Abstract
Although most patients with chronic myeloid leukemia (CML) have the same initial molecular abnormality, the BCR-ABL fusion gene, the duration of chronic phase (CP) varies widely. To identify the possible molecular basis of this heterogeneity, we studied CD34+ cells collected at diagnosis from 68 patients with CML-CP. By using oligonucleotide microarray screening, we performed gene-expression profiling on 2 subsets of patients, one comprising patients with an “aggressive disease” who developed blastic transformation (BT) within 3 years of diagnosis (n = 10) and, at the other extreme, patients with an “indolent disease” whose BT occurred 7 or more years from diagnosis (n = 9). This screening revealed 20 genes differentially expressed in patients with aggressive and indolent disease, which were validated by quantitative reverse transcriptase/polymerase chain reaction (Q-RT/PCR). A multivariate Cox regression model identified the combination of low CD7 expression with high expression of proteinase 3 or elastase as associated with longer survival in the complete cohort of 68 patients. This differential pattern of gene expression probably reflects the intrinsic heterogeneity of the disease; if so, assessing expression levels of selected genes at diagnosis may be valuable in predicting duration of survival in patients treated with imatinib and the newer tyrosine kinase inhibitors.
Introduction
Chronic myeloid leukemia (CML) is characterized cytogenetically by a reciprocal t(9;22)(q34;q11) chromosomal translocation which results in the formation of the BCR-ABL oncogene. This encodes a constitutively activated tyrosine kinase1 responsible for the clinical features of the leukemia.2,3 CML typically presents in chronic phase (CP), when the disease responds well to treatments such as hydroxyurea, interferon-α, or the new tyrosine kinase inhibitor, imatinib. Before the imatinib era, CP lasted on average 4 to 5 years before progressing to blastic transformation (BT), which usually proved fatal within a few months. Thus, until recently the median survival for patients treated predominantly with interferon-α was around 5 to 6 years,4 but imatinib, now widely regarded as the best initial therapy for CML in CP, promises to prolong life very substantially.5 However, despite the very encouraging responses to imatinib, there is good evidence that it does not eradicate all leukemic progenitor cells,6-8 this in effect means that allogeneic stem cell transplantation (SCT), ideally performed in the early stage of the disease, is still the only curative treatment for CML.
Despite a consistent molecular finding, CML has always exhibited a marked heterogeneity in the duration of CP,4 and preliminary experience suggests that this heterogeneity is also seen in patients treated with imatinib.5 Various attempts have been made to determine prognosis for individual patients at the time of diagnosis in CP. Thus, the Sokal and Hasford prognostic scores9,10 proved moderately useful in predicting the duration of survival for individual patients treated with busulfan or interferon-α, respectively. The Sokal score also serves to discriminate survival without progression in patients treated with imatinib11 and may still be useful to guide therapeutic decisions.12 Deletions adjacent to the ABL-BCR junction on the derivative 9q+ chromosome may be associated with an adverse prognosis,13 but only about 10% of patients with CML have this additional cytogenetic abnormality. Telomere lengths, which are already shorter in CML compared with normal cells, have been reported to be further shortened in patients in CP with early disease progression14 or a more rapid onset of BT.15,16
We investigated the hypothesis that differential expression of specific genes at the time of diagnosis may underlie the intrinsic heterogeneity of CML, in which case profiling the expression of selected genes would be useful to predict the duration of survival even in patients treated with tyrosine kinase inhibitors. Using oligonucleotide microarrays, we identified 20 genes whose expression significantly differentiated between patients with aggressive versus indolent disease in an initial cohort of 19 patients diagnosed in CP. We then confirmed these findings using quantitative real-time reverse transcription/polymerase chain reaction (Q-RT/PCR) and correlated gene expression with patient survival using a multivariate Cox regression model on 68 patients with CML. We found that the combination of a low expression level of CD7, with a high expression of proteinase 3 (PR-3) or elastase (ELA2), was highly predictive of a longer survival.
Patients, materials, and methods
Patients
Patients with Philadelphia (Ph) chromosome–positive CML whose nucleated cells were collected by leukapheresis and cryopreserved within 3 months of diagnosis before start of treatment were eligible for this study if the stored cells were still viable and if adequate clinical data were available. Informed consent for the use of these cells for research had been obtained according to the requirements of the Hammersmith, Queen Charlotte, and Chelsea and Acton Hospitals Research Ethics Committee (London, United Kingdom), and, where possible, in accordance with the Declaration of Helsinki. Diagnosis of CML-CP was based on clinical parameters and morphology of blood and bone marrow17 and was confirmed by a karyotype exhibiting the t(9;22)(q34;q11) translocation with a single Ph chromosome. Patients were excluded if they (1) had variant translocations or additional cytogenetic abnormalities at diagnosis and (2) were in long-term remission after allogeneic SCT. Out of more than 400 patients, a cohort of 68 fulfilled all of these criteria; each of these 68 patients had been diagnosed with CML-CP and underwent leukapheresis within 3 months of diagnosis between October 1979 and July 2001. The majority of exclusions were due to the unavailability of cells collected and stored within 3 months of diagnosis. The median age at diagnosis of the selected patients was 45.2 years (range, 17.6-68.3 years). The male-to-female ratio was 1.8:1 (44 males, 24 females). The majority of patients were diagnosed in the era before imatinib and treated with hydroxyurea and/or interferon-α. Patient characteristics are shown in Table 1. Their median survival was 8.7 years. Patients who developed BT within 3 years of diagnosis were defined as having aggressive disease (n = 18), whereas those who survived for longer than 7 years prior to the onset of BT were defined as having indolent disease (n = 23). Patients who had survived between 3 and 7 years without developing BT were categorized as having intermediate disease (n = 27). The relatively low median age of patients in this series reflects the pattern of tertiary referral to the Hammersmith Hospital in London.
Sample preparation and oligonucleotide microarray methods
Mononuclear cells from cryopreserved material were isolated by density gradient centrifugation (Lymphoprep; Nycomed, Oslo, Norway), and CD34+ cells were selected by binding to immunomagnetic beads (Mini-MACS [magnetic-activated cell sorting]; Miltenyi Biotech, Bergisch-Gerbach, Germany). The mean CD34+ purity was 92% ± 5.5%. Total RNA was extracted from CD34+ cells using the Qiagen RNeasy kit (Qiagen, Crawley, United Kingdom). Of the 68 samples, only 19 (10 aggressive and 9 indolent CML) yielded sufficient RNA for gene expression profiling on HG-U133A GeneChip arrays (Affymetrix, Santa Clara, CA). The detailed protocol for sample preparation and microarray processing is available on line.18 The 19 samples used for gene expression profiling were also analyzed for confirmation of the presence of a single Ph chromosome and for screening for a deletion of the 9q+ derivative by fluorescence in situ hybridization using the BCR-ABL LSI dual-color dual-fusion translocation probes (Vysis, Downers Grove, IL).
Microarray data analysis
Microarray expression measures were generated from 3 data analysis packages: MAS 5.0 (Affymetrix, Santa Clara, CA),19 dChip20 (www.dchip.org), and RMA (Robust Multichip Average (www.bioconductor.org)),21 which use different data extraction and normalization algorithms, and were loaded into GeneSpring 5.0 software (Silicon Genetics, Redwood City, CA). Expression measures derived from MAS 5.0 were normalized by using the median expression values of each array and each probe set in the experiment. Normalized expression measures generated from both MAS 5.0 and dChip (which use an invariant set normalization method) were then filtered by excluding probe sets with (1) expression measures within 2-fold of the local background in all arrays and (2) greater than 75% of arrays having “Absent” calls as assigned by the software. The expression measures between the indolent and aggressive CML groups were then compared using the Mann-Whitney U test. Expression measures from the 2 patient groups derived from RMA, which had undergone quantile normalization, were compared using a 2-tailed Student t test. A P value of less than .05 was considered significant and identified genes as differentially expressed between the 2 groups only if confirmed by all 3 analysis packages. In addition to minimizing the inclusion of probe sets identified because of an inherent bias present in a given algorithm, this methodology is also expected to allow the implementation of different measures to minimize nonbiologically relevant noise. Hierarchical clustering of the probe sets representing the most discriminatory genes was performed with HCE software (Human-Computer Interaction Lab, University of Maryland, College Park; www.cs.umd.edu/hcil/hce).
Probe sequences of each probe set on the array identified as differentially regulated between the aggressive and indolent CML groups were obtained from the Affymetrix website (http://www.affymetrix.com) and aligned with the most updated version of the mRNA sequence stored in the Reference Sequence (RefSeq) database curated by the National Center for Biotechnology Information (Bethesda, MD). Probe sets containing more than 2 (of 11) probe sequences mismatched with the consensus mRNA were considered unreliable for gene expression analysis.
Q-RT/PCR amplification
RNA was treated with DNase I (Invitrogen, Paisley, United Kingdom) to eliminate genomic DNA, and random hexamer primed cDNA was synthesized according to standard methods. Expression of the 20 genes identified by microarray screening as differentially regulated in the aggressive and indolent groups was assessed by Q-RT/PCR using the ABI PRISM 7700 sequence detection system (Applied Biosystems, Foster City, CA). All Q-RT/PCR reactions were performed on 25 μL volume (Tables 2 and 3). The ABI Assays-on-demand TaqMan probe-and-primer reagents were used according to the manufacturer's instructions for 14 genes, ELA2, PR-3, STARD1, AZU1, EDN, DF, MBP, CG, CLC, GFI1, CXCR4, HIST1H2BG, HTM4, and CEBPA. Fluorogenic LUX primers (Invitrogen) were used for MPO, CD7, SMA4, FUT4, ECP, and HM74 according to the manufacturer's instructions. ABL expression was used as the endogenous cDNA quantity control for all samples.22 Its expression was measured using the Q-RT/PCR core kit (Eurogentec, Romsey, United Kingdom), 300 nM primers and 200 nM probe.
Flow cytometry
Fluorescence-activated cell sorting (FACS) was performed with the FACS-Calibur flow cytometer and analyzed using CellQuestPRO software (Becton Dickinson, Cowley, Oxford, United Kingdom). Lineage and maturation markers were assessed on CD34+ cells from 16 of the 19 samples included in the microarray experiments. The panel of monoclonal antibodies consisted of fluorescein isothiocyanate (FITC)–conjugated lineage markers CD2, CD19, CD14, CD16, CD61, and glycophorin A (Caltag, Burlingame, CA); phycoerythrin (PE)–conjugated CD38; peridinin chlorophyll protein (PerCP)–conjugated CD34; and allophycocyanin (APC)–conjugated CD7. A minimum of 5 × 105 cells were labeled with 2 μL each of the FITC-conjugated lineage cocktail, CD38-PE and CD7-APC and 10 μL CD34-PerCP, incubated on ice for 30 minutes, and subsequently washed in phosphate-buffered saline (PBS) prior to analysis. A minimum of 100 000 events were acquired.
For proteinase 3 (PR-3) intracellular staining, CD34+ cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature, permeabilized with 0.3% saponin in 0.5% bovine serum albumin–supplemented PBS, and stained with 3 μL murine CLB-12.8 clone (a kind gift from Dr Y. M. van der Geld, Groningen, The Netherlands) for 1 hour at room temperature. After washing in 0.3% saponin PBS, the cells were incubated for 30 minutes at room temperature with 2 μL goat-anti–mouse Ig conjugated to FITC. For identification of cells with dual CD7 and CD34 surface markers, CD34+ cells were incubated on ice for 30 minutes with 2 μL CD7-FITC (Caltag) and 10 μL CD34-PerCP (Becton Dickinson) or 2 μL CD34-PE (Caltag) conjugated monoclonal antibodies. After washing in PBS, a minimum of 30 000 events were acquired.
Statistical methods
The results obtained from microarray and Q-RT/PCR assays were compared by Spearman ρ. Survival curves were calculated by using the Kaplan-Meier method, and groups were compared using the log-rank test. Patients were divided into groups by using Q-RT/PCR or protein expression values (FACS measurement) delineated by the median, upper, or lower quartile cutoffs. Genes with discriminating expression values identified from the univariate analysis with P values of less than .20 were entered into a Cox regression analysis, and a forward and backward stepping procedure was used to find the best model to predict survival. All quoted P values are from 2-sided tests, with values less than .05 considered significant.
Results
Standard prognostic indicators
Sokal prognostic scores were calculated, and comparisons were made between the 3 prognostic groups, but in this group of patients there were no differences in survival probabilities (P = .90) (Table 1). The derivative 9q+ deletion13 was found in only 1 patient of the 19 included for microarray screening. There was no difference in the proportion of the CD34+ subsets of CD38+Lin+, CD38+Lin-, or CD38-Lin- cells between patients in the indolent as compared with the aggressive CML groups (data not shown).
Gene expression profile of CML CD34+ cells
Microarray files are available from http://www.ebi.ac.uk/arrayexpress/query/login. The expression measures of 32 probe sets, from a total of 22 283 present on the HG-U133A GeneChip array, were identified as significantly different between the indolent and aggressive CML groups using the combination of MAS 5.0, dChip, and RMA software. Subsequently, the exclusion of probe sets with sequences mismatching public mRNA databases and those bearing gene sequences which were Y-chromosome associated (excluded as there were more men in the aggressive CML group) yielded 25 probe sets, representing 20 genes (Table 4). Hierarchical clustering using the expression of these genes demonstrated the demarcation between indolent and aggressive CML groups (Figure 1).
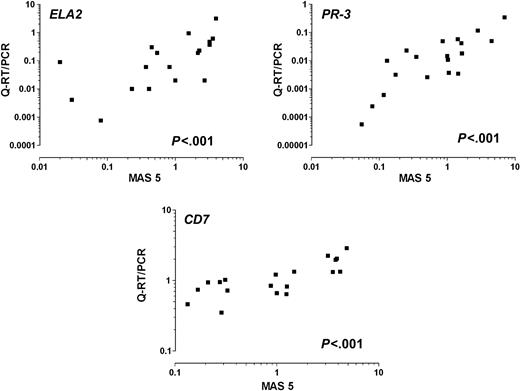
There was good correlation between the measurements of gene expression by microarray hybridization and Q-RT/PCR amplification, with 18 of the 20 genes having P values less than .005, STARD1 P = .033, and only FUT4 with poor correlation (P = .21) (Figure 2).
Association between gene expression and survival
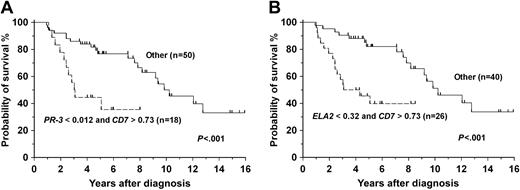
Univariate analyses of survival using expression measures from Q-RT/PCR for each of the 20 genes identified STARD1, MBP, ELA2, AZU1, PR-3, CD7, and SMA4 as being associated with differential survival in the entire cohort of 68 patients (Table 5). In Cox regression analyses, a backward-stepping approach found high expression of CD7 (relative risk [RR], 2.42; 95% confidence interval [CI], 1.16-5.03) and low expression of ELA2 (RR, 3.26; 95% CI, 1.28-8.32) to be associated with poor survival, whereas a forward stepping method disclosed high expression of CD7 (RR, 2.28; 95% CI, 1.11-4.67) and low expression of PR3 (RR, 2.30; 95% CI, 1.11-4.78). Combinations of these variables enabled patients with poor risk to be identified (Figure 3).
Hierarchical clustering using microarray expression measures on 20 most significantly different genes, represented by 25 probe sets, in 19 patient samples. Ind indicates indolent CML; Agg, aggressive CML. Red squares indicate genes that were overexpressed, and green squares indicate genes that were underexpressed. Note that in 4 cases individual genes were studied with more than one probe set.
Hierarchical clustering using microarray expression measures on 20 most significantly different genes, represented by 25 probe sets, in 19 patient samples. Ind indicates indolent CML; Agg, aggressive CML. Red squares indicate genes that were overexpressed, and green squares indicate genes that were underexpressed. Note that in 4 cases individual genes were studied with more than one probe set.
Representative scatter plots showing a good correlation between the expression of ELA2, PR-3, and CD7 measured by microarray (MAS 5.0, using an arbitrary scale) and Q-RT/PCR methods (P values refer to Spearman ρ).
Representative scatter plots showing a good correlation between the expression of ELA2, PR-3, and CD7 measured by microarray (MAS 5.0, using an arbitrary scale) and Q-RT/PCR methods (P values refer to Spearman ρ).
Measurement of gene expression at the protein level by FACS confirmed that the percentage of CD34+ cells with PR-3 (n = 25) and CD7 (n = 29) expression was also of prognostic value by a univariate analysis (P = .029 and P = .031, respectively) (Figure 4). Patients who had greater than 11% CD34+ cells expressing surface CD7 protein at diagnosis had a poor survival. Conversely, patients with greater than 2% CD34+ cells expressing cytoplasmic PR-3 had a superior survival.
Discussion
The prognosis for patients with CML depends on the stage of the disease at presentation,9 but, even for patients diagnosed in CP, there is significant variability in survival.4,23 Many efforts have in the past been made to predict survival for individual patients diagnosed in CP but neither the Sokal9 nor the Hasford10 systems, both currently in common use, provide the prognostic precision necessary to make important therapeutic decisions, notably to decide for a given patient whether to offer initial treatment with antileukemia drugs or to proceed immediately to allogeneic SCT. Moreover, the ability to characterize a patient's leukemia before starting treatment with a tyrosine kinase inhibitor would contribute to the decision whether to change therapy if the patient's subsequent response were suboptimal. Here, we compared gene expression in CD34+ cells collected at diagnosis from patients with CML who proved subsequently to have either relatively short or relatively long survivals and showed that 20 genes were differentially regulated. In particular, patients whose CD34+ cells had increased expression of CD7 in combination with a low expression of either ELA2 or PR-3 had a poor survival probability. Furthermore, the prognostic information derived from the differential level of expression of these genes in patients with CML was confirmed at the protein level for the 2 genes that could be tested by FACS analysis, namely PR-3 and CD7.
The adverse influence on prognosis of increased numbers of CD34+ cells with increased CD7 expression in patients with CML in CP has been suggested previously.24,25 Recently, Kosugi et al26 have shown that CD34+CD7+ cells in CML may be involved in clonal maintenance and evolution. Furthermore, PR-3 and neutrophil elastase (NE), which are encoded by the PR-3 and ELA2 genes, respectively, have also been implicated as possible prognostic indicators in CML.27,28 Both proteins are serine proteases that accumulate in the primary azurophilic granules of granulocytes and are overexpressed in CML as compared with normal progenitors. Patients with CML have cytotoxic T lymphocytes (CTLs) against PR1, a nonapeptide common to PR-329 and NE,28 and the degree of cytotoxicity correlates with the amount of PR-3 expression.30 Moreover the presence of PR1-directed CTLs has been correlated with clinical responses to interferon-α and allogeneic SCT.27 Our study provides the first demonstration that higher levels of ELA2 and PR-3 expression are in fact associated with a longer survival and suggests that in some patients with CML an immune-mediated effect, possibly involving CTLs directed against overexpressed antigens such as NE and PR-3, may render the chronic phase of CML relatively indolent and thereby reduce the risk of progression to BT.
Probability curves based on Cox regression analysis models. A backward-stepping procedure (B) yielded a model with the combination of low ELA2 and high CD7 as predictive of poor survival (P < .001). A forward-stepping procedure (A) yielded a model with the combination of low PR-3 and high CD7 as predictive of poor survival (P < .001). Values of genes represent the Q-RT/PCR expression as a ratio of the gene of interest to the ABL control gene.
Probability curves based on Cox regression analysis models. A backward-stepping procedure (B) yielded a model with the combination of low ELA2 and high CD7 as predictive of poor survival (P < .001). A forward-stepping procedure (A) yielded a model with the combination of low PR-3 and high CD7 as predictive of poor survival (P < .001). Values of genes represent the Q-RT/PCR expression as a ratio of the gene of interest to the ABL control gene.
Microarray screening is currently the most powerful tool to identify differences in the transcriptome between 2 or more populations of cells/individuals. In the case of CML, the use of CD34+ progenitors, which are better representatives of the clonogenic leukemia population than mixed mononuclear or total leukocyte fractions,31,32 should provide a profile more biologically reflective of disease phenotype in individual patients. Our preliminary experiments, which have been confirmed by others,33 demonstrated substantial differences in gene expression profile between mononuclear cells and CD34+ progenitors from the same patients, which were a consequence of the different cell populations present in the mononuclear pool (data not shown). We reasoned that to avoid the potential difficulties in interpreting data from highly variable mixed cell populations in total leukocytes between individuals, the use of CD34+ progenitors for gene profiling in our study was preferred. This reasoning has been strengthened by a recent report that expression profiling of unselected total leukocytes was unhelpful in differentiating 2 populations of patients with CML in CP with varying responses to imatinib treatment.32 Although the CD34+ progenitor pool is still a relatively heterogeneous population of cells at differing stages of “stemness,” we detected no significant differences in the composition of CD38+Lin+, CD38+Lin-, or CD38-Lin- cells between the 2 groups of patients. We concede that our results may still reflect a heterogeneity in the maturation profile of the bulk of the CD34+ population in patients with CML at diagnosis, as distinct from differences in equivalent leukemia cells between different patients, but even this heterogeneity may be of prognostic significance. We were not able to ascertain the proportion of BCR-ABL–negative CD34+ cells in all the patients included in this study because of the lack of biologic material. However, it is expected that at least 70% of CD34+ cells would be leukemic from previously published observations26,31 of similarly unmanipulated CD34+ cells from untreated patients with CML CP at diagnosis. Our choice of the 2 extremes of disease patterns defined by the duration of CP from diagnosis to the onset of BT aimed at maximizing the identification of genes underlying the extreme prognostic heterogeneity characteristic of CML. Because all patients in our cohort had the same chromosomal and molecular abnormality, the gene profiles of the groups under comparison were expected to have a greater homogeneity than, for example, those with acute leukemias, whereby a variety of chromosomal abnormalities are present.34,35 We increased the stringency of our analysis by obtaining gene expression measures using 3 separate microarray data analysis packages, MAS 5.0,19 dChip,20 and RMA,21 to identify the most consistently discriminatory genes. Our data must be interpreted with some caution because we had insufficient patient numbers to divide our cohort into “training set” versus “testing set” groups for separate microarray tests. However, even if this had been possible, the results might not have been entirely conclusive because, as recently pointed out by Michiels et al,36 even in large patient series, which genes are identified as prognostically important is highly dependent on the actual samples included in the training set. Taken together, our data could therefore be used as a guide to ensure that the genes we have identified are included in future prospective studies involving larger numbers of patients.
Probabilities of survival using CD7 and PR-3 protein expression. The median protein expression by FACS is used to segregate the patients into 2 groups. The expression of both CD7 (A) (n = 29) and PR-3 (B) (n = 25) are significant predictors of survival (P = .031 and P = .029, respectively).
Probabilities of survival using CD7 and PR-3 protein expression. The median protein expression by FACS is used to segregate the patients into 2 groups. The expression of both CD7 (A) (n = 29) and PR-3 (B) (n = 25) are significant predictors of survival (P = .031 and P = .029, respectively).
The majority of patients included in this study were treated with agents in the era before imatinib, and it is possible that initial treatment of patients with CML with this tyrosine kinase inhibitor will eliminate the heterogeneity in survival observed with prior therapy. However, data from the prospective studies of imatinib5 and from single center experience37 reveal that significant variability still exists in both response to imatinib and rate of relapse among patients in CP with similar hematologic and clinical features.5,11 These differences in response rates are shown to translate into heterogeneity in progression-free survival.11 The mechanisms underlying this heterogeneity in treatment response are still unknown. Recently, the combination of the Sokal score and the concentration of imatinib producing a 50% decrease in the level of its Crkl substrate was shown to have a strong predictive value in a subset of patients.12 These results, taken together with those presented here, suggest that the heterogeneity in treatment response and associated progression-free survival are related to the biology of the CML cell in individual patients. This, in turn, may be governed by transcriptional or metabolic pathways inherent to the CD34+ progenitor population at diagnosis.
Previous microarray studies involving patients with CML have compared the gene signatures of normal with leukemic cells, or of cells from different stages of CML.38,39 These approaches were not able to identify subtle gene expression differences within the patients in CP and did not take into account possible differences in disease tempo between the patients studied. We aimed to avoid this issue by using samples within 3 months of diagnosis and excluded patients who, despite fulfilling morphologic CP criteria, had additional cytogenetic abnormalities. Patients in long-term remission after allogeneic SCT were also excluded to eliminate bias toward the good prognostic groups attributable to the benefits of a successful allograft. Despite the relatively small numbers of patient samples included in this study, we obtained clear-cut results. Thus, we believe that the expression levels of ELA2, PR-3, and CD7 in progenitor cells from CML at diagnosis may constitute important new prognostic markers. For practical purposes, expression of these genes could readily be measured at the RNA or protein levels by Q-RT/PCR or immunophenotyping of CD34+ cells, laboratory assays which are now widely available in clinical practice. Our results offer a timely opportunity to exploit the prognostic markers here identified in the design of prospective trials of imatinib and of the promising new tyrosine kinase inhibitors.40-44 They also reinforce the rationale for treating patients who have not responded optimally to Abl tyrosine kinase inhibitors by vaccination with relevant peptides, and these studies are now being planned.
Prepublished online as Blood First Edition Paper, September 6, 2005; DOI 10.1182/blood-2005-05-2155.
Supported by grants from Malaysia Public Services Department, Lady Tata Memorial Trust, Hammersmith Hospitals Trust Research Committee (A.S.M.Y. and J.V.M.), and the Leukaemia Research Fund, United Kingdom (J.V.M.).
A.S.M.Y. collected the patient data, processed all samples, performed all experiments, analyzed the microarray data and wrote the report. R.M.S. performed the statistical analyses and helped write the report. J.F.A. and J.M.G. provided clinical care, recorded clinical data, provided advice on the design of the study, and commented on the manuscript. J.V.M. conceived and designed the study, supervised its execution, and helped write the report. The corresponding author (J.V.M.) had full access to all the data in the study and had final responsibility for the decision to submit for publication.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the staff of the Stem Cell Laboratory and the CSC/IC Microarray Centre, Hammersmith Hospital, for help with identification of stored cells and practical assistance with the microarray studies, and Dr Eric Hoffman and Sara Hilmer, Research Center for Genetic Medicine at the Children's National Medical Center, Washington, DC, for advice on microarray data analysis.