left panel), bone marrow, and spleen (right panel) were analyzed by competitive PCR for the presence of targeted (fl) and deleted (Δ) PU.1 alleles. The antibody combinations used and the rate of deletion calculated by densitometry are as follows: fetal liver (CD19+B220+, 73%); bone marrow: pre-BI (B220+ckit+, 59%), pre-BII large (B220+c-Kit-CD25+, forward scatter [FSC] large, 86%), pre-BII small (B220+c-Kit-CD25+, FSC small, 86%), immature B (B220loIgM+, 90%), and mature recirculating (B220hiIgM+, 98%); spleen: transitional 1 (CD23-IgM+-CD21lo, 98%), transitional 2 (CD23+IgMhiCD21hi, 96%), follicular (CD23+IgMloCD21lo, 92%), and marginal zone (CD23-IgM+CD21hi, 100%). (D) Loss of PU.1 in B cells from PU.1Δ/ΔCD19 mice. Total protein extracts were made from IgM+B220+ cells sorted by flow cytometry from the spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. The presence of PU.1 and β-actin was assessed by Western hybridization.
Conditional deletion of PU.1. (A) PU.1 conditional allele. The PU.1 targeting construct containing exon 5 of PU.1 surrounded by LoxP sites (▴) and a reporter cassette flanked by 2 Frt sites (•) containing both the internal ribosome entry site-green fluorescent protein (IRESGFP) and the phosphoglycerate kinase (PGK) promoter driving the selection marker Neomycin (Neo) was introduced into the PU.1 locus by homologous recombination to generate the PU.1gfp allele. When crossed to mice harboring flp recombinase, the reporter/selection cassette is lost, generating PU.1fl allele. Exon 5 of PU.1 is deleted by crossing with mice carrying Cre recombinase, creating the allele PU.1Δ. (B) Expression of lineage markers in E18.5 fetal livers. Following isolation, fetal livers were treated with red cell lysis buffer and analyzed by flow cytometry for the presence of B cells (CD19+B220+) and monocytes/granulocytes (Gr-1+Mac-1+). (C) Deletion efficiency of PU.1fl/fl alleles in the B-cell fractions of PU.1Δ/ΔCD19 mice. Isolated B-cell fractions from the fetal liver (left panel), bone marrow, and spleen (right panel) were analyzed by competitive PCR for the presence of targeted (fl) and deleted (Δ) PU.1 alleles. The antibody combinations used and the rate of deletion calculated by densitometry are as follows: fetal liver (CD19+B220+, 73%); bone marrow: pre-BI (B220+ckit+, 59%), pre-BII large (B220+c-Kit-CD25+, forward scatter [FSC] large, 86%), pre-BII small (B220+c-Kit-CD25+, FSC small, 86%), immature B (B220loIgM+, 90%), and mature recirculating (B220hiIgM+, 98%); spleen: transitional 1 (CD23-IgM+-CD21lo, 98%), transitional 2 (CD23+IgMhiCD21hi, 96%), follicular (CD23+IgMloCD21lo, 92%), and marginal zone (CD23-IgM+CD21hi, 100%). (D) Loss of PU.1 in B cells from PU.1Δ/ΔCD19 mice. Total protein extracts were made from IgM+B220+ cells sorted by flow cytometry from the spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. The presence of PU.1 and β-actin was assessed by Western hybridization.
To determine whether the lack of B cells was a result of a requirement of PU.1 for the development of the B lineage or rather a more widespread failure of hematopoietic progenitors, we crossed the PU.1fl/fl allele to CD19-Cre mice, which provided a B cell-specific source of Cre. The mice generated by this cross are termed PU.1Δ/ΔCD,19 referring to deletion of PU.1 in CD19-expressing cells. The appropriate control, which is mice harboring Cre in one CD19 locus, and both wild-type PU.1 alleles are termed PU.1+/+CD19. Surprisingly, the PU.1Δ/ΔCD19 embryos had normal numbers of B cells in the fetal liver at E18.5 despite having a frequency of exon-5 deletion of approximately 70% (Figure 1B), suggesting that PU.1 was not essential for the development of cells committed to the B lineage. The presence of B cells in E18.5 fetal liver led us to investigate whether they were also present in adult PU.1Δ/ΔCD19 mice. Table 1 shows that PU.1Δ/ΔCD19 mice have cells in each of the previously defined B-cell fractions in the bone marrow, spleen, lymph nodes, and peritoneum. The total number of cells present in the various fractions was, however, slightly perturbed compared with controls with enhanced numbers of pre-BII and T1/T2 cells but reduced numbers of mature recirculating/follicular B cells. In contrast to a recent publication, the B cell-specific isoform of CD45, B220, was found to be expressed at normal levels on adult B cells from PU.1Δ/ΔCD19 mice as well as in E18.5 embryos (data not shown and Figure 1B).8
To ensure the B cells observed in PU.1Δ/ΔCD19 mice had deleted the conditional PU.1 allele, we sorted cells from each fraction and subjected them to PCR analysis. Figure 1C shows that at the pre-BII stage 86% of the PCR product was from the deleted allele, a frequency that rose to more than 90% for subsequent developmental stages, indicating that most cells in each fraction are deleted. When IgM+CD19+ cells were sorted from the spleen and subjected to Western analysis, very low levels of PU.1 protein were detected from cells of the PU.1Δ/ΔCD19 mice. These data confirm that relatively normal B lymphopoiesis occurs in the absence of PU.1.
Expression of EBF and IL-7Rα in PU.1Δ/ΔCD19 pre-BII cells
PU.1 has been suggested to directly activate the expression of Il7r9 and Ebf1,8 both of which are required for normal B-cell development.7,10 The observation that B cells from PU.1Δ/ΔCD19 mice express B220, another putative PU.1 target gene,8 prompted us to investigate if the expression of either Ilr7 or Ebf1 was effected by PU.1 deletion. To do this, we analyzed expression levels in pre-BII cells sorted from bone marrow, because most cells in this population have deleted the PU.1 alleles (Figure 1C). The levels of IL-7Rα as measured by both FACS and RT-PCR were found to be the same in PU.1Δ/ΔCD19 and PU.1+/+CD19 pre-BII cells (Figure 2). This suggests that at this stage in B-cell development PU.1 is not required for the expression of IL-7R. Ebf1 mRNA levels, however, were markedly decreased in PU.1Δ/ΔCD19 B cells, confirming that PU.1 regulates Ebf1 in pre-BII cells. The deletion of PU.1 appeared to have little or no impact on other genes required for B-cell development, including Spib, Pax5, B29, mb1, or Rag2; however, the expression of both VpreB and Rag2 was reduced in mice lacking PU.1 (Figure 2B).
Resting levels of immunoglobulin isotypes and response to T-dependent immunization of PU.1Δ/ΔCD19 mice
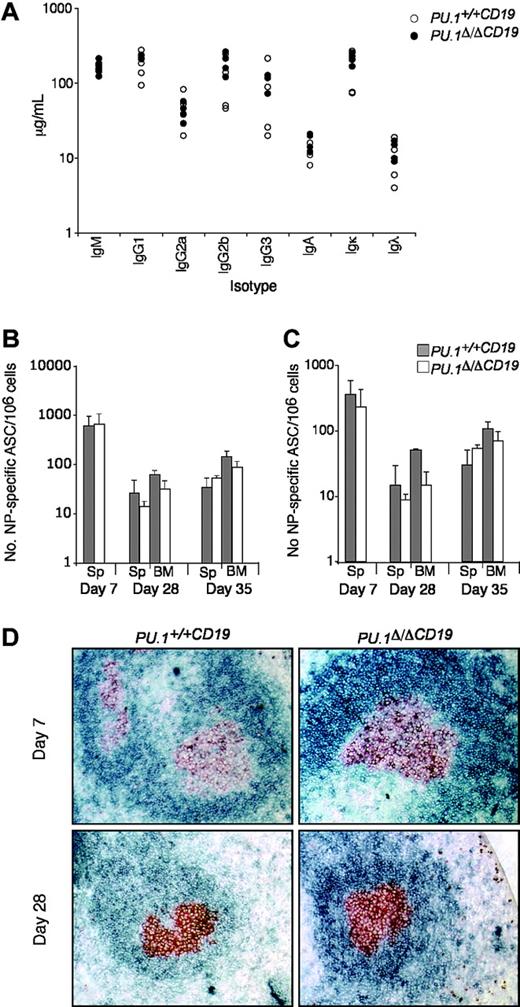
Several reports have implicated PU.1 in the function of B cells, because it is thought to be part of the mechanism controlling the expression of IgH, Igκ, and Igλ as well as switching and secretion of the immunoglobulin isotypes.15,33 As a first step to analyze whether the loss of PU.1 influences B-cell function, we measured the relative abundance of the various immunoglobulin isotypes in untreated mice. Both the total amount and ratios of the various isotypes were found to be substantially the same between PU.1Δ/ΔCD19 and PU.1+/+CD19 sera (Figure 3).
PU.1 has also been implicated in the maintenance of the T-dependent immune response, because crossing Spib-/- mice onto a PU.1+/- background increased apoptosis in germinal center (GC) B cells over that observed in Spib-/- mice, resulting in the failure of GCs to form.21 To test if the complete loss of PU.1 alone effected the T-dependent immune response, we assessed the ability of PU.1Δ/ΔCD19 and control mice to respond to immunization with the hapten nitrophenyl linked to NP-KLH, which typically generates IgG1λ ASCs.30
Following immunization, the number of NP-specific IgG1+ ASCs in the spleen and bone marrow was assessed at day 7, 28, and 35 (following boosting on day 28). Figure 3B-C shows that NP-ASCs were present at all time points in PU.1Δ/ΔCD19 mice and that somatic hypermutation had occurred to generate high-affinity ASCs. There was a small reduction in the total number of NP-ASCs in PU.1Δ/ΔCD19 mice but, in contrast to the previously reported lack of GCs in PU.1+/-Spib-/- mice, immunohistochemistry demonstrated the presence of normal GC in PU.1Δ/ΔCD19 mice at both day 7 and 28 (Figure 3D). The presence of normal levels of immunoglobulin isotypes in untreated PU.1Δ/ΔCD19 mice and their ability to make and maintain a T cell-dependent response implies the lack of PU.1 in B cells does not alter their normal function.
The effect of PU.1 deletion on IL-7Rα and EBF expression in pre-BII cells. (A) IL-7Rα expression on pre-BII cells. Pre-BII cells (B220+c-Kit-CD25+) from the bone marrow of PU.1Δ/ΔCD19 and PU.1+/+CD19 were analyzed for the expression of IL-7Rα by FACS. (B) Gene expression in pre-BII cells. Pre-BII cells were sorted by FACS, total RNA extracted, and gene expression analyzed by RT-PCR.
The effect of PU.1 deletion on IL-7Rα and EBF expression in pre-BII cells. (A) IL-7Rα expression on pre-BII cells. Pre-BII cells (B220+c-Kit-CD25+) from the bone marrow of PU.1Δ/ΔCD19 and PU.1+/+CD19 were analyzed for the expression of IL-7Rα by FACS. (B) Gene expression in pre-BII cells. Pre-BII cells were sorted by FACS, total RNA extracted, and gene expression analyzed by RT-PCR.
In vitro anti-IgM stimulation of PU.1Δ/ΔCD19 B cells
PU.1 and Spi-B have been implicated in the survival of B cells during an immune reaction by directly up-regulating the expression of the NF-κB family member c-rel, which has a prosurvival function in response to BCR signaling.22 We were interested to investigate whether the reduction in NP-specific ASCs observed in PU.1Δ/ΔCD19 mice was due to increased apoptosis and whether this was the result of a failure to up-regulate crel. Small resting B cells were isolated from PU.1Δ/ΔCD19 and PU.1+/+CD19 spleens and stimulated by cross-linking IgM. The proliferation of PU.1Δ/ΔCD19 B cells was measured by [3H]thymidine incorporation and found to be significantly reduced compared with the PU.1+/+CD19 cells over 3 days (Figure 4A). This reduced proliferation resulted from enhanced levels of apoptosis compared with controls in the first 24 hours following stimulation with anti-IgM (Figure 4B). A similar increased level of apoptosis was previously observed in PU.1+/-Spib-/- B cells; however, in that case the expression of crel, a critical regulator of B-cell proliferation, was significantly reduced.22 In contrast, PU.1Δ/ΔCD19 B cells maintained expression of crel when stimulated with anti-IgM (Figure 4C) and did not have reduced levels of other antiapoptotic genes, including BclXL, Bcl2, and A1.
The effect of PU.1 deletion on B-cell function. (A) Immunoglobulin isotype levels in untreated mice. Sera from 8- to 12-week-old mice (n = 6 for each genotype) were analyzed for levels of IgM, IgG1, IgG2a, IgG2b, IgG3, IgA, Igκ, and Igλ by ELISA. PU.1+/+CD19 and PU.1Δ/ΔCD19 are shown in black and white circles, respectively. (B-C) Number of NP-specific IgG1 ASCs in bone marrow and spleen. PU.1+/+CD19 and PU.1Δ/ΔCD19 mice were immunized with 100 μg NP-KLH precipitated in alum and boosted at day 28 with 20 μg soluble NP-KLH. On the indicated day, spleens (Sp) and bone marrow (BM) were harvested and a known number of cells were cultured overnight on ELISPOT plates with high (B) or low (C) conjugation ratio NP-specific coat. Bound antibody was detected by incubating with IgG1-HRP and spots revealed with AEC substrate. The number of PU.1+/+CD19 and PU.1Δ/ΔCD19 ASCs are shown in gray and white, respectively. Numbers represent the mean ± standard deviation. (D) Germinal center formation. Spleens from mice were collected at day 7 and 28 following immunization. Sections were cut and stained for the germinal center B cells (GL7, red/brown) and follicular B cells (IgD, blue).
The effect of PU.1 deletion on B-cell function. (A) Immunoglobulin isotype levels in untreated mice. Sera from 8- to 12-week-old mice (n = 6 for each genotype) were analyzed for levels of IgM, IgG1, IgG2a, IgG2b, IgG3, IgA, Igκ, and Igλ by ELISA. PU.1+/+CD19 and PU.1Δ/ΔCD19 are shown in black and white circles, respectively. (B-C) Number of NP-specific IgG1 ASCs in bone marrow and spleen. PU.1+/+CD19 and PU.1Δ/ΔCD19 mice were immunized with 100 μg NP-KLH precipitated in alum and boosted at day 28 with 20 μg soluble NP-KLH. On the indicated day, spleens (Sp) and bone marrow (BM) were harvested and a known number of cells were cultured overnight on ELISPOT plates with high (B) or low (C) conjugation ratio NP-specific coat. Bound antibody was detected by incubating with IgG1-HRP and spots revealed with AEC substrate. The number of PU.1+/+CD19 and PU.1Δ/ΔCD19 ASCs are shown in gray and white, respectively. Numbers represent the mean ± standard deviation. (D) Germinal center formation. Spleens from mice were collected at day 7 and 28 following immunization. Sections were cut and stained for the germinal center B cells (GL7, red/brown) and follicular B cells (IgD, blue).
The ability of the PU.1Δ/ΔCD19 B cells to respond to immunization by forming GCs and producing long-lived antigen-specific cells capable of secreting high-affinity antibody suggests that the apoptosis observed in vitro upon IgM cross-linking is overcome in vivo during T-dependent response. A possible explanation for this may be that T-cell help and particularly the effects of IL-4 counteract the apoptotic effects of BCR cross-linking. To test this hypothesis, we stimulated B cells with anti-IgM in the presence of IL-4 and measured the level of apoptosis observed over 30 hours. Figure 4B shows that apoptosis is significantly reduced when anti-IgM is combined with IL-4, to the extent that the PU.1Δ/ΔCD19 B cells are more viable than the PU.1+/+CD19 cells stimulated with anti-IgM alone. This result conflicts with data from PU.1+/-Spib-/- B cells that are not afforded protection from apoptosis by IL-4,21 suggesting a different molecular mechanism controlling BCR signal-induced apoptosis though most likely not involving A1, Bcl2, or BclXL.
Enhanced apoptosis of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. (A) Reduced proliferation of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Small resting naive B cells were isolated from the spleen and cultured in the presence of 10 μg/mL F(ab′)2 anti-IgM. Cultures were pulsed with 1 μCi (37 Bq) [3H]thymidine for 6 hours and thymidine incorporation assessed. Mean counts per minute (CPM) ± standard deviation is shown. PU.1+/+CD19 is shown in black circles and a solid line and PU.1Δ/ΔCD19 in white with a dashed line. (B) Enhanced apoptosis in PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Cells were cultured in the presence of 10 μg/mL anti-IgM or IL-4 or in combination for the indicated time, the number of viable cells (annexin V negative, propidium iodide-negative) assessed by FACS, and the data converted to the percentage of the initial culture that remained viable at that time point. Numbers represent the mean ± standard deviation. (C) The expression of antiapoptotic genes in PU.1Δ/ΔCD19 and PU.1+/+CD19 BCR-stimulated B cells. Total RNA was isolated from cells cultured for 24 hours and assessed for the expression of antiapoptotic genes by RT-PCR.
Enhanced apoptosis of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. (A) Reduced proliferation of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Small resting naive B cells were isolated from the spleen and cultured in the presence of 10 μg/mL F(ab′)2 anti-IgM. Cultures were pulsed with 1 μCi (37 Bq) [3H]thymidine for 6 hours and thymidine incorporation assessed. Mean counts per minute (CPM) ± standard deviation is shown. PU.1+/+CD19 is shown in black circles and a solid line and PU.1Δ/ΔCD19 in white with a dashed line. (B) Enhanced apoptosis in PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Cells were cultured in the presence of 10 μg/mL anti-IgM or IL-4 or in combination for the indicated time, the number of viable cells (annexin V negative, propidium iodide-negative) assessed by FACS, and the data converted to the percentage of the initial culture that remained viable at that time point. Numbers represent the mean ± standard deviation. (C) The expression of antiapoptotic genes in PU.1Δ/ΔCD19 and PU.1+/+CD19 BCR-stimulated B cells. Total RNA was isolated from cells cultured for 24 hours and assessed for the expression of antiapoptotic genes by RT-PCR.
In vitro anti-CD40 and LPS stimulation of PU.1Δ/ΔCD19 B cells
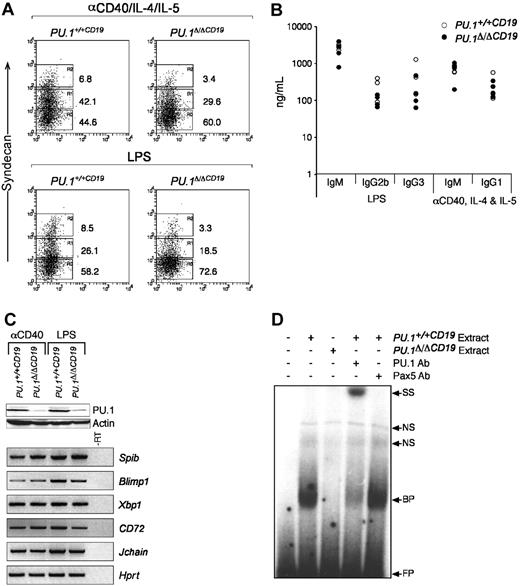
PU.1 has been suggested to regulate several components of the machinery required for the expression and secretion of immunoglobulin15-18,33 and directly interacts with PU.1 interacting partner (PIP)/interferon regulatory factor-4 (PIP/IRF4), a factor essential for immunoglobulin production.34 The data outlined in Figure 3 show normal ratios of the different isotypes in the serum of PU.1Δ/ΔCD19 mice, suggesting B cells that lack PU.1 are able to switch and secrete immunologlobulin. The homeostatic mechanisms that maintain immunoglobulin levels in the serum (such as the rate of clearance) may, however, be compensating for any functional abnormalities in PU.1Δ/ΔCD19 B cells. To investigate this possibility, we isolated naive B cells and cultured them in vitro with LPS or anti-CD40/IL-4/IL-5 and measured their capacity to differentiate into ASCs, which was first assessed by analyzing the expression of the marker of ASC CD138 (syndecan-1). In the case of both LPS and anti-CD40 an up-regulation of syndecan-1 was observed, although the percentage of positive PU.1Δ/ΔCD19 cells was lower in both conditions. To test whether switching and secretion occurred normally, we quantified the immunoglobulin secreted by ELISA. Figure 5 demonstrates that the absence of PU.1 had little effect on the levels of immunoglobulin switch recombination and antibody secretion induced in the presence of either LPS or anti-CD40. These data suggest that while the levels of syndecan-1 are reduced, the function of ASCs is not impaired by the lack of PU.1, and it therefore does not appear to play an essential role in the expression, switching, or secretion of immunoglobulin from stimulated B cells.
The in vitro development of comparatively normal ASCs from PU.1Δ/ΔCD19 B cells suggests that PU.1 is not essential for the expression of the machinery required for immunoglobulin secretion and switching. To test this hypothesis we analyzed the expression of genes critical for ASC function by RT-PCR. Figure 5C shows that genes essential for the development and function of ASCs, including Blimp1, Xbp1, and the purported direct PU.1 targets CD72 and Jchain, are expressed at normal levels in cells that Western blotting confirmed expressed greatly reduced levels of PU.1 protein.
PU.1Δ/ΔCD19 B-cell response to anti-CD40 and LPS. (A) Syndecan-1 expression in B-cell cultures. Small naive B cells were isolated from spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice and cultured with either anti-CD40/IL-4/IL-5 or LPS for 3 days, after which the expression of syndecan-1 was assessed. (B) Expression of immunoglobulin isotypes by anti-CD40/IL-4/IL-5- or LPS-stimulated PU.1+/+CD19 and PU.1Δ/ΔCD19 B cells. B cells from PU.1+/+CD19 and PU.1Δ/ΔCD19 mice were cultured for 3 days, washed, counted, and a known number of cells reseeded and cultured overnight in fresh media containing no stimuli. The supernatants were then harvested and the level of the indicated immunoglobulin isotypes measured by ELISA. (C) Western and RT-PCR analysis of gene expression in PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. (Top) Western hybridization of PU.1 in cells following 3 days of stimulation with anti-CD40/IL-4/IL-5 or LPS; β-actin is shown as a loading control. (Bottom) RT-PCR analysis of gene expression in PU.1+/+CD19 and PU.1Δ/ΔCD19 cells after 3 days of stimulation. Hprt is shown as a loading control, and a sample of PU.1+/+CD19 minus RT was used a negative control. (D) Electrophoretic mobility shift assays. Nuclear extracts were made from PU.1+/+CD19 and PU.1Δ/ΔCD19 B cells stimulated with LPS and incubated with a radiolabeled oligonucleotide representing the high-affinity PU.1 binding site from the SV40 promoter. Binding of PU.1 was confirmed by the addition of anti-PU.1 and anti-Pax5 antibodies. FP indicates free probe; BP, bound probe; NS, nonspecific; and SS, supershift.
PU.1Δ/ΔCD19 B-cell response to anti-CD40 and LPS. (A) Syndecan-1 expression in B-cell cultures. Small naive B cells were isolated from spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice and cultured with either anti-CD40/IL-4/IL-5 or LPS for 3 days, after which the expression of syndecan-1 was assessed. (B) Expression of immunoglobulin isotypes by anti-CD40/IL-4/IL-5- or LPS-stimulated PU.1+/+CD19 and PU.1Δ/ΔCD19 B cells. B cells from PU.1+/+CD19 and PU.1Δ/ΔCD19 mice were cultured for 3 days, washed, counted, and a known number of cells reseeded and cultured overnight in fresh media containing no stimuli. The supernatants were then harvested and the level of the indicated immunoglobulin isotypes measured by ELISA. (C) Western and RT-PCR analysis of gene expression in PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. (Top) Western hybridization of PU.1 in cells following 3 days of stimulation with anti-CD40/IL-4/IL-5 or LPS; β-actin is shown as a loading control. (Bottom) RT-PCR analysis of gene expression in PU.1+/+CD19 and PU.1Δ/ΔCD19 cells after 3 days of stimulation. Hprt is shown as a loading control, and a sample of PU.1+/+CD19 minus RT was used a negative control. (D) Electrophoretic mobility shift assays. Nuclear extracts were made from PU.1+/+CD19 and PU.1Δ/ΔCD19 B cells stimulated with LPS and incubated with a radiolabeled oligonucleotide representing the high-affinity PU.1 binding site from the SV40 promoter. Binding of PU.1 was confirmed by the addition of anti-PU.1 and anti-Pax5 antibodies. FP indicates free probe; BP, bound probe; NS, nonspecific; and SS, supershift.
The absence of a severe phenotype in B cells lacking PU.1 may be interpreted as there being a functional redundancy between PU.1 and some other factor. An obvious candidate for this factor is Spi-B, because it has been shown to compensate for PU.1 in some instances23 ; however, we have not observed any up-regulation of Spib mRNA in the absence of PU.1. In an attempt to assess whether Spi-B substitutes for PU.1 in B cells, we made nuclear extracts from LPS-stimulated PU.1Δ/ΔCD19 and PU.1+/+CD19 cells and assessed the level of PU.1-like DNA binding activity by EMSA (Figure 5D). These experiments used a binding site in the SV40 promoter, which has a high affinity for PU.116,35 and presumably for any molecule able to functionally substitute for it. Extracts from PU.1+/+CD19 cells interacted strongly with the probe, whereas no equivalent DNA binding activity was observed with extracts from PU.1Δ/ΔCD19 cells. The band obtained from PU.1+/+CD19 extracts was confirmed as PU.1 by the addition of an anti-PU.1 antibody, which resulted in a supershift, whereas an anti-Spi-B antibody had no effect (data not shown). Taken together the data in this study suggest that PU.1 is dispensible for the development and function of B lymphocytes from the pre-B stage onward.
Discussion
B lymphopoiesis is a multistep highly regulated process beginning with the HSC and ending with the production of B lymphocytes that are competent to respond to antigen by differentiating into plasma cells capable of secreting massive amounts of high-affinity immunoglobulin. Two knock-out and numerous molecular biology studies have implicated the transcription factor PU.1 as being critical for key stages in this entire process, from the initial commitment of progenitors to the B lineage to the expression and secretion of immunoglobulin. In the work presented here, we have made use of an inducible PU.1 deletion allele to rigorously test this entrenched view.
The phenotype of a true PU.1 null mouse has been the point of some conjecture because the 2 alternate deletion strategies adopted resulted in some significant differences; however, in both cases the B-cell lineage was severely perturbed.2,3 Scott et al2 reported no B lymphocytes, whereas McKercher et al3 observed a population of aberrant B220+ cells that did not correspond to any known B-cell subset. In the present study, the complete deletion of the inducible PU.1 allele resulted in the loss of B cells along with all other lymphoid and myeloid cells within the fetal liver and death between E18.5 and birth. This phenotype is similar to that of Scott et al,2 supporting the conclusion that their approach best represents a true PU.1 null.
The hematopoiesis-wide defects caused by PU.1 deletion prompted us to investigate whether there was a specific failure of B lymphopoiesis or rather if the loss of B cells was due to a more catastrophic failure in hematopoietic progenitors. Deletion of PU.1 in the B lineage using a CD19-Cre transgene had no pronounced effect on development, with B cells present in the fetal liver and the adult bone marrow, spleen, lymph nodes, and peritoneum. This implies that after the point when CD19 transcription begins, when cells are committed to the B lineage, PU.1 is not essential for further development. It is interesting that within the pre-BI fraction of PU.1Δ/ΔCD19 mice, PCR analysis demonstrated that 50% of the PU.1 alleles were deleted; however, this jumped to 95% by the subsequent pre-BII large stage. A previous study has reported Cre-mediated deletion rates of only 50% in pre-B cells,36 suggesting that lacking PU.1 may provide a developmental advantage at the pre-BI to pre-BII large transition. The data presented here do not address the requirement for PU.1 at the initial stages of commitment to the lymphoid and B lineages, because deletion driven by CD19-Cre begins after commitment has occurred.
PU.1 has been suggested to regulate the expression of B2208 and, in agreement with this, no B220+ cells were observed in the fetal liver of PU.1Δ/Δdeleter mice. Deletion of PU.1 after B-cell commitment, however, had no effect on B220 expression, demonstrating that PU.1 function is not required to maintain its expression. A recent study in the authors' laboratory has shown that within 14 days of deleting PU.1 using the MX-Cre/polyIC inducible system, IL-7R expressing common lymphoid progenitors was no longer detectable and that, when PU.1 was deleted in chimeric mice made up of wild-type and PU.1fl/fl cells, no lymphocytes were produced from the deleted progenitors.25 Furthermore, the expression of all splice variants of CD45 is lost,25 apart from that seen on long-lived mature B cells, a finding in agreement with Medina et al,8 Anderson et al,37 and the data presented here.
We propose that these apparently different effects are the result of PU.1 normally functioning to initiate or create the conditions necessary for the expression of genes such as CD45 in the earliest hematopoietic cells but subsequently playing little or no role in maintenance of their expression. This would explain the apparent paradox of the widespread hematopoietic defects caused by unconditional deletion of PU.1 versus the relatively normal differentiation when deletion of PU.1 is targeted to the B-cell lineage. Another gene that may fall into this category is Ilr7, because a report by DeKoter et al9 has shown that PU.1 directly activates its expression; however, we have shown here that deletion of PU.1 after commitment to the B lineage has no effect on Ilr7 expression.
The transcription factor EBF has also been shown to be regulated by PU.1 and can substitute efficiently for PU.1 by driving in vitro B-cell development from PU.1-/- FLPs.8 We show here a contemporaneous reduction in Ebf1 levels when PU.1 is deleted, suggesting that unlike B220 or Ilr7, the activity of PU.1 is required for EBF expression at the pre-BII stage. Although there is a slight perturbation in the makeup of the B-cell fractions in PU.1Δ/ΔCD19 mice, the reduction in Ebf1 beyond the pre-BII stage does not appear to have any significant deleterious effect on B lymphopoiesis. This is despite a reduction in the levels Rag1 expression and that of the surrogate light chain gene VpreB. VpreB has been shown to be a direct target of EBF in both mice and humans,38-40 explaining why its expression is reduced in PU.1Δ/ΔCD19 mice.
The lethality of deleting PU.1 and the failure of mutant fetal liver cells to reconstitute adult bone marrow has precluded studying if there is a requirement for PU.1 activity during B-cell function. There have been, however, several studies intimating a key role for PU.1, because it has been shown to bind regulatory elements within genes required for the function of B cells such as IgH,15 Igκ,16 Igλ,17 J chain,18 and CD72.19 A further link between PU.1 activity and B-cell function came when a single copy of PU.1 was knocked out in mice already deficient for the related factor Spi-B.21 This combination resulted in the intensification of BCR cross-linking-mediated apoptosis observed in Spi-B-deficient mice and the corresponding complete failure of GCs to form. A follow-up study demonstrated that the phenotype was due a lack of up-regulation of the prosurvival gene c-rel in PU.1+/-Spib-/- B cells.22
To assess the effect of deleting PU.1 alone on an immune response, we immunized mice with the well-characterized immunogen NP-KLH, which elicits a high-affinity anti-NP IgG1λ response.41 We found that contrary to expectation, PU.1-deficient B cells were able to respond to antigen by producing high-affinity ASCs and forming GCs. It appears PU.1 does have a role in the end stages of B lymphopoiesis, because there was a reduction in the overall number of ASCs in the PU.1Δ/ΔCD19 mice, which in vitro studies suggested was due to increased apoptosis. The observation that PU.1Δ/ΔCD19 B cells stimulated in vitro with anti-IgM were seen to express crel, albeit at reduced levels, along with other protective antiapoptotic genes and the results showing that IL-4 is able to protect PU.1Δ/ΔCD19 B cells from BCR-mediated apoptosis suggest the increased apoptosis is operating through a mechanism different from that reported for PU.1+/-Spib-/- B cells.22
An immediate and obvious retort to the lack of phenotype in B cells lacking PU.1 is that another factor—most likely Spi-B—is functionally substituting for PU.1. Indeed, the DNA binding specificities of the 2 factors have been suggested to be virtually identical.42 The argument in favor of redundancy between PU.1 and Spib is provided some validity by the observation that Spib can partially substitute for PU.1 in myeloid development but not lymphoid development,23 a somewhat enigmatic finding given that Spi-B is normally only expressed in B and T cells and not the myeloid lineages.43 The data presented here provide 2 pieces of evidence to argue against a complete functional redundancy between Spi-B and PU.1. Firstly, the expression of Ebf1, a known PU.1 target gene, is reduced in PU.1-deficient pre-BII cells despite continued expression of Spib. Secondly, gel shift assays performed with an oligonucleotide from the SV40 promoter that has been shown to have a high affinity for PU.116,35 and presumably any factor capable of functionally substituting for it suggest that there is no PU.1-like binding activity in PU.1Δ/ΔCD19 B cells and that Spi-B, if present, is not substituting for it.
The data presented in this paper support the surprising conclusion that PU.1 is not required to generate functional B cells from the pre-B stage onward and rather that the failure of B lymphocytes to form in PU.1-/- mice is a result of defects in progenitor cells rather than the commitment of cells to the B lineage. In addition, despite a substantial archive of in vitro data purporting an essential role for PU.1 in the regulation of immunoglobulin expression, switching, and secretion, we have found that the deletion of PU.1 did not substantially affect the function of ASCs.
Prepublished online as Blood First Edition Paper, June 2, 2005; DOI 10.1182/blood-2005-01-0283.
Supported by the Cancer Council Victoria and The National Health and Medical Research Council of Australia. S.L.N. is the Walter and Eliza Hall Institute Metcalf Fellow.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors express their gratitude to Jaclyn Carneli for expert animal husbandry.

![Figure 1. Conditional deletion of PU.1. (A) PU.1 conditional allele. The PU.1 targeting construct containing exon 5 of PU.1 surrounded by LoxP sites (▴) and a reporter cassette flanked by 2 Frt sites (•) containing both the internal ribosome entry site-green fluorescent protein (IRESGFP) and the phosphoglycerate kinase (PGK) promoter driving the selection marker Neomycin (Neo) was introduced into the PU.1 locus by homologous recombination to generate the PU.1gfp allele. When crossed to mice harboring flp recombinase, the reporter/selection cassette is lost, generating PU.1fl allele. Exon 5 of PU.1 is deleted by crossing with mice carrying Cre recombinase, creating the allele PU.1Δ. (B) Expression of lineage markers in E18.5 fetal livers. Following isolation, fetal livers were treated with red cell lysis buffer and analyzed by flow cytometry for the presence of B cells (CD19+B220+) and monocytes/granulocytes (Gr-1+Mac-1+). (C) Deletion efficiency of PU.1fl/fl alleles in the B-cell fractions of PU.1Δ/ΔCD19 mice. Isolated B-cell fractions from the fetal liver (left panel), bone marrow, and spleen (right panel) were analyzed by competitive PCR for the presence of targeted (fl) and deleted (Δ) PU.1 alleles. The antibody combinations used and the rate of deletion calculated by densitometry are as follows: fetal liver (CD19+B220+, 73%); bone marrow: pre-BI (B220+ckit+, 59%), pre-BII large (B220+c-Kit-CD25+, forward scatter [FSC] large, 86%), pre-BII small (B220+c-Kit-CD25+, FSC small, 86%), immature B (B220loIgM+, 90%), and mature recirculating (B220hiIgM+, 98%); spleen: transitional 1 (CD23-IgM+-CD21lo, 98%), transitional 2 (CD23+IgMhiCD21hi, 96%), follicular (CD23+IgMloCD21lo, 92%), and marginal zone (CD23-IgM+CD21hi, 100%). (D) Loss of PU.1 in B cells from PU.1Δ/ΔCD19 mice. Total protein extracts were made from IgM+B220+ cells sorted by flow cytometry from the spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. The presence of PU.1 and β-actin was assessed by Western hybridization.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2005-01-0283/6/m_zh80180584170001.jpeg?Expires=1766023222&Signature=WYDygi8IjMovZslq3g6elG74JbLOAJ43~2if5owPnQR-4bZqd8JT4RZ4amuyj-GHETgzSnA1bc8TdrKl~3K-bNIrYxX7ZzAacS~KpUuaaPsFmEPQI104ScwDXlGVbd9-AHW2bXt8YksKkSYoGElrp5rkkweXB7e2pVUzePn7Yn0nHDs8OJ5GdIqVlihcoLCm~~AY5xfMGX-jDgiJeZhp9I2BhFZ7PFuwcAWXKOQ9wDYvvCqW7OxFv54nQPRvpXblv8ge-g10hlhjvrj0rt2-TA02fQGR7ur2ZREHvDnT0ksBsZRgKrv9fqim1RWe0XyMj-GslwdXQmggaTrsqXvSgg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Enhanced apoptosis of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. (A) Reduced proliferation of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Small resting naive B cells were isolated from the spleen and cultured in the presence of 10 μg/mL F(ab′)2 anti-IgM. Cultures were pulsed with 1 μCi (37 Bq) [3H]thymidine for 6 hours and thymidine incorporation assessed. Mean counts per minute (CPM) ± standard deviation is shown. PU.1+/+CD19 is shown in black circles and a solid line and PU.1Δ/ΔCD19 in white with a dashed line. (B) Enhanced apoptosis in PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Cells were cultured in the presence of 10 μg/mL anti-IgM or IL-4 or in combination for the indicated time, the number of viable cells (annexin V negative, propidium iodide-negative) assessed by FACS, and the data converted to the percentage of the initial culture that remained viable at that time point. Numbers represent the mean ± standard deviation. (C) The expression of antiapoptotic genes in PU.1Δ/ΔCD19 and PU.1+/+CD19 BCR-stimulated B cells. Total RNA was isolated from cells cultured for 24 hours and assessed for the expression of antiapoptotic genes by RT-PCR.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2005-01-0283/6/m_zh80180584170004.jpeg?Expires=1766023222&Signature=JsSvqRwuLqobvX9PVcsJMX6UD0Fc23GnNZbGAMOX4C0Bwr0j-0mYIddRgVoK5CgvkONL0DTPmFXT4eVSTkETaqvzpetQ3iMrXZns0NjplpCcgfyF9uTBYz0HWPIKrXPvDLHC4Q33TbE6DT8DHreadqDZq8zUkE3QPLXxgsaGxFVpDk9xvbecXyGJ6fO7MtGNG8QqTycUlbmRe0yQ~g1zI4ZeszC4ATvttVDmApr9UMVg~Q-rW961BMEw135~uzjTUbdu3sSy0HKLpzZ6VHU7TyTGPovdtTxymPbobAAY5jvDvtRkHEE8Q131Yj7t25cguBZJtGDXtJzATFfpvvFymw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Conditional deletion of PU.1. (A) PU.1 conditional allele. The PU.1 targeting construct containing exon 5 of PU.1 surrounded by LoxP sites (▴) and a reporter cassette flanked by 2 Frt sites (•) containing both the internal ribosome entry site-green fluorescent protein (IRESGFP) and the phosphoglycerate kinase (PGK) promoter driving the selection marker Neomycin (Neo) was introduced into the PU.1 locus by homologous recombination to generate the PU.1gfp allele. When crossed to mice harboring flp recombinase, the reporter/selection cassette is lost, generating PU.1fl allele. Exon 5 of PU.1 is deleted by crossing with mice carrying Cre recombinase, creating the allele PU.1Δ. (B) Expression of lineage markers in E18.5 fetal livers. Following isolation, fetal livers were treated with red cell lysis buffer and analyzed by flow cytometry for the presence of B cells (CD19+B220+) and monocytes/granulocytes (Gr-1+Mac-1+). (C) Deletion efficiency of PU.1fl/fl alleles in the B-cell fractions of PU.1Δ/ΔCD19 mice. Isolated B-cell fractions from the fetal liver (left panel), bone marrow, and spleen (right panel) were analyzed by competitive PCR for the presence of targeted (fl) and deleted (Δ) PU.1 alleles. The antibody combinations used and the rate of deletion calculated by densitometry are as follows: fetal liver (CD19+B220+, 73%); bone marrow: pre-BI (B220+ckit+, 59%), pre-BII large (B220+c-Kit-CD25+, forward scatter [FSC] large, 86%), pre-BII small (B220+c-Kit-CD25+, FSC small, 86%), immature B (B220loIgM+, 90%), and mature recirculating (B220hiIgM+, 98%); spleen: transitional 1 (CD23-IgM+-CD21lo, 98%), transitional 2 (CD23+IgMhiCD21hi, 96%), follicular (CD23+IgMloCD21lo, 92%), and marginal zone (CD23-IgM+CD21hi, 100%). (D) Loss of PU.1 in B cells from PU.1Δ/ΔCD19 mice. Total protein extracts were made from IgM+B220+ cells sorted by flow cytometry from the spleens of PU.1+/+CD19 and PU.1Δ/ΔCD19 mice. The presence of PU.1 and β-actin was assessed by Western hybridization.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2005-01-0283/6/m_zh80180584170001.jpeg?Expires=1766188595&Signature=n3kaA1otVh~mTDOYvFWkbRCpVW-CqGNMSLidN3kZbxzrbNIRfFWrxt8vKiLC4HKYoSvheRJMsBtzahG41R2O4ktFv5G3iYthGaqV33gSIHfm6o4AOoK73UVIrefuuuXC0-ZRRuTObmPbW88m1VL3ZfTXNJ8xLgiV9IqP~JKi~Sw2tvUbkIkghm7Gq3cg2~OwGGjLHG~hN07AhDH~rMILkAZKC09JFZWsG7vEiKDPNFZQtjI0MX94pud2cVgK1Rf~hDXe5XhvR77MT-Lh4cAqPegdC2d8UdcPbkmYq5FNPEOU0XwbRi7KIHon-oaL8~FAkPZlvC4m8yKA6reOAWfLkw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 4. Enhanced apoptosis of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. (A) Reduced proliferation of PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Small resting naive B cells were isolated from the spleen and cultured in the presence of 10 μg/mL F(ab′)2 anti-IgM. Cultures were pulsed with 1 μCi (37 Bq) [3H]thymidine for 6 hours and thymidine incorporation assessed. Mean counts per minute (CPM) ± standard deviation is shown. PU.1+/+CD19 is shown in black circles and a solid line and PU.1Δ/ΔCD19 in white with a dashed line. (B) Enhanced apoptosis in PU.1Δ/ΔCD19 B cells upon BCR cross-linking. Cells were cultured in the presence of 10 μg/mL anti-IgM or IL-4 or in combination for the indicated time, the number of viable cells (annexin V negative, propidium iodide-negative) assessed by FACS, and the data converted to the percentage of the initial culture that remained viable at that time point. Numbers represent the mean ± standard deviation. (C) The expression of antiapoptotic genes in PU.1Δ/ΔCD19 and PU.1+/+CD19 BCR-stimulated B cells. Total RNA was isolated from cells cultured for 24 hours and assessed for the expression of antiapoptotic genes by RT-PCR.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/106/6/10.1182_blood-2005-01-0283/6/m_zh80180584170004.jpeg?Expires=1766188595&Signature=4Wlc4~wpQoS~tt7uo4yEkeqKk-IWm~B5x58EabkUWHMWllulb8ZmJfYIvfRoUD0kXcHNOiXONiVG1bu8DIloxphoDj7XHuioPDRCnm~SXXUjJUJBdSCg8FcyebGxFIh5ghS9zyEihmuugV5Z80C5uhP-kBUuAf8DiIuLahiWWvDlFSFdMFpGC9BqJ2U4GkSuSNvUCY~VuOhtKRtTNL1MCBrJh-Bmq6R88W7Eez4FhjUVZDqa~7vDnZVgoWTnMNFD0Rm7PSdvSyS9hMJUQZz5nstYkHjkkWqDtT7KV1KMCjdHf-s4~H4q2qAwa3ClkrQRaOfA4IQqQw3slHzaDn0POg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)