Abstract
Characterization of the antigens recognized by tumor-reactive T cells isolated from patients successfully treated with allogeneic HLA-matched hematopoietic stem cell transplantation (SCT) can lead to the identification of clinically relevant target molecules. We isolated tumor-reactive cytotoxic CD8+ T-cell (CTL) clones from a patient successfully treated with donor lymphocyte infusion for relapsed multiple myeloma after allogeneic HLA-matched SCT. Using cDNA expression cloning, the target molecule of an HLA-B7–restricted CTL clone was identified. The CTL clone recognized a minor histocompatibility antigen produced by a single nucleotide polymorphism (SNP) in the angiogenic endothelial-cell growth factor-1 (ECGF1) gene also known as thymidine phosphorylase. The SNP leads to an Arg-to-His substitution in an alternatively translated peptide that is recognized by the CTL. The ECGF1 gene is predominantly expressed in hematopoietic cells, although low expression can also be detected in other tissues. The patient from whom this CTL clone was isolated had mild graft-versus-host disease despite high numbers of circulating ECGF-1–specific T cells as detected by tetramer staining. Because solid tumors expressing ECGF-1 could also be lysed by the CTL, ECGF-1 is an interesting target for immunotherapy of both hematologic and solid tumors.
Introduction
The identification of tumor-associated antigens and the growing understanding of tumor-specific immune responses provide new possibilities to develop cellular immunotherapy as a strategy for the treatment of cancer. However, the results of many clinical trials have been disappointing because clinical responses were observed in only a limited number of patients. Vaccination protocols have not led to improvement in overall survival of patients with cancer.1,2 The main impediment of these vaccination strategies is that in most cases nonmutated self-proteins were targeted. In patients, T cells specific for these self-antigens are probably anergic, tolerized, or of low affinity because of peripheral or central selection processes.
High-avidity T-cell responses capable of eradicating hematologic tumors can be generated in an allogeneic setting. In hematologic malignancies, allogeneic HLA-matched hematopoietic stem cell transplantation (SCT) provides a platform for allogeneic immunotherapy as a result of the induction of T-cell–mediated graft-versus-tumor (GVT) immune responses. The clinical potency of the GVT reactivity has been demonstrated by the induction of complete remissions by administration of donor lymphocyte infusion (DLI) in patients with relapsed leukemia after allogeneic SCT.3-5 Immunotherapy in an allogeneic setting enables induction of effective T-cell responses because T cells of donor origin are not selected for low reactivity against self-antigens of the recipient. Therefore, high-affinity T cells against tumor- or recipient-specific antigens can be found in the T-cell inoculum administered to the patient during or after SCT. The main targets of the tumor-reactive T-cell responses are polymorphic proteins for which donor and recipient are disparate, designated minor histocompatibility antigens (mHag)6 or overexpressed proteins such as proteinase-3.7
Induction of GVT reactivity may coincide with the development of graft-versus-host disease (GVHD), especially when immune responses are directed against mHags that are broadly expressed in various tissues. GVT can be separated from GVHD by induction of T cells against target structures specific for or overexpressed in tumor cells. In addition, antigens for which expression is restricted to cells of hematopoietic origin, such as HA-1,8 HA-2,9,10 and BCL2A1,11 may serve as specific targets for GVT. T cells specific for these antigens will destroy both malignant and normal cells of the hematopoietic system of recipient origin. Because after allogeneic SCT hematopoietic stem cells have been replaced by donor-derived cells that are not recognized by these T cells, normal donor hematopoiesis in the patient will not be affected.
Appropriate antigens for tumor-associated T-cell responses that play a role in vivo can be identified by analysis of patients with good clinical responses after allogeneic hematopoietic SCT. Characterization of the target structures of the T-cell responses in patients with relapsed hematologic cancers that respond to DLI with no or limited GVHD may result in the identification of clinically relevant tumor-specific targets for immunotherapy of cancer. Recently, we isolated tumor-reactive CD8+ cytotoxic T-cell (CTL) clones from peripheral blood of a patient successfully treated with DLI for relapsed multiple myeloma (MM) after HLA-matched allogeneic SCT.12 In this study, we describe the identification of a polymorphism in the angiogenic endothelial-cell growth factor-1 (ECGF-1), also known as thymidine phosphorylase, as the target structure recognized by one of the CTL clones. Because the polymorphic ECGF1 gene is predominantly expressed in cells of hematopoietic origin and overexpressed in various solid tumors, it can be further developed as a target for immunotherapy of cancer.
Materials and methods
Cell culture
After informed consent was obtained, the mHAg-specific HLA-B7–restricted CD8+ CTL clone RDR173 was isolated from peripheral blood of a patient with relapsed MM responding to DLI after HLA-matched allogeneic SCT using the IFN-γ secretion assay.12 The study was approved by the Leiden University Medical Center (The Netherlands) institutional review board. CTL clone RDR173 was cultured by stimulation with irradiated (50 Gy) allogeneic peripheral blood mononuclear cells (PBMCs) and patient-derived EBV-transformed B cells in IMDM (Cambrex, Verviers, Belgium) supplemented with 5% human ABO serum, 5% fetal bovine serum (FBS), 100 U/mL IL-2 (Chiron, Amsterdam, the Netherlands), and 0.8 μg/mL phytohemagglutinin (PHA; Murex Biotec Limited, Dartford, United Kingdom). EBV-transformed lymphoblastoid cell lines (EBV-LCLs), COS-7 cells, and melanoma cell lines were maintained in IMDM (Cambrex) containing 8% FBS.
CFSE-based cytotoxicity assay
To determine lysis by CTL clone RDR173 of malignant cells in a heterogenous population of target cells, a CFSE-based cytotoxicity assay was performed as described before.13 Briefly, PBMCs, bone marrow cells, or tumor cells were labeled with 2.5 μM CFSE and incubated with T-cell clone RDR173 (E/T ratio, 3:1) at 37°C. After 24 or 48 hours, MM and chronic myeloid leukemia (CML) cells were stained with PE-labeled anti-CD138 and anti-CD34 antibodies, respectively. Melanoma cells retrovirally transduced with HLA-B7 were stained with APC-labeled antibodies against the marker gene NGFR. To exclude dead cells, propidium iodide was added prior to analysis. To allow quantitative analysis, 104 Flow-Count Fluorospheres (Coulter, Miami, FL) were added, and samples were directly analyzed by flow cytometry.
IFN-γ ELISA
EBV-LCL cells were seeded at 3 × 104 cells/well together with 5 × 103 T cells in 96-well round-bottom plate. For blocking studies, target cells were preincubated with antibodies against HLA class I (w6/32), HLA class II (PdV5.2), or HLA-B/C alleles (B1.23.2) for 30 minutes at room temperature. After 24 hours of incubation at 37°C, release of IFN-γ in the supernatant was measured by using standard enzyme-linked immunosorbent assay (ELISA; Sanguin, Amsterdam, the Netherlands).
Analysis of TCRVα and TCRVβ chains
Construction of cDNA library
A cDNA library was constructed using the SuperScript Choice System kit (Invitrogen, Breda, the Netherlands). RNA was isolated from the patient-derived EBV-LCL using Trizol (Invitrogen) according to the manufacturer's instructions and further purified using the RNA Cleanup protocol of the RNeasy kit (QIAGEN, Venlo, the Netherlands). Poly (A)+ mRNA was isolated using the PolyATract mRNA Isolation System (Promega, Leiden, the Netherlands), and converted to cDNA using an oligo-d(T) primer containing a NotI restriction site at the 5′ end. After double-strand cDNA synthesis, the cDNA was ligated to BamHI-EcoRI adapters (Stratagene, La Jolla, CA), digested with NotI, and size-fractionated using column chromatography. Fractions containing the largest cDNA fragments were ligated into BamHI and NotI sites of the pCR3.1 expression vector (Invitrogen). Ligation products were transformed into Escherichia coli Top10 bacteria, and transformed clones were selected with ampicillin. The library was divided into pools of approximately 40 cDNA clones. Each pool was amplified in liquid culture for 4 hours, and plasmid DNA was isolated using QIAprep 96 Turbo Miniprep Kit (QIAGEN).
Transfection of COS-7 cells and CTL stimulation assay
To identify the gene encoding the epitope that is recognized by CTL clone RDR173, COS-7 cells were transfected with the cDNA library and tested for recognition. Briefly, COS-7 cells retrovirally transduced with the HLA-B7 cDNA (COS/HLA-B7) were plated in a flat-bottom 96-well plate at 2 × 104 cells/well and incubated overnight at 37°C. Approximately 100 ng plasmid DNA from each pool was mixed with 0.8 μL lipofectamine (Invitrogen) in 50 μL OptiMEM medium (Invitrogen). Transfection mixtures were incubated for 30 minutes at room temperature and added to the COS/HLA-B7 cells. After 5 hours of incubation at 37°C, 50 μL IMDM/10% FBS was added. Twenty-four hours after transfection, 3 × 103 CD8+ CTLs were added to the transfected COS/HLA-B7 cells. After 24 hours of coculture at 37°C, the release of IFN-γ in the supernatant was determined by using standard ELISA (Sanguin, Amsterdam, the Netherlands).
Deletion constructs of ECGF-1
Deletion constructs of ECGF-1 were obtained by reverse transcriptase (RT)–PCR using ECGF-1–specific forward primers containing BamHI or BglII restriction sites and ECGF-1–specific reverse primers containing NotI restriction sites. PCR products were digested with the restriction enzymes and ligated into the PCR3.1 expression vector.
Tetrameric HLA class I/peptide complexes and flow cytometric analysis
PE-conjugated tetrameric complexes of HLA-B7 molecules with the LB-ECGF-1H peptide RPHAIRRPLAL were constructed as previously described16 with minor modifications. For cytometric analysis and fluorescence-activated cell sorting (FACSort) experiments, cells were labeled with tetrameric complexes for 2 hours at 4°C in RPMI without phenol, supplemented with 2% FBS. Cells were counterstained with FITC-labeled antibodies against CD4 (Caltag, San Francisco, CA) and CD40 (Serotec, Oxford, United Kingdom) or CD8 (Caltag).
Screening for population frequency
RNA was isolated from PBMCs by using Trizol (Invitrogen) according to the manufacturer's instructions and transcribed to cDNA using M-MLV reverse transcriptase (Invitrogen). PCR was performed in 50 μL reaction mixture containing 1 × buffer, 300 nM forward primer (5′-TATAAGATCTGCCACCATGGGCGCAGAGCTGCTGGTC-3′), 300 nM reverse primer (5′-TACGCGGCCGCTTATTGCTGCGGCGGCAGAAC-3′), 500 μM dNTP, GC-rich solution (Roche, Indianapolis, IN), 1 U PWO Superyield DNA polymerase (Roche). Amplification using the following PCR program 2 minutes at 95°C, 30 cycles of 15 seconds at 95°C, 30 seconds at 55°C, 1 minute at 72°C, and final elongation for 7 minutes at 72°C resulted in a fragment of 223 bp (base pairs) that was digested with 10 U BssHII for 2 hours at 50°C and analyzed on a 2% agarose gel. Digestion of PCR product-derived from the LB-ECGF-1R allele results in bands of 69 and 154 bp, whereas amplification and digestion of the LB-ECGF-1H allele results in a single band of 223 bp.
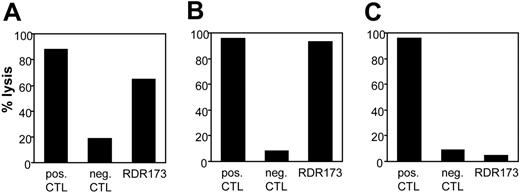
CTL clone RDR173 lyses tumor cells and hematopoietic cells. Lysis of patient-derived tumor cells (A), EBV-LCL cells (B), and donor-derived EBV-LCL cells (C) was tested in a CFSE-based cytotoxicity assay. Target cells were labeled with CFSE and incubated for 48 hours with CTL clone RDR173 (E/T ratio, 3:1). As a positive control, target cells were incubated with an HLA-A2–specific alloreactive CTL clone (pos CTL), and as negative control cells were incubated with a male-specific CTL clone (neg CTL).
CTL clone RDR173 lyses tumor cells and hematopoietic cells. Lysis of patient-derived tumor cells (A), EBV-LCL cells (B), and donor-derived EBV-LCL cells (C) was tested in a CFSE-based cytotoxicity assay. Target cells were labeled with CFSE and incubated for 48 hours with CTL clone RDR173 (E/T ratio, 3:1). As a positive control, target cells were incubated with an HLA-A2–specific alloreactive CTL clone (pos CTL), and as negative control cells were incubated with a male-specific CTL clone (neg CTL).
Quantitative RT-PCR
Quantitative real-time PCR was performed for the commercially available Human Blood fraction MTC Panel, Human MTC Panel I and II (BD Biosciences, Alphen aan de Rijn, the Netherlands). Using Primer Express software (Applied Biosystems, Foster City, CA), forward primer 5′-CGGAATCCTATATGCAGCCAG-3′, reverse primer 5′-CCCTCCGAACTTAACGTCCA-3′, and probe 5′-(TET)-TGCCACTCATCACAGCCTCCATTCTC-(TAMRA)-3′ were designed for ECGF-1. PCR was performed in 50 μL reaction mixture containing 1 × Taqman buffer, 300 nM of each primer, 120 nM probe, 200 μM dNTP, 3 mM MgCl2, 1.25 U AmpliTaq Gold polymerase, and 10 to 100 ng cDNA sample. PCR amplification was also performed for the housekeeping gene porphobilogen deaminase (PBGD) using forward primer 5′-GGCAATGCGGC-TGCAA-3′, reverse primer 5′-GGGTACCCACGCGAATCAC-3′, and probe 5′(TET)-CTCATCTTTGGGCTGTTTTCTTCCGCC-(TAMRA)-3′. For PBGD, PCR was performed in 50 μL reaction mixture containing 1 × Taqman-buffer, 300 nM of each primer, 120 nM probe, 200 μM dNTP, 4 mM MgCl2, 1.25 U AmpliTaq Gold polymerase, and 10 to 100 ng cDNA sample. Amplification was started with 10 minutes at 95°C, followed by 55 cycles of 15 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 60°C. Expression of ECGF-1 was normalized to expression of the housekeeping gene. Differential expression was calculated according to the 2-ΔΔCT method.17
Results
Isolation and characterization of tumor-reactive T-cell clone RDR173
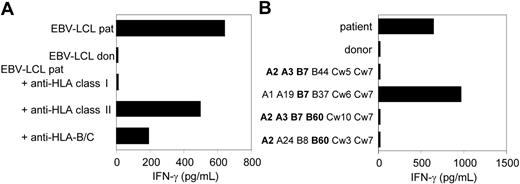
As previously described, we isolated from the peripheral blood of a patient successfully treated with DLI for relapsed MM CD8+ CTL clones by direct cloning of T cells that produced IFN-γ on stimulation with irradiated bone marrow cells harvested from the patient before SCT.12 CTL clones recognizing several distinct antigens in the context of various HLA class I alleles were isolated, including CTL clone RDR173. The tumor-reactivity of CTL clone RDR173 was demonstrated by the recognition of MM cells in the patient's bone marrow from the moment of relapse using a CFSE-based cytotoxicity assay.13 To identify the tumor cells in the heterogeneous cell population, malignant cells were stained with PE-labeled anti-CD138 antibodies. As shown in Figure 1A, patient-derived malignant MM cells were lysed by CTL clone RDR173 similar to a control anti–HLA-A2–specific CTL clone, whereas they were not lysed by a male-specific, HLA-A1–restricted CTL clone (negative control CTL clone). In addition to tumor cells, CTL clone RDR173 also lysed the patient-derived EBV-LCL cells (Figure 1B), whereas donor-derived EBV-LCL cells were not recognized (Figure 1C). Similar results were obtained for patient- and donor-derived PHA-blasts (data not shown). The differential recognition of patient- and donor-derived cells indicated that CTL clone RDR173 was directed against a mHag. Recognition of patient-derived EBV-LCL cells could be blocked by antibodies against HLA class I and HLA-B/C alleles as shown in Figure 2A. Screening a panel of EBV-LCL cells resulted in the recognition of an EBV-LCL cell line expressing only HLA-B7 in common with the patient, demonstrating that the epitope is presented by HLA-B7. The differential recognition of HLA-B7–positive EBV-LCL cells in the panel confirmed that CTL clone RDR173 recognized an mHag. To demonstrate the clonality of clone RDR173, the TCR α and β chains of CTL clone RDR137 were determined by using TCR VB-specific monoclonal antibodies and TCR AV and BV family-specific oligonucleotides and sequencing. Because only one TCR α (AV2S2A1) and one TCR β (BV8S2A1T) could be detected, the tumor-reactive CTL clone RDR173 was demonstrated to be a monoclonal CTL recognizing an mHag presented in HLA-B7.
CTL clone RDR173 recognizes an mHag presented in HLA-B7. (A) EBV-LCL cells preincubated with antibodies against HLA class I, HLA class II, or HLA B and C alleles were tested for recognition by CTL clone RDR173. (B) EBV-LCL cells expressing one or more HLA class I alleles in common with the patient (A2, A3, B7, B60, Cw3) were incubated with CTL clone RDR173 for 24 hours. Release of IFN-γ was determined by using ELISA. HLA alleles that are shared with the patient are depicted in bold.
CTL clone RDR173 recognizes an mHag presented in HLA-B7. (A) EBV-LCL cells preincubated with antibodies against HLA class I, HLA class II, or HLA B and C alleles were tested for recognition by CTL clone RDR173. (B) EBV-LCL cells expressing one or more HLA class I alleles in common with the patient (A2, A3, B7, B60, Cw3) were incubated with CTL clone RDR173 for 24 hours. Release of IFN-γ was determined by using ELISA. HLA alleles that are shared with the patient are depicted in bold.
Identification of the epitope recognized by CD8+ CTL clone RDR173
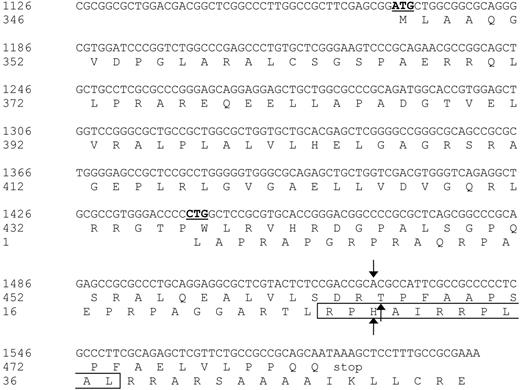
To identify the antigen that is recognized by CTL clone RDR173, cDNA expression cloning was performed based on the procedure as described by De Plaen et al.18 Briefly, a library of approximately 4 × 106 cDNA clones ranging from 400 bp to more than 3 kb (kilobase) in size, was constructed from EBV-LCL cells derived from the patient. A total of 960 plasmid pools consisting of approximately 40 cDNAs from the library were transfected into COS-7 cells that were retrovirally transduced with HLA-B7 cDNA (COS/HLA-B7). Transfected COS/HLA-B7 cells were tested for recognition by CTL clone RDR173. One cDNA pool stimulated IFN-γ production of the CTL and subcloning of this pool resulted in the isolation of a plasmid containing a 475-bp cDNA insert that was recognized by CTL clone RDR173 after transfection in COS/HLA-B7 cells. A BLAST search revealed that this cDNA insert was identical to the 3′ part (nt 1126-1600) of endothelial-cell growth factor-1 (ECGF-1, also known as platelet-derived-ECGF or thymidine phosphorylase) cDNA.44 The incomplete cDNA fragment of ECGF-1 that we isolated from the cDNA library contained an internal ATG codon (nt 1169-1171) from which translation results in a protein identical to the C-terminal 137 amino acids of ECGF-1. To examine whether the epitope recognized by CTL clone RDR173 could also be produced by full-length ECGF-1, ECGF-1 mRNA derived from the patient and the donor was cloned. COS/HLA-B7 cells transfected with full-length ECGF-1 from the patient were recognized by CTL clone RDR173, whereas full-length ECGF-1 derived from the donor was not recognized, demonstrating that ECGF-1 encodes the mHag recognized by CTL clone RDR173. Comparing ECGF-1 cDNA from the patient with donor-derived ECGF-1 cDNA showed that these cDNA sequences were completely identical except for a G-to-A transition at nt 1526.
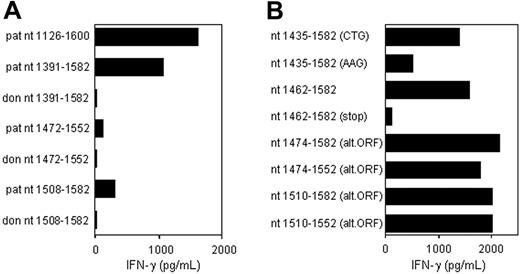
CTL clone RDR173 recognizes an epitope translated in an alternative ORF. (A) Deletion constructs derived from patient and donor ECGF-1 cDNA were tested for recognition by CTL clone RDR173 on transfection into COS/HLA-B7 cells. The smaller fragments were hardly recognized by CTL clone RDR173, although they contained the polymorphism. (B) To investigate whether the epitope was encoded by an alternative ORF construct with the possible alternative CTG start codon at positions 1441 to 1443 mutated into an AAG codon, constructs with a stop codon introduced in the alternative ORF, and deletion constructs with Kozak-ATG sequences in the alternative ORF were transfected into COS/HLA-B7. Transfected cells were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ was measured by ELISA.
CTL clone RDR173 recognizes an epitope translated in an alternative ORF. (A) Deletion constructs derived from patient and donor ECGF-1 cDNA were tested for recognition by CTL clone RDR173 on transfection into COS/HLA-B7 cells. The smaller fragments were hardly recognized by CTL clone RDR173, although they contained the polymorphism. (B) To investigate whether the epitope was encoded by an alternative ORF construct with the possible alternative CTG start codon at positions 1441 to 1443 mutated into an AAG codon, constructs with a stop codon introduced in the alternative ORF, and deletion constructs with Kozak-ATG sequences in the alternative ORF were transfected into COS/HLA-B7. Transfected cells were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ was measured by ELISA.
To determine whether this nonsynonymous single nucleotide polymorphism (SNP) encoded the epitope recognized by CTL clone RDR173, constructs were made containing nt 1391 to 1582, 1472 to 1552, and 1508 to 1582 of the cDNA insert, respectively, proceeded by Kozak-ATG sequences to ensure translation. The construct containing nt 1391 to 1582 was clearly recognized by CTL clone RDR173 on transfection in COS/HLA-B7 (Figure 3A). However, the smaller fragments containing nt 1472 to 1552 and 1508 to 1582 were hardly recognized. Even if the Ala-to-Thr substitution caused by the polymorphism is not present in the epitope itself, but affects antigen processing, eg proteasomal cleavage, at least one of these constructs was expected to be recognized by CTL clone RDR173. Donor-derived fragments of ECGF-1 were not recognized by CTL clone RDR173, confirming that the G-to-A transition at position 1526 was likely to be involved in the differential expression or recognition of the epitope.
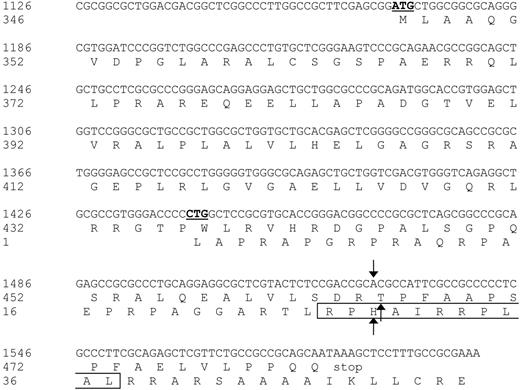
Nucleotide sequence and protein translation of the partial ECGF-1 cDNA isolated from the cDNA library. The internal ATG codon of the primary ECGF-1 ORF and the CTG start codon of the alternative ORF are depicted in bold and underlined. The SNP and amino acid substitutions are indicated with arrows. The CTL epitope recognized by CTL clone RDR173 is boxed. Numbers are relative to the full-length ECGF cDNA and protein.
Nucleotide sequence and protein translation of the partial ECGF-1 cDNA isolated from the cDNA library. The internal ATG codon of the primary ECGF-1 ORF and the CTG start codon of the alternative ORF are depicted in bold and underlined. The SNP and amino acid substitutions are indicated with arrows. The CTL epitope recognized by CTL clone RDR173 is boxed. Numbers are relative to the full-length ECGF cDNA and protein.
Analysis of nt 1391 to 1472 of the cDNA insert revealed the presence of a CTG codon (nt 1441-1443) capable of serving as an alternative start codon. Translation initiated by this CTG codon would result in a protein encoded by another open reading frame (ORF) than ECGF-1 in which the SNP at nt 1526 caused an Arg-to-His substitution (Figure 4). To investigate whether the epitope recognized by CTL clone RDR173 was encoded by the alternative ORF, we made a construct in which the CTG codon was mutated to AAG. Transfection of COS/HLA-B7 cells with this construct demonstrated decreased recognition (Figure 3A), suggesting that translation starting at the CTG at nt 1441 to 1443 resulted in production of the epitope recognized by CTL clone RDR173. In addition, a construct was made in which we introduced a stop codon by a single nucleotide mutation (CAG into TAG) after the CTG start codon. The regular ECGF-1 ORF was not affected by this mutation (CTC into CTT, both coding for leucine residue). As shown in Figure 3B, the introduction of a stop codon in the alternative ORF diminished recognition by CTL clone RDR173, confirming that the epitope was not located in the regular ECGF-1 protein but was derived from alternative translation.
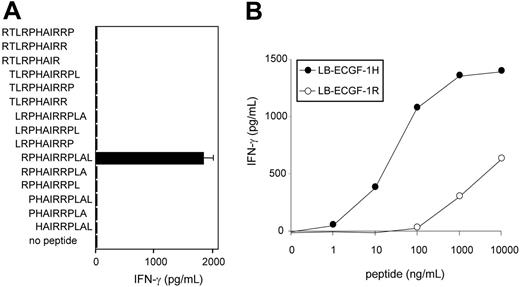
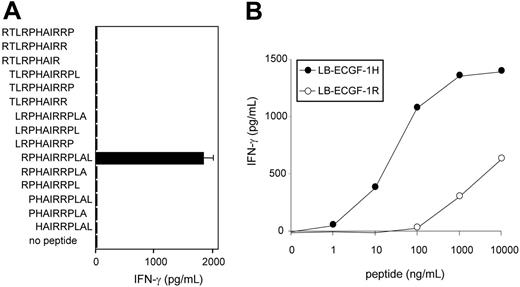
To further determine the location of the epitope recognized by CTL clone RDR173, smaller cDNA fragments with Kozak-ATG sequences in all 3 ORFs were constructed and tested for recognition. Because the fragment containing nt 1510 to 1552 translated in the third ORF was still recognized by CTL clone RDR173 (Figure 3B), the epitope was located within the 14–amino acid sequence encoded by this fragment. To identify the minimal peptide recognized by CTL clone RDR173, overlapping 9-, 10-, and 11-mer peptides were synthesized, pulsed on donor-derived EBV-LCL cells, and tested for recognition. Figure 5A demonstrates that the 11-mer RPHAIRRPLAL peptide is the epitope recognized by CTL clone RDR173 (LB-ECGF-1H).
CTL clone RDR173 recognizes the minimal epitope RPHAIRRPLAL. (A) Overlapping 9-, 10-, and 11-mer peptides (1 μg/mL) were pulsed on donor-derived EBV-LCL cells for 2 hours at 37°C. Peptide-pulsed EBV-LCLs were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ in the supernatant was measured by ELISA (shown as mean value ± SD). (B) Increasing concentrations of the patient-derived peptide RPHAIRRPLAL (•) and the donor-derived peptide RPRAIRRPLAL (○) were pulsed on donor-derived EBV-LCL cells for 2 hours at 37°C. Peptide-pulsed EBV-LCLs were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ in the supernatant was measured by ELISA.
CTL clone RDR173 recognizes the minimal epitope RPHAIRRPLAL. (A) Overlapping 9-, 10-, and 11-mer peptides (1 μg/mL) were pulsed on donor-derived EBV-LCL cells for 2 hours at 37°C. Peptide-pulsed EBV-LCLs were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ in the supernatant was measured by ELISA (shown as mean value ± SD). (B) Increasing concentrations of the patient-derived peptide RPHAIRRPLAL (•) and the donor-derived peptide RPRAIRRPLAL (○) were pulsed on donor-derived EBV-LCL cells for 2 hours at 37°C. Peptide-pulsed EBV-LCLs were cocultured with CTL clone RDR173 for 24 hours. Release of IFN-γ in the supernatant was measured by ELISA.
To reveal whether the differential recognition of recipient and donor cells by CTL clone RDR173 is due to differences in presentation of the epitope or recognition by the T cells, the donor-derived peptide RPRAIRRPLAL (LB-ECGF-1R) was synthesized. EBV-LCL cells pulsed with the LB-ECGF-1R peptide were not recognized by CTL clone RDR173 (Figure 5B). Because LB-ECGF-1H and LB-ECGF-1R bind to HLA-B7 with comparable affinities (EC50 of 14.2 and 26.2, respectively; data not shown), these data indicate that the amino acid substitution is critical for TCR recognition and affects a TCR contact residue.
Analysis of LB-ECGF-1H–specific immune response in vivo
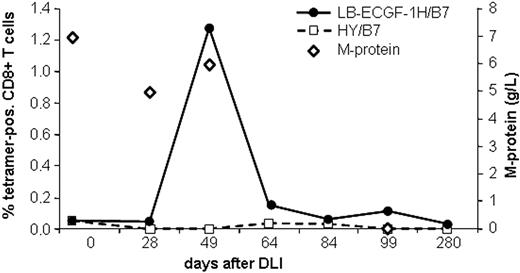
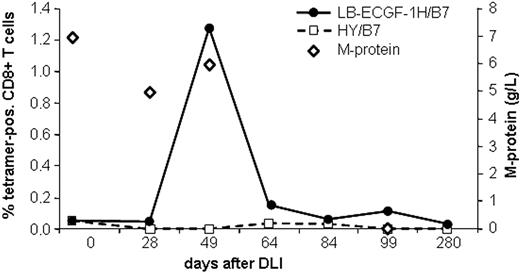
The magnitude of the LB-ECGF-1H–specific immune response in the peripheral blood of the patients with MM responding to DLI was analyzed using tetrameric complexes. LB-ECGF-1H–specific T cells were not detectable (< 0.05%) before DLI or in the DLI material. At the time of the clinical response (day 49), 1.3% of the CD8+ T cells bound the HLA-B7/LB-ECGF-1H tetramer (Figure 6). At day 64 still low levels of LB-ECGF-1H–specific CD8+ T cells were detectable. Nonspecific T-cell activation or staining with tetramers after DLI was excluded because HLA-B7–restricted HY-specific T cells were not detected in this male-to-female transplantation. Five weeks after the peak in tetramer-positive cells, the M-protein was decreased below detection level, and myeloma cells could not be detected in the bone marrow of the patient. Tetramer-positive T cells were cloned by FACSsorting, and TCR α and β chains were determined from proliferating, HLA-B7/LB-ECGF-1H–positive clones. All clones expressed the same TCR α and β chain as clone RDR173, indicating that the LB-ECGF-1H–specific T cells in the peripheral blood of the patients were derived from a monoclonal T-cell response.
Analysis of the LB-ECGF-1H–specific immune response in vivo. PBMCs from the patient obtained at various time points before and after DLI were stained with anti-CD8 antibody and LB-ECGF-1H/B7 tetramers (•) or, as a control, HY/B7 tetramers (□). M-protein (⋄) is expressed in grams per liter in peripheral blood.
Analysis of the LB-ECGF-1H–specific immune response in vivo. PBMCs from the patient obtained at various time points before and after DLI were stained with anti-CD8 antibody and LB-ECGF-1H/B7 tetramers (•) or, as a control, HY/B7 tetramers (□). M-protein (⋄) is expressed in grams per liter in peripheral blood.
Population frequency of LB-ECGF-1H/R alleles
The disruption of a BssHII restriction site by the G-to-A transition at position 1526 was used to determine the population frequency of the LB-ECGF-1H and LB-ECGF-1R alleles. A 223-bp cDNA fragment was amplified from 70 unrelated individuals and digested with BssHII. PCR products obtained from the LB-ECGF-1H allele could not be cut by BssHII, whereas digestion of LB-ECGF-1R–derived PCR products resulted in 2 bands of 69 and 154 bp. Sixty-two persons were homozygous for the LB-ECGF-1R allele, whereas 8 individuals were heterozygous for this polymorphism, indicating a frequency of 11% LB-ECGF-1H–positive individuals in the population. Individuals who were homozygous for the LB-ECGF-1H allele were not detected.
Expression of ECGF-1 in normal tissue and malignancies
Analysis of the expression pattern of ECGF-1 in a microarray study of gene expression across tissues (http://symatlas.gnf.org/SymAt-las/; see Su et al19 ) showed high expression of ECGF-1 in several cells of the hematopoietic system, in particular CD14+ monocytes and dendritic cells. Furthermore, the ECGF1 gene was reported to be present in cDNA derived from lung, liver, and heart. To confirm these data, we performed quantitative real-time RT-PCR for ECGF-1. In accordance with the DNA microarray study, we detected the highest levels of ECGF1 expression in CD14+ cells (Table 1). However, we also detected ECGF1 expression in other PBMCs, like CD4+, CD8+, and CD19+ cells. Furthermore, ECGF1 was detected in lung, spleen, liver, and thymus. In all the other tissues, ECGF1 gene expression was low or absent.
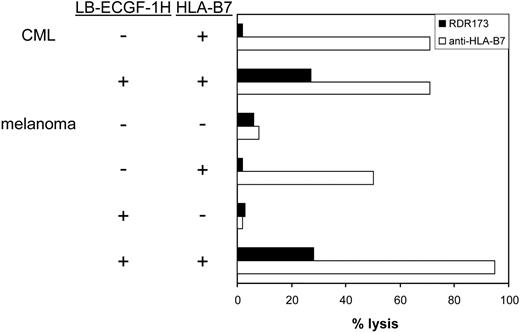
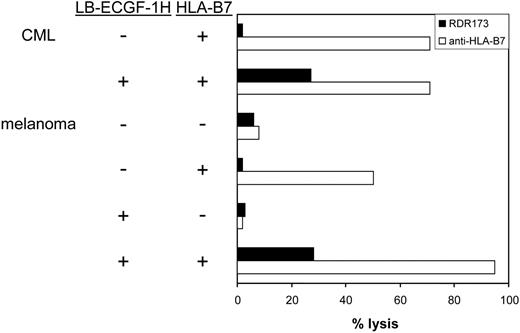
CTL clone RDR173 lyses LB-ECGF-1H–positive tumor cells. HLA-B7–expressing CML cells were labeled with CFSE and incubated with CTL clone RDR173. Melanoma cells were transduced with HLA-B7 and tested for recognition by CTL clone RDR173 (▪) in a CFSE-based cytotoxicity assay. Control cells were transduced with retroviral construct containing only the marker gene. Target cells were labeled with CFSE and incubated for 48 hours with CTL clone RDR173 (E/T ratio, 1:1). Target cells were also incubated with an HLA-B7–specific alloreactive T-cell clone (□) to demonstrate functional HLA-B7 expression.
CTL clone RDR173 lyses LB-ECGF-1H–positive tumor cells. HLA-B7–expressing CML cells were labeled with CFSE and incubated with CTL clone RDR173. Melanoma cells were transduced with HLA-B7 and tested for recognition by CTL clone RDR173 (▪) in a CFSE-based cytotoxicity assay. Control cells were transduced with retroviral construct containing only the marker gene. Target cells were labeled with CFSE and incubated for 48 hours with CTL clone RDR173 (E/T ratio, 1:1). Target cells were also incubated with an HLA-B7–specific alloreactive T-cell clone (□) to demonstrate functional HLA-B7 expression.
In hematologic malignancies such as multiple myeloma (n = 3), CML (n = 5), acute myeloid leukemia (AML; n = 5), and acute lymphoid leukemia (ALL; n = 4) and in cell lines derived from solid tumors such as melanoma (n = 13), breast carcinoma (n = 4), colon carcinoma (n = 3), and ovarian carcinoma (n = 2), ECGF-1 mRNA could be detected (data not shown). The levels of expression were variable from hardly detectable to expression levels comparable to PBMCs (data not shown). To investigate whether tumor cells other than MM cells could also be recognized by CTL clone RDR173, CML cells from an HLA-B7–expressing, LB-ECGF-1H–positive patient were tested for recognition by CTL clone RDR173. Furthermore, LB-ECGF-1H–positive melanoma cells in which low expression of the ECGF1 gene could be detected were tested for recognition by CTL clone RDR173. Because the melanoma cell line is HLA-B7 negative, these cells were retrovirally transduced with HLA-B7. As shown in Figure 7, HLA-B7–expressing, LB-ECGF-1H–positive CML, and melanoma cells were lysed by CTL clone RDR173, whereas HLA-B7–expressing, LB-ECGF-1R–positive cells were not lysed by CTL clone RDR173. Similar results were obtained when release of IFN-γ was determined (data not shown). These data demonstrate that the LB-ECGF-1H–specific CTL clone RDR173 can be reactive against both hematologic malignancies and solid tumors.
Discussion
Isolation and characterization of tumor-reactive T cells from a patient with relapsed MM who was successfully treated with DLI after allogeneic SCT revealed that these T cells were directed against the angiogenic factor ECGF-1. The epitope recognized by the T cells was encoded by a single nucleotide polymorphism (SNP), resulting in an ECGF-1 mHag (LB-ECGF-1H). This SNP has previously been reported as a mutation possibly associated with the autosomal recessive disease mitochondrial neurogastrointestinal encephalomyopathy (MNGIE).20 The G-to-A transition in the ECGF1 cDNA leads to an Arg-to-His substitution in an alternatively translated peptide that is recognized by the CTL. Recently, other T-cell epitopes obtained from untranslated regions or encoded by alternative ORF have been described (reviewed by Shastri et al21 ). Most of these unexpected epitopes are derived from tumor and viral antigens, but the ECGF-1–derived epitope described in the present study and the recently identified epitope in the 5′ UTR of TMSB4Y22 demonstrate that mHag can also be derived from unexpected translational products.
Interestingly, the T cells directed against the angiogenic factor ECGF-1 were isolated from a patient with MM showing complete remission after cellular immunotherapy. Bone marrow angiogenesis has been found to be an adverse prognostic factor in multiple myeloma,23-25 explaining the success of new treatment strategies with antiangiogenic drugs such as thalidomide.26 Therefore, generation of T-cell responses to polymorphisms in ECGF1 may be a potent combination of direct antitumor immunotherapy and antiangiogenic treatment. The SNP at nt 1526 may not be the only mHag in ECGF1. Several other SNPs have been reported in the ECGF1 gene that may lead to polymorphic peptides that are differentially expressed in recipients and donors in allogeneic SCT.
On the basis of the successes in hematologic malignancies, allogeneic SCT is also under investigation for the treatment of solid tumors. Beneficial effects of allogeneic SCT have been demonstrated in patients with renal cell carcinoma, breast carcinoma, and colon carcinoma, but little is known about the antigens that play a role in GVT reactivity.27 Interestingly, expression of ECGF-1 (also known as thymidine phosphorylase) has been shown in our study and by others in many solid tumors, such as breast carcinoma28 renal cell carcinoma,29 colorectal cancer,30 cervical carcinoma,31 and ovarian carcinoma.32 ECGF-1 was detected in neoplastic or stromal cells, but the cell type that expressed ECGF-1 varied between tumors, even between tumors of the same histologic origin. Consistent with the expression of ECGF-1 in hematopoietic cells such as CD14+ cells, ECGF-1 expression was also detected in tumor-infiltrating cells. Exposure of tumor cells to stress factors, like hypoxia, hypoglycemia, inflammatory factors, but also chemotherapy and radiotherapy, stimulates the expression of ECGF-1.33-37 As in multiple myeloma, ECGF-1 overexpression in solid tumors is correlated with intratumoral microvessel density and poor prognosis. In addition to the angiogenic activity, ECGF-1 enhances resistance of tumor cells to apoptosis induced by hypoxia,38,39 Fas,40,41 or cisplatin.42 The up-regulated expression and correlation with improved tumor survival makes the polymorphic ECGF-1 an interesting target for immunotherapy of solid tumors as well.
In bone marrow–derived cancers, the most attractive candidates for cellular immunotherapy based on allogeneic SCT are antigens solely expressed in hematopoietic cells. T cells specific for these antigens will destroy cells of the hematopoietic system of recipient origin, but, because hematopoiesis has been taken over by donor-derived cells, this will not be hazardous for the patient. Using gene expression profiling and real-time RT-PCR, expression of the ECGF1 gene was detected predominantly in cells of hematopoietic origin. However, ECGF1 cDNA was also detected in some normal tissues, such as liver and lung. Immunohistochemical analysis demonstrated that the expression of ECGF-1 in these tissues was due mainly to the presence of macrophages, although moderate or weak expression in stromal cells could also be detected.43 Whether low levels of ECGF-1 expression in these cells have implications for the development of GVHD reactivity when the ECGF-1 will be used in immunotherapy has to be further analyzed. The patient from whom the LB-ECGF-1H–specific T cells were isolated had grade II GVHD of the skin and liver, responding to prednisone at a dose of 1 mg/kg per day despite the presence of high numbers of circulating LB-ECGF-1H–specific T cells as detected by tetramer staining. The patient is in complete remission and in good clinical condition without GVHD more than 5 years after DLI. This observation indicates that ECGF-1–specific T cells may be useful for therapeutic purposes without major detrimental side effects.
In conclusion, the angiogenic factor ECGF-1 has been identified as the target of a profound T-cell response in a patient successfully treated with DLI for relapsed MM after allogeneic SCT. The epitope is encoded by a polymorphism in the ECGF1 gene that results in an amino acid substitution in an alternatively translated peptide. In addition, several other polymorphisms in ECGF1 have been reported that also may serve as epitopes for T-cell responses. Because ECGF-1 is expressed in both hematologic and solid malignancies and is involved in tumor cell survival, it may not only be a potential target for immunotherapy of hematologic malignancies but also for solid tumors.
Prepublished online as Blood First Edition Paper, February 23, 2006; DOI 10.1182/blood-2005-09-3883.
Supported by AlloStem (EU grant 503319) and by the Dutch Cancer Society (grant NKB 99-2028).
E.H.S., M.W.H., E.D.M., R.W., and J.H.F.F. designed the study. E.H.S., M.W.H., E.D.M., S.A.P.L-H., F.M.K., M.G.D.K., I.J., W.A.E.M., and J.H.F.F. performed the research. M.R.S. provided and analyzed the patient's materials and provided the clinical analysis. E.H.S., R.W., and J.H.F.F. wrote the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.