Abstract
Mast cells mediate both IgE-dependent allergic reactions and protective responses against acute infections, possibly through the activation of Toll-like receptors (TLRs). We find that antigen interacts synergistically with TLR2 and TLR4 ligands to markedly enhance production of cytokines in murine mast cell lines. However, the TLR ligands neither stimulated degranulation and release of arachidonic acid nor influenced such responses to antigen, probably because these ligands failed to generate a necessary calcium signal. The enhanced cytokine production could be attributed to synergistic activation of mitogen-activated protein kinases in addition to the engagement of a more effective repertoire of transcription factors for cytokine gene transcription. The synergistic interactions of TLR ligands and antigen might have relevance to the exacerbation of IgE-mediated allergic diseases by infectious agents.
Introduction
Immunoglobulin E (IgE)–dependent atopic diseases such as allergic rhinitis, asthma, and anaphylactic reactions affect, respectively, 20%, 7%, and 2% of the population in the United States.1,2 The respiratory manifestations of these diseases may be exacerbated by acute respiratory infections from influenza, rhino- and syncytial viruses in humans3,4 and animals,5,6 as well as by lipopolysaccharide (LPS), which is a ubiquitous environmental contaminant from Gram-negative bacteria.7,8 These pathogens may act through Toll-like receptors (TLRs).9,10 TLR4 appears to play an essential role in the immune response to respiratory syncytial virus in the human lung,11,12 and ligands for TLR2 and TLR4 augment inflammatory responses to inhaled antigen in the mouse lung.13,14 Thirteen TLRs have been described to date, each of which recognize specific pathogen-associated molecular patterns.15,16 Cooperative interactions between TLRs allow further discrimination. For example, a widely used synthetic ligand, tripalmitoyl Cys-Ser-(Lys)4 (P3C),17 has preference for TLR2/TLR1 heterodimers, while bacterial peptidoglycans (PGN) and macrophage-activating lipopeptide from Mycoplasma fermentans (MALP2) have preference for TLR2/TLR6 heterodimers.
TLRs interact with ligands as homodimers or, as noted for TLR2, heterodimers, and thus recruit cytosolic adaptor molecules that include myeloid differentiation protein 88 (MyD88), MyD88-adaptor like/TIR-associated protein (MAL/TIRAP), Toll-receptor–associated activator of interferon (TRIF), and Toll-receptor–associated molecule (TRAM).16,18,19 The recruitment of MyD88 by TLR2 and TLR4, the focus of the present study, results in sequential activation of protein kinase interleukin-1 (IL-1) receptor-associated kinases (IRAKs) and transforming growth factor-β–activated kinase (TAK1). TAK1, in turn, regulates activation of mitogen-activated protein (MAP) kinases and the transcription factor, nuclear factor κB (NF-κB), with ensuing production of inflammatory cytokines. TLR4 also activates a MyD88-independent pathway through TRAM and TRIF that uses interferon (IFN)–regulatory factor (IRF3) and NF-κB to induce production of IFN-β. In addition, TLRs engage signaling molecules that are commonly used by other types of receptors, such as phosphatidylinositol 3-kinase (PI3K), AKT, and possibly Src kinases, in addition to the MAP kinases.15 However, the contribution of these “common” signaling molecules and their downstream targets to the overall response to TLR ligands is uncertain.
IgE-dependent allergic diseases are initiated by multivalent binding of allergens to IgE that is bound to receptors with high affinity for IgE (FcϵRI) on mast cells.20 Mast cells also express functional TLRs,21 which may account for the protection conferred by mast cells against bacterial and parasitic infections in animal models.22,23 Activated mast cells release an array of potent inflammatory mediators by rapid discharge of preformed mediators in granules, the generation of inflammatory lipids from arachidonic acid, and the production of numerous Th2-type cytokines and chemokines.23 All these responses are evoked by allergens via FcϵRI, while stimulation of mast cells via TLR2 and TLR4 results primarily in generation of cytokines such as IL-4, IL-5, IL-6, IL-10, IL-13, and TNFα.24 There are reports that TLR2 ligands also induce degranulation and generation of leukotriene C4,22,25,26 but current models for TLR signaling do not define a clear mechanism for these responses, and such responses have not been observed in all studies.27 Although less well studied, TLR1, TLR3, TLR6, TLR7, TLR8, and TLR9 are variably expressed in mast cells of different origins.25,28-30
Stimulation of mast cells through FcϵR1 leads to activation of Src kinases and Syk. The ensuing phosphorylation of adaptor proteins enables recruitment of other signaling molecules and propagation of downstream signals.31 Signaling molecules so recruited include phosphatidylinositol 3-kinase (PI3K), phospholipase (PL) Cγ1/2, and Ras. A PLC-mediated increase in intracellular levels of free Ca2+ and activation of protein kinase C provide essential signals for degranulation,20,32 whereas the activation of the Ras/Raf/MAP kinase pathway results in production of arachidonic acid.33 These signaling pathways also regulate activation of transcription factors such as NF-κB, c-Jun, c-Fos, ATF2, and NFAT, to promote cytokine-gene transcription (Hundley et al34 and citations therein). The repertoire of activated transcription factors is expanded further on co-stimulation of mast cells with antigen and Kit ligand (stem cell factor). As a consequence, the production of cytokines is markedly augmented in a synergistic manner.34
Here we show that FcϵRI also interacts synergistically with TLR2 and TLR4 in several rodent mast cell lines. Both types of receptor use unique as well as common signaling pathways, which in combination reinforce the activation of cytokine-related transcription factors. This results in up to 20-fold enhancement in production of cytokines, an enhancement that may contribute to the exacerbation of allergic symptoms by infectious agents.
Materials and methods
Materials
Reagents were obtained from the following sources: media and culture reagents from Invitrogen/GIBCO (Carlsbad, CA); recombinant mouse IL-3 from Pepro Tech (Rocky Hill, NJ); monoclonal anti–DNP IgE antibody and the antigen, dinitrophenylated human serum albumin (DNP-HSA), lipopolysaccharide (Escherichia coli 055:B5), and peptidoglycan (Staphylococcus aureus) from Sigma (St Louis, MO); Pam3Cys-Ser-(Lys)4 3HCl and MALP2 from EMC microcollections GmbH (Tuebingen, Germany); myelin basic protein and antibodies against c-Fos from Upstate (Lake Placid, NY); antibody against IRAK1 from Santa Cruz Biotechnology (Santa Cruz, CA); polyclonal antibodies that detect activating phosphorylations of Akt(Ser473), ERK-2 (Thr202/Tyr204), JNK (Thr183/Tyr185),35 p38 MAP kinase (Thr180/Tyr182),36 c-Jun (Ser63),37 ATF-2 (Thr71),38 STAT1 (Ser727), STAT1 (Tyr701), STAT3 (Ser727), STAT3 (Tyr705), STAT5 (Tyr694), STAT6 (Tyr641),39-41 and NF-κB p65 (Ser536)42 as well as the proteins themselves from Cell Signaling Technology (Beverly, ME); [14C]arachidonic acid from PerkinElmer Life and Analytical Sciences (Boston, MA). LPS from other strains of E coli and P3C from other companies were found to have similar activities as those described here for the products described in this section. All other chemicals were molecular biology grade from several sources.
Culture of mast cell lines and experimental conditions
MC/9 were grown in suspension in 75-mm2 flask (3 × 105 cells/mL) in RPMI 1640 supplemented with 15% fetal calf serum, 30 ng/mL IL-3, 2 mM glutamine, 100 μM nonessential amino acids, 10 μM 2-mercaptoethanol, and 1 mM sodium pyruvate. Primary bone marrow–derived mast cells (BMMCs) were obtained from C57BL/6 mice by previously described procedures43 and grown in the same medium as MC/9 cells. A spontaneously transformed IL-3–dependent BMMC culture line (tBMMC) and RBL-2H3 cells were cultured as in previous studies.44 Cultures were sensitized to DNP-HSA by incubation at 37°C for 18 hours with 50 ng/mL DNP-IgE of FcϵR1 by IgE. Cultures were stimulated with concentrations of ligands that achieved maximal or near maximal responses (data not shown) except as indicated for experiments where concentrations were varied.
Measurement of degranulation and release of [14C]arachidonic acid
For these measurements, cultures were washed and medium replaced with a PIPES-buffered saline medium before stimulation.33 Degranulation was determined by measurement of release of the granule marker β-hexosaminidase by use of a colorimetric assay in which release of p-nitrophenol from p-nitrophenyl-N-acetyl-β-d-glucosaminide is measured. Values were expressed as the percent of intracellular β-hexosaminidase that was released into the medium.32 Release of radiolabeled arachidonic acid was determined in cultures that had been labeled to equilibrium by incubation overnight with [14C]arachidonic acid (0.1 μCi [0.0037 MBq]/mL medium) and then washed. The amount of radiolabel released into the medium was expressed as a percent of intracellular [14C]lipids in nonstimulated cells.33
Assay of cytokine mRNA and protein
Individual cytokines (TNFα, IL-6, and IL-13) were determined by enzyme-linked immunosorbent assay (ELISA) kits (Biosource, Camarillo, CA). For these determinations, cultures were incubated for 3 hours, unless stated otherwise, with the added stimulants in complete growth medium for measurement of release of cytokines into the medium. To examine the array of cytokines produced, a mouse cytokine antibody array (Ray Biotech, Norcross, GA) was used. A reference standard, biotin-labeled IgG on each array allowed determination of the relative changes in cytokine concentration.
Measurement of cytosolic Ca2+
Cells were incubated with 2:M Fura-2 AM ester (Molecular Probes, Eugene, OR) for 30 minutes in 96-well black culture plates (CulturPlate-96 F, Perkin Elmer Life Sciences; 5 × 104 cells/0.1 mL/well). Fluorescence was monitored in a Wallac VICTOR2 plate reader (Perkin Elmer Life Sciences) with alternating excitation at 340 nm and 380 nm and an emission wavelength set at 510 nm. Data were corrected for background fluorescence of cells that did not contain Fura-2, and the ratio of fluorescence at 340 nm and at 380 nm was determined.
Assay of IRAK activity
Immunoprecipitated IRAK1 was incubated at 30°C for 30 minutes with 1 μg of bacterially expressed myelin basic protein (MBP) as substrate in 20 μL of a kinase buffer: 10 mM HEPES (pH7.4), 1 mM dithiothreitol, 5 mM MgCl2, and 5 μCi (0.185 MBq) [γ-32/P]-ATP (3000 Ci/mmol [111 Bq/mmol]). Samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the dried gels were then exposed to X-ray film.
Immunoblotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and then lysed (30 minutes) in 0.1 mL lysis buffer: 20 mM HEPES, pH 7.3, 1% Triton X-100, 10% glycerol, 12.5 mM sodium pyrophosphate, 10 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 30 μg/mL leupeptin, 30 μg/mL aprotinin, and 25 mM p-nitrophenyl phosphate. Proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes for immunoblotting with the indicated primary antibodies. Immunoblots were visualized by chemiluminescence (Amersham, Piscataway, NJ) and quantitated by densitometry (Molecular Dynamics, Piscataway, NJ).
Expression levels and DNA-binding activity of transcription factors
A nuclear extraction kit (Active Motif, Carlsbad, CA) was used to prepare nuclear extracts. Activated transcription factors were identified by use of oligonucleotide arrays (TranSignal Protein/DNA Arrays, Panomics, Redwood City, CA) according to the manufacturer's instructions. Promotor binding activity was determined for NFATc1, NF-κB, and c-Fos by use of TransAM ELISA-based kits (Active Motif). In this procedure, nuclear extracts were added to multiwell plates with an immobilized oligonucleotide that contained the consensus binding site for the given transcription factor. The quantity of bound transcription factor was then determined by use of a primary antibody and a secondary antibody conjugated to horseradish peroxidase in an ELISA-type assay.
Results
TLR ligands fail to stimulate degranulation and release of arachidonic acid
The TLR2/TLR1 ligand, P3C, the TLR2/TLR6 ligands, PGN and MALP2, and the TLR4 ligand LPS were tested for their ability to stimulate degranulation and release of [14C]arachidonic acid. None of these ligands stimulated degranulation or release of [14C]arachidonic acid by themselves (Figure 1A,B) nor augmented such responses to antigen (Figure 1C-F) in MC/9 cells and BMMCs. Similar negative results were obtained in response to high concentrations of LPS (up to 10 μg/mL) and P3C (up to 100 μg/mL), and after prior exposure of cells to conditioned media or growth factors, namely, pokeweed mitogen-stimulated spleen-conditioned medium in the case of primary BMMCs22 and IL-3, IL-4, IFNγ, WEHI-conditioned media, or stem cell factor in the case of MC/9 cells (data not shown). P3C, PGN, and LPS also failed to stimulate or augment degranulation and release of radiolabeled arachidonic acid in RBL-2H3 cells and the transformed cell line, tBMMC (data not shown).
P3C and LPS act in synergy with antigen to augment production of cytokines
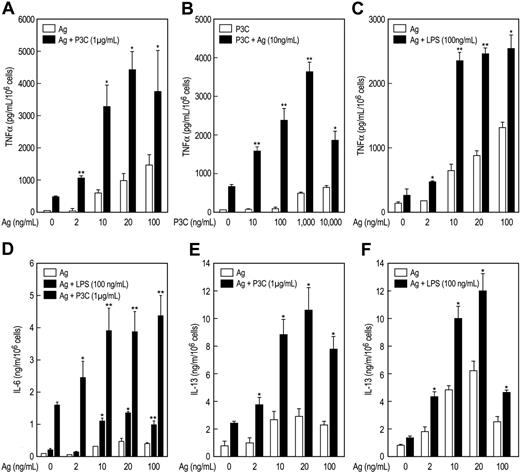
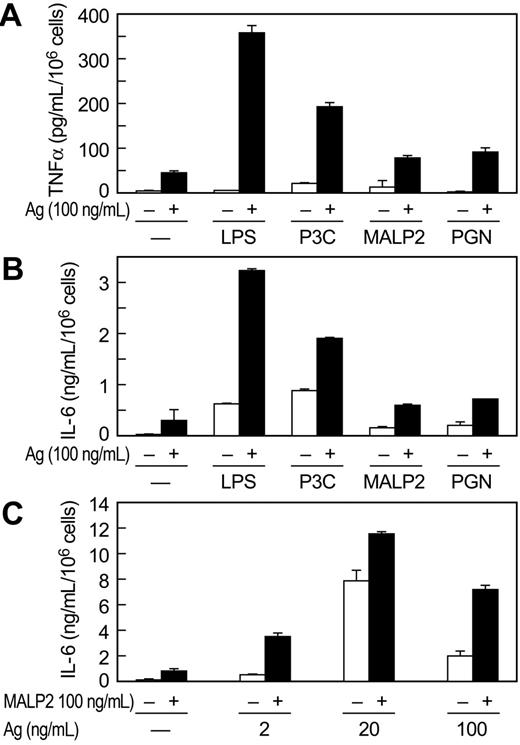
P3C and LPS stimulated release of TNFα from MC/9 cells but to a lesser extent than antigen. However, the combination of P3C or LPS with antigen markedly augmented release of TNFα (Figure 2). This augmentation was evident at concentrations of antigen (Figure 2A,C) or P3C (Figure 2B) that by themselves elicited minimal or no production of TNFα. At concentrations where the individual stimulants caused some production of TNFα, the antigen-induced response was augmented by up to 20-fold on co-stimulation with TLR ligand (data from all experiments). Therefore, antigen and the TLR ligands appeared to act in a mutually synergistic manner or even potentiate one another at threshold concentrations. The TNFα was apparently newly generated, as kinetic studies revealed no detectable release of TNFα 10 minutes after addition of stimulants (data not shown). Synergy or potentiation between antigen and P3C or LPS was observed also in the production of IL-6 (Figure 2D) and IL-13 (Figure 2E,F). As was true for TNFα production, P3C was more effective in augmenting production of IL-6 and IL-13 than LPS. Studies with MALP2 (10 and 100 ng/mL) in MC/9 cells also indicated that this ligand acted in synergy with antigen to increase production of TNFα from 3.7- to 4.4-fold and IL-13 from 3.1- to 5.3-fold over that expected from the sum of responses to the individual stimulants (data not shown).
TLR ligands do not affect degranulation and release of [14C]arachidonic acid. IgE-primed MC/9 cells and BMMCs were exposed to vehicle (NS), antigen (Ag; 20 ng/mL), PGN (100 μg/mL), P3C (1 μg/mL), LPS (100 ng/mL), or MALP2 (100 μg/mL) (A-B) or with the indicated concentrations of Ag or P3C plus 1 μg/mL P3C (C) or 20 ng/mL Ag (D) for 20 minutes for measurement of released β-hexosaminidase. (C-F) Vehicle was substituted for antigen in the samples designated “0.” Virtually identical results were obtained with LPS (100 ng/mL), PGN (100 μg/mL), and MALP-2 (100 ng/mL) as those shown in panel C (data not shown). For measurement of production of [14C]arachidonic acid, cells were incubated with [14C]arachidonic acid (1 μCi/mL [0.037 MBq]) for 18 hours before stimulation with antigen in combination with P3C (1 μg/mL) for 20 minutes (E,F). Values are the mean ± SEM from 4 experiments and are expressed as percent release of intracellular β-hexosaminidase or as a percent of total intracellular [14C]lipids that were released into the medium as [14C]arachidonic acid.
TLR ligands do not affect degranulation and release of [14C]arachidonic acid. IgE-primed MC/9 cells and BMMCs were exposed to vehicle (NS), antigen (Ag; 20 ng/mL), PGN (100 μg/mL), P3C (1 μg/mL), LPS (100 ng/mL), or MALP2 (100 μg/mL) (A-B) or with the indicated concentrations of Ag or P3C plus 1 μg/mL P3C (C) or 20 ng/mL Ag (D) for 20 minutes for measurement of released β-hexosaminidase. (C-F) Vehicle was substituted for antigen in the samples designated “0.” Virtually identical results were obtained with LPS (100 ng/mL), PGN (100 μg/mL), and MALP-2 (100 ng/mL) as those shown in panel C (data not shown). For measurement of production of [14C]arachidonic acid, cells were incubated with [14C]arachidonic acid (1 μCi/mL [0.037 MBq]) for 18 hours before stimulation with antigen in combination with P3C (1 μg/mL) for 20 minutes (E,F). Values are the mean ± SEM from 4 experiments and are expressed as percent release of intracellular β-hexosaminidase or as a percent of total intracellular [14C]lipids that were released into the medium as [14C]arachidonic acid.
Antigen and the TLR ligands also interacted synergistically in promoting release of TNFα and IL-6 from primary BMMC cultures derived from C57BL/6 mice (Figure 3). Production of TNFα and IL-6 was significantly augmented by the combination of antigen and LPS and P3C (Figure 3A,B). The TLR2/6 ligands PGN and MALP2 also acted in synergy with antigen in stimulating cytokine production but were less active in this regard than LPS and P3C (Figure 3A,B). We note, however, that synergy was most pronounced at threshold and supramaximal concentrations of antigen, where responses to antigen are minimal, and least pronounced at optimal concentrations of antigen. This was so for MALP2 (Figure 3C), PGN, LPS, and P3C in studies conducted in BMMCs as well as MC/9 cells (data not shown). In contrast to the findings in MC/9 cells, LPS was more effective than P3C in augmenting production of cytokines in antigen-stimulated BMMCs (Figure 3A,B). LPS is reported to be less, equi-, or considerably more potent than TLR2 ligands in stimulating cytokine production, depending on the study and mast cell line,22,26,27,29,30,45 a possible reflection of the phenotypic variability of mast cells.
Studies with antibody arrays revealed that in addition to TNFα, IL-6, and IL-13, antigen stimulated production of IL-12 p70, TCA-3, monocyte chemoattractant protein (MCP1), and macrophage inflammatory factor (MIP2) in MC/9 cells. In combination with P3C or LPS, production of these cytokines was consistently enhanced to levels several-fold greater than would be expected from additive responses (data not shown).
P3C and LPS act in synergy with antigen to markedly potentiate production of cytokines in MC/9 cells. IgE-primed MC/9 cells were stimulated in complete growth medium with the indicated concentrations of antigen (Ag), P3C, and LPS, individually or in the combinations indicated. Levels of TNFα (A-C), IL-6 (D), and IL-13 (E,F) were determined by ELISA 3 hours (A-E) or 6 hours (F) after addition of stimulants. Vehicle was substituted Ag (A,C,D,E, and F) or PC3 (B) where the concentration is designated as 0. Values are the mean ± SEM from 3 cultures. Asterisks indicate significant increase in response as compared to the responses to antigen (*P < .05; **P < .001). Identical results were obtained in 2 other experiments.
P3C and LPS act in synergy with antigen to markedly potentiate production of cytokines in MC/9 cells. IgE-primed MC/9 cells were stimulated in complete growth medium with the indicated concentrations of antigen (Ag), P3C, and LPS, individually or in the combinations indicated. Levels of TNFα (A-C), IL-6 (D), and IL-13 (E,F) were determined by ELISA 3 hours (A-E) or 6 hours (F) after addition of stimulants. Vehicle was substituted Ag (A,C,D,E, and F) or PC3 (B) where the concentration is designated as 0. Values are the mean ± SEM from 3 cultures. Asterisks indicate significant increase in response as compared to the responses to antigen (*P < .05; **P < .001). Identical results were obtained in 2 other experiments.
TLR ligands potentiate production of cytokines in BMMCs. IgE-primed cultures of BMMCs from C57BL/6 mice were incubated for 3 hours with vehicle (–), the indicated concentrations of antigen (Ag), 100 ng/mL LPS, 1 μg/mL P3C, 100 ng/mL MALP2, or 100 μg/mL PGN, individually or in combination. TNFα (A) and IL-6 (B, C) were determined by ELISA. A different source of C57BL/6 mice was used for the experiments shown in panel C. Values are the mean ± SEM from 3 cultures and are representative of 3 experiments.
TLR ligands potentiate production of cytokines in BMMCs. IgE-primed cultures of BMMCs from C57BL/6 mice were incubated for 3 hours with vehicle (–), the indicated concentrations of antigen (Ag), 100 ng/mL LPS, 1 μg/mL P3C, 100 ng/mL MALP2, or 100 μg/mL PGN, individually or in combination. TNFα (A) and IL-6 (B, C) were determined by ELISA. A different source of C57BL/6 mice was used for the experiments shown in panel C. Values are the mean ± SEM from 3 cultures and are representative of 3 experiments.
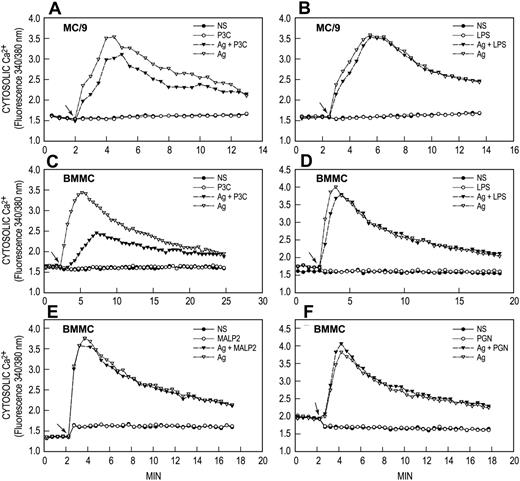
Induction of a calcium signal by antigen but not by TLR ligands
FcϵRI and TLR-related signaling processes were examined to identify possible mechanisms for the synergy between antigen and TLR ligands. The calcium signal was examined first because an increase in concentration of cytosolic Ca2+ is an essential signal for degranulation and for release of arachidonic acid in antigen-stimulated mast cells.20 Figure 4 shows typical results for P3C, LPS, MALP2, and PGN, individually and in combination with antigen. Antigen by itself caused an increase in concentration of cytosolic Ca2+, which reached a maximum 2 to 3 minutes after addition of antigen and then slowly declined in both MC9 cells (Figure 4A,B) and BMMCs (Figure 4C,D). None of the TLR ligands induced an increased cytosolic Ca2+ or enhanced the antigen-induced calcium signal. Indeed, P3C attenuated the calcium response to antigen in MC/9 cells and BMMCs by a mechanism yet to be determined. The absence of a calcium signal most likely accounts for the inability of the TLR ligands to influence degranulation or release of arachidonic acid in our studies.
Activation of protein kinases
Analysis by use of cDNA arrays indicated that MC/9 cells possessed message for TLR2 and TLR4 as well as TLR3, TLR5, TLR6, and all of the associated signaling molecules such as MyD88, MAL/TIRAP, TRAM, TRIF, IRAK1, and TAK1 (data not shown). Examination of IRAK 1 revealed that P3C and LPS induced robust activation of this kinase as determined by an in vitro kinase assay of immunoprecipitated IRAK 1. Antigen, in contrast, did not stimulate IRAK 1 activity and, in fact, partially attenuated IRAK-1 activation when added in combination with TLR ligands (Figure 5A).
TLR ligands do not generate calcium signals. IgE-primed MC/9 cells and BMMCs were loaded with Fura-2 and then stimulated or not (NS) with 20 ng/mL antigen (Ag), 1 μg/mL P3C, 100 ng/mL LPS, 100 ng/mL MALP2, or 10 μg/mL PGN, individually or in combination as indicated. Arrows indicate time of addition of stimulant(s). Values are expressed as the ratio of Fura-2 fluorescence at 510 nm when cells were excited alternately at 340 nm and 380 nm. The results are representative of 3 or more separate experiments.
TLR ligands do not generate calcium signals. IgE-primed MC/9 cells and BMMCs were loaded with Fura-2 and then stimulated or not (NS) with 20 ng/mL antigen (Ag), 1 μg/mL P3C, 100 ng/mL LPS, 100 ng/mL MALP2, or 10 μg/mL PGN, individually or in combination as indicated. Arrows indicate time of addition of stimulant(s). Values are expressed as the ratio of Fura-2 fluorescence at 510 nm when cells were excited alternately at 340 nm and 380 nm. The results are representative of 3 or more separate experiments.
Other kinases were activated by antigen and the TLR ligands. Activating phosphorylations of the MAP kinases Erk1/2, JNK, and p38 MAP kinase, as well as the phosphatidylinositol 3-kinase–dependent phosphorylation of Akt, were apparent 15 minutes after stimulation with antigen. Activating phosphorylations of JNK and p38 MAP kinase also were apparent 30 to 60 minutes after stimulation with LPS or P3C. However, LPS and P3C had no discernable effect on phosphorylation of Erk1/2 and Akt. Representative blots of these phosphorylations are shown in Figure 5B. When cells were co-stimulated with antigen and LPS or P3C, phosphorylation of JNK and, to a lesser extent, p38 MAP kinase, was enhanced. The extent of phosphorylation of JNK was up to 5-fold greater than would be expected from the additive responses to the individual stimulants (Figure 4C, 30- and 60-minute time points), while the phosphorylation of p38 MAP kinase was either additive or slightly more than additive 30 and 60 minutes after co-stimulation (data not shown, but see example in Figure 5B). No significant enhancement was observed in the phosphorylation of Erk and Akt during co-stimulation.
Activation of protein kinases by TLR ligands and antigen and effects of MAP kinase inhibitors on TNFα production in MC/9 cells. IgE-primed MC/9 cells were stimulated or not (NS) for the periods indicated with 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), individually or in combination. (A) IRAK1 was immunoprecipitated and assayed for kinase activity by incubation with [γ-32/P]ATP and myelin basic protein (MBP). The product, 32P-labeled MBP, was separated and detected by electrophoresis and autoradiography. (B) Immunoblots were prepared also from cell extracts and then probed with antibodies that detected either the indicated activated phosphorylated protein kinase or the protein itself. The blots are representative of blots from at least 3 separate experiments. (C) The relative densities of the doubly phosphorylated (Thr183/Tyr185)–JNK band were determined by densitometry. The values are the mean ± SEM of 4 experiments that were terminated 15, 30, and 60 minutes after addition of ligand(s). (D) IgE-primed MC/9 cells were incubated with vehicle, 25 μM SP 600125, 10 μM SB 203580, or 30 μM PD 98059 for 1 hour before addition of 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen, individually or in combination. TNFα was assayed by ELISA 3 hours thereafter. ND indicates not detectable. Values are mean ± SEM of 4 cultures and are from 1 of 2 identical experiments.
Activation of protein kinases by TLR ligands and antigen and effects of MAP kinase inhibitors on TNFα production in MC/9 cells. IgE-primed MC/9 cells were stimulated or not (NS) for the periods indicated with 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), individually or in combination. (A) IRAK1 was immunoprecipitated and assayed for kinase activity by incubation with [γ-32/P]ATP and myelin basic protein (MBP). The product, 32P-labeled MBP, was separated and detected by electrophoresis and autoradiography. (B) Immunoblots were prepared also from cell extracts and then probed with antibodies that detected either the indicated activated phosphorylated protein kinase or the protein itself. The blots are representative of blots from at least 3 separate experiments. (C) The relative densities of the doubly phosphorylated (Thr183/Tyr185)–JNK band were determined by densitometry. The values are the mean ± SEM of 4 experiments that were terminated 15, 30, and 60 minutes after addition of ligand(s). (D) IgE-primed MC/9 cells were incubated with vehicle, 25 μM SP 600125, 10 μM SB 203580, or 30 μM PD 98059 for 1 hour before addition of 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen, individually or in combination. TNFα was assayed by ELISA 3 hours thereafter. ND indicates not detectable. Values are mean ± SEM of 4 cultures and are from 1 of 2 identical experiments.
Although the above studies suggested that signals transduced through JNK and p38 MAP kinase were enhanced by co-stimulation, further studies with inhibitors suggested that production TNFα was dependent, to some degree, on all 3 MAP kinases (Figure 5D). For example, the JNK inhibitor SP 60012546 substantially suppressed TNFα production whether cells were stimulated with TLR ligands, antigen, or the combination of stimulants. The p38 MAP kinase inhibitor, SB203580,47 completely abrogated the responses to the TLR ligands, but it had minimal or insignificant effect on the TNFα response to antigen. PD 98059, an inhibitor of the MEK-1/Erk-pathway,48 partially suppressed production of TNFα in response to all stimulants, individually or in combination.
Induction and activation of transcription factors
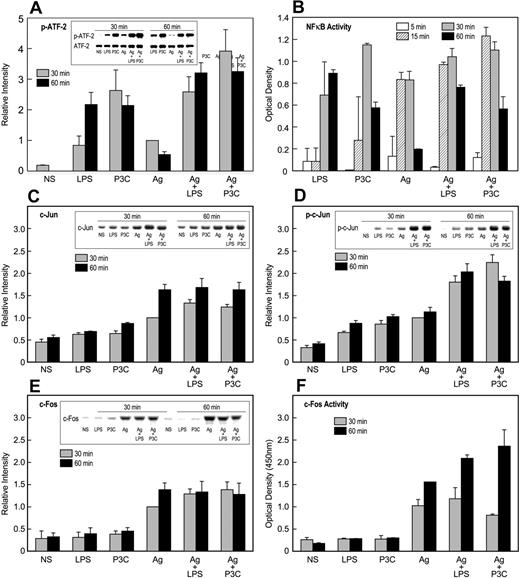
ATF-2, AP-1 binding proteins (ie, c-Jun and c-Fos), and NF-κB were initially investigated because these factors are activated via TLR2 or TLR4.49,50 In addition, ATF-2 and c-Jun are the downstream targets of p38 MAP kinase and JNK, respectively. We observed that ATF-2 was most strongly activated by TLR ligands, while c-Jun and c-Fos were activated primarily by antigen. NF-κB was activated to the same extent by TLR ligands and antigen. Typical experimental results were as follows.
In regard to ATF-2, LPS and P3C induced robust and persistent phosphorylation of ATF-2 as compared to antigen, and in combination with antigen, this phosphorylation was augmented in an additive manner (Figure 6A). Stimulation also resulted in small increases in levels of ATF-2 protein, but these increases were relatively minor compared to the changes in phosphorylation (Figure 6A). NF-κB was activated to the same extent whether activated through TLR ligands or antigen as indicated by its phosphorylation (Figure 5A) and DNA-binding activity (Figure 6B). However, these responses were not significantly augmented, either additively or synergistically, when stimulants were used in combination (Figure 6A,B).
Activation of transcription factors by TLR ligands and antigen. IgE-primed MC/9 cells were stimulated or not (NS) for the periods indicated with 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), individually or in combination. Immunoblots were prepared from cell extracts and then probed for the indicated transcription factors or their activated phosphorylated forms with appropriate antibodies. Typical blots and their relative densities (mean ± SEM of values from 3 experiments) are shown for phosphorylated (Thr71)–ATF-2 (A), c-Jun (C), phosphorylated (Ser63)–c-Jun (D), and c-Fos (E). In addition, nuclear extracts were assayed for NF-κB (B) and c-Fos (F) oligonucleotide-binding activities by use of a commercial kit. The values are the mean ± SEM from 3 cultures. Identical results were obtained in a second experiment.
Activation of transcription factors by TLR ligands and antigen. IgE-primed MC/9 cells were stimulated or not (NS) for the periods indicated with 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), individually or in combination. Immunoblots were prepared from cell extracts and then probed for the indicated transcription factors or their activated phosphorylated forms with appropriate antibodies. Typical blots and their relative densities (mean ± SEM of values from 3 experiments) are shown for phosphorylated (Thr71)–ATF-2 (A), c-Jun (C), phosphorylated (Ser63)–c-Jun (D), and c-Fos (E). In addition, nuclear extracts were assayed for NF-κB (B) and c-Fos (F) oligonucleotide-binding activities by use of a commercial kit. The values are the mean ± SEM from 3 cultures. Identical results were obtained in a second experiment.
Substantial augmentation of responses was noted in the induction and phosphorylation of proteins of the AP-1 complex. Analysis of mRNA indicated that antigen and, minimally so, TLR ligands caused increased message for c-Jun, Jun-B, Jun-D, and c-Fos (data not shown). Further examination of c-Jun and c-Fos (Figure 6) revealed that LPS and P3C, in contrast to antigen, failed to induce significant increases in levels of c-Jun (Figure 6C) and c-Fos (Figure 6E) protein. All 3 stimulants elicited some phosphorylation of c-Jun, but this response was augmented more than 2-fold by the combination of antigen with LPS or P3C (Figure 6D). Studies to determine the state of phosphorylation of c-Fos were unsuccessful. As an alternate strategy, the DNA-binding activity of c-Fos (presumably as the c-Jun/c-Fos heterodimer) was determined, and the data indicated that LPS and P3C failed to stimulate binding activity. However, these ligands markedly potentiated activity when cells were co-stimulated with antigen (Figure 6F).
We also have investigated the phosphorylation of the signal transducer and activator of transcription (STAT) family of proteins because of the potential paracrine effects of type I IFNs that might be produced as a consequence of TLR signaling through TRAM and TRIF.51 No consistent increase in phosphorylation of STAT3, STAT5, or STAT6 was observed by immunoblotting (see “Materials and methods” for list of antibodies tested) with any stimulant or combination of stimulants in MC/9 cells (data not shown). A minimal increase was observed in serine and tyrosine phosphorylation of STAT1 in response to stimulation by TLR ligands and antigen (data not shown). However, no augmentation was noted with the combination of stimulants. Use of DNA-binding arrays also indicated minimal, if any, increase in binding of STATs to DNA consensus sequences during stimulation of the cells.
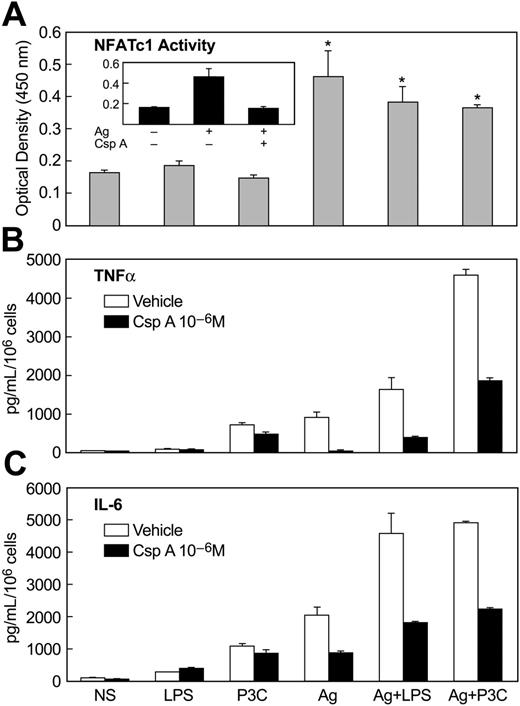
Finally, we examined the calcium/calcineurin-dependent activation of NFAT because of its activation and presumed role in mediating transcription of cytokine genes in antigen-stimulated mast cells.52 In contrast to antigen, LPS and P3C failed to stimulate NF-AT DNA-binding activity or to enhance this activity upon co-stimulation of MC/9 cells with antigen and LPS or P3C (Figure 7A). This result was consistent with the lack of a calcium signal in LPS or P3C-stimulated cells (Figure 4). As expected, the stimulation of NF-AT DNA-binding activity by antigen was blocked by prior treatment of cells with the calcineurin inhibitor cyclosporin A (Figure 7A, inset). Cyclosporin A also suppressed the production of TNFα (Figure 7B) and IL-6 (Figure 7C) in antigen-stimulated cells, while it had minimal effect on production of these cytokines in cells stimulated with LPS or P3C. In addition, cyclosporine A markedly attenuated the augmented production of TNFα and IL-6 in cells co-stimulated with antigen and LPS or P3C (Figure 7B,C). These results implied that activation of NF-AT was essential for production of TNFα and IL-6 in response to antigen stimulation but not in response to the TLR ligands. However, the activation of NF-AT by antigen could act in synergy with TLR-induced signals to augment cytokine production. Presumably, the TLR ligands activate signals that can substitute for and act in synergy with NF-AT.
Activation of NF-AT by antigen and inhibitory effects of cyclosporine-A on NF-AT activation and production of cytokines. IgE-primed MC/9 cells were stimulated or not (NS) in complete growth medium for 30 minutes for measurement of NF-AT oligonucleotide-binding activity (A) or for 3 hours for measurement of TNFα (B) and IL-6 (C). Stimulants included 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), alone or in combination. Where indicated, cultures were incubated with 1 μM cyclosporine A (CsA) for 60 minutes before addition of stimulants. The values are the mean ± SEM of 3 cultures. Identical results were obtained in 2 other experiments.
Activation of NF-AT by antigen and inhibitory effects of cyclosporine-A on NF-AT activation and production of cytokines. IgE-primed MC/9 cells were stimulated or not (NS) in complete growth medium for 30 minutes for measurement of NF-AT oligonucleotide-binding activity (A) or for 3 hours for measurement of TNFα (B) and IL-6 (C). Stimulants included 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), alone or in combination. Where indicated, cultures were incubated with 1 μM cyclosporine A (CsA) for 60 minutes before addition of stimulants. The values are the mean ± SEM of 3 cultures. Identical results were obtained in 2 other experiments.
Discussion
TLRs not only enable activation of inflammatory cells of the innate immune system, but they may also regulate the subsequent development of acquired immunity by promoting maturation of antigen-presenting cells (reviewed in Basu and Fenton9 ). There is evidence that the type of TLR engaged may direct development toward either T-helper type 1 (Th1) or Th2 adaptive immune responses. TLR ligands may thus influence the asthmatic condition by regulating T-cell function if Th1 cells are protective and Th2 cells are detrimental in the development of asthma as proposed by some workers.10 However, the possibility remains that TLRs act more directly by interacting synergistically with other receptor systems that promote inflammatory responses. To examine this possibility, we focused on mast cells because these cells express FcϵRI as well as TLRs, which, respectively, participate in allergic diseases and protection against pathogens.24 Moreover, co-stimulation of BMMCs through FcϵR1 and TLR4 is reported to augment production of cytokines.53 Our data show that TLR2 and TLR4 ligands substantially enhance antigen-induced production of cytokines (Figures 2-3) and the associated signals, without affecting degranulation or production of arachidonic acid (Figure 1) in MC/9 cells and primary cultures of BMMCs. If this scenario is relevant to atopic disease, activation of TLRs would have the most pronounced effect at late stages of the inflammatory response to antigen that coincides with the production of inflammatory cytokines by mast cells.54,55
The pattern of signals activated correlates with the responses noted in the previous paragraph. An increase in levels of cytosolic Ca2+ is an essential signal for degranulation20 and production of arachidonic acid via phospholipase A256 in mast cells. The fact that TLR ligands elicited none of these responses in MC/9 cells, BMMCs, and other mast cell lines is consistent with this requirement for a calcium signal. This is in contrast to stem cell factor, which elicits a calcium signal and enhances antigen-induced degranulation as well as cytokine production.34
Nevertheless, TLR ligands stimulate activating phosphorylations of the MAP kinases (Figure 5), all of which appear to regulate to some degree the production of TNFα (Figure 5D). Most notable was the synergistic enhancement of the phosphorylation of JNK (Figure 5C) and the additive phosphorylation of p38 MAP kinase (Figure 5B) when cells were stimulated with TLR ligands in combination with antigen. This pattern correlated with the downstream activating phosphorylations of transcription factors by MAP kinases, namely, c-Jun and ATF-2,57 as determined by use of phosphorylation-specific antibodies. The phosphorylation c-Jun (Figure 6D) and ATF-2 (Figure 6A) was most pronounced after stimulation with the combination of TLR ligands and antigen.
The transcriptional activity of c-Jun also is dependent on dimerization with itself or other bZIP proteins that permit binding to relevant promotor sites, such as the binding of the c-Jun/c-Fos dimer to AP1. Of note, antigen but not TLR ligands induced synthesis of c-Fos (Figure 6E) and binding of c-Fos to AP1 (Figure 6F), presumably as the c-Jun/c-Fos dimer. The enhancement of this binding activity by TLR ligands was likely due to the augmented phosphorylation of c-Jun when cells were co-stimulated with TLR ligands and antigen.
The net effect was that some transcription factors were activated by both types of stimulants (NF-κB), while others were activated most robustly by either antigen (c-Jun/c-Fos and NFAT) or TLR ligands (ATF2). Co-stimulation with antigen and TLR ligands resulted in additive or synergistic activation of ATF-2, c-Jun, and c-Fos. Therefore, co-stimulation not only broadened the array of transcription factors activated but also reinforced the activation of some of these factors. The same results were obtained by use of DNA-binding arrays (data not shown). The array data revealed that in addition, E twenty-six (Ets) proteins were activated by all stimulants, Sp1 by antigen only, and as noted earlier, STAT proteins were minimally phosphorylated by TLR ligands and antigen.
Optimal transcription of genes for cytokines such as TNFα, IL-6, and IL-13 requires the coordinate actions of a repertoire of transcription factors. The identified consensus binding sites in the TNFα gene loci, for example, include Ets, cyclic AMP regulatory element (CRE), AP1, AP2, Sp1, and multiple sites for NFAT and NF-κB (κ1, κ2, and κ3).58-60 Of relevance to our findings, ATF-2 and c-Jun act cooperatively with NFAT family proteins on the adjacent CRE/κ3 sites,61 and transcriptional activity is further enhanced on co-stimulation with Sp1.58 Similar interaction occurs between the Jun (CRE) site and a nearby Ets binding site.62 The promotor region of IL-6 also contains interacting consensus sites for AP1, CRE, C/EBP, and NF-κB,63,64 and cooperative interactions between NFAT and AP1 have been noted for the transcription of other cytokine genes, including IL-13.65 Although little is known about the combinatorial requirements for cytokine gene transcription in mast cells, our results suggest that optimal activation of the required repertoire of transcription factors is best achieved on co-stimulation of mast cells with antigen and the TLR ligands.
An interesting paradox is that activation of NFAT is absolutely essential for cytokine gene transcription following antigen stimulation, but this is not so for TLR stimulation. This suggests that the TLR ligands activate effective but unidentified transcription signal(s) in the absence of NFAT activation. TLR-mediated cytokine production has been linked to activation of NF-κB and IFNβ-inducible genes, but RNase protection assays revealed no significant increase in IFNβ mRNA during the time course of our experiments (data not shown). Therefore, we believe it unlikely that the activation of IFNβ-inducible genes by TLR ligands15 was a significant factor in the synergistic interactions described here.
Although our studies encompass a variety of rodent cell lines and conditions, it should be noted that mast cells exhibit some variation in phenotype according to anatomical location, species, and culture conditions.66 Such variability might account for the expression of TLR4 in human peripheral blood–derived mast cells29 but not in human cord blood–derived mast cells.25 Nevertheless, the latter cells can be made to express TLR4 after priming with IL-4.26 Because of reports that TLR2 ligands elicit degranulation and generation of leukotriene C4 in some22,25,26 but not all27 studies, we examined the effects of various cytokines that can affect mast cell maturation and differentiation. These included stem cell factor, IL3, IL-4, and IFNγ.66 As noted earlier, we failed to induce degranulation and release of arachidonic acid when these cytokines were included in the growth medium, either individually or in combination. Therefore, we were unsuccessful in defining culture conditions that might explain the discrepancy in results among different laboratories.
However, if the synergy observed between antigen and TLR ligands in cultured mast cell lines has clinical relevance, there are implications in the treatment of atopic disease during infection. Apart from use of antiviral agents or the development of inhibitors of TLR activation, we note that low concentrations of glucocorticoids effectively suppress cytokine production in mast cells in response to not only antigen44 but also to TLR2 and TLR4 ligands (H.Q., unpublished data, June 2005). Therefore, glucocorticoids may serve a double purpose in the treatment of asthmatic patients during the course of airway infections.
Prepublished online as Blood First Edition Paper, September 20, 2005; DOI 10.1182/blood-2005-06-2271.
Supported in part by Intramural Research Program, National Institutes of Health, and the National Heart, Lung, and Blood Institute; and National Institutes of Health grant 1 RO1 TW006612-01 (M.V.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Alasdair M. Gilfillan for his suggestions in the preparation of this manuscript.

![Figure 1. TLR ligands do not affect degranulation and release of [14C]arachidonic acid. IgE-primed MC/9 cells and BMMCs were exposed to vehicle (NS), antigen (Ag; 20 ng/mL), PGN (100 μg/mL), P3C (1 μg/mL), LPS (100 ng/mL), or MALP2 (100 μg/mL) (A-B) or with the indicated concentrations of Ag or P3C plus 1 μg/mL P3C (C) or 20 ng/mL Ag (D) for 20 minutes for measurement of released β-hexosaminidase. (C-F) Vehicle was substituted for antigen in the samples designated “0.” Virtually identical results were obtained with LPS (100 ng/mL), PGN (100 μg/mL), and MALP-2 (100 ng/mL) as those shown in panel C (data not shown). For measurement of production of [14C]arachidonic acid, cells were incubated with [14C]arachidonic acid (1 μCi/mL [0.037 MBq]) for 18 hours before stimulation with antigen in combination with P3C (1 μg/mL) for 20 minutes (E,F). Values are the mean ± SEM from 4 experiments and are expressed as percent release of intracellular β-hexosaminidase or as a percent of total intracellular [14C]lipids that were released into the medium as [14C]arachidonic acid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-06-2271/4/m_zh80020689620001.jpeg?Expires=1768616001&Signature=K1EFi1Vv58ggwezpfomxoi-EHFqA8c-N3uAjfGfTOqwwPUWLc1AzO938zDMYzRnICXeLJbWm9XpOBFtGJJFFeg0fr6CL8y7XiOyyEf0N3foK7Y3etWTky3MBRA7MSGfWra7zaM~qvQbsVTML2XGBRYTSPx6kiGv-truhJZM1NCuVqGpudX6RZzumiaDA39Z5usyz-nfgGmGQWMF3bPZxFvi0XaoFXVXUs3qoCkgjucoTZGrXs~gAbVJlB2V0KICtuZT~ql~b2deCLqIkh~UwluKdgmVfCqjLF9fVYdSTUbogx77pQ7JYz3TBNO0j3zr0alPtfjk-3AwYrVbY8ioOOg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Activation of protein kinases by TLR ligands and antigen and effects of MAP kinase inhibitors on TNFα production in MC/9 cells. IgE-primed MC/9 cells were stimulated or not (NS) for the periods indicated with 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen (Ag), individually or in combination. (A) IRAK1 was immunoprecipitated and assayed for kinase activity by incubation with [γ-32/P]ATP and myelin basic protein (MBP). The product, 32P-labeled MBP, was separated and detected by electrophoresis and autoradiography. (B) Immunoblots were prepared also from cell extracts and then probed with antibodies that detected either the indicated activated phosphorylated protein kinase or the protein itself. The blots are representative of blots from at least 3 separate experiments. (C) The relative densities of the doubly phosphorylated (Thr183/Tyr185)–JNK band were determined by densitometry. The values are the mean ± SEM of 4 experiments that were terminated 15, 30, and 60 minutes after addition of ligand(s). (D) IgE-primed MC/9 cells were incubated with vehicle, 25 μM SP 600125, 10 μM SB 203580, or 30 μM PD 98059 for 1 hour before addition of 100 ng/mL LPS, 1 μg/mL P3C, and 20 ng/mL antigen, individually or in combination. TNFα was assayed by ELISA 3 hours thereafter. ND indicates not detectable. Values are mean ± SEM of 4 cultures and are from 1 of 2 identical experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-06-2271/4/m_zh80020689620005.jpeg?Expires=1768616001&Signature=RLCCCxonZ7M3ncFIM4huBYDdsbT3Ge17X9uapKpfXQ-z8Cjg2kxKRMb7REN6yAqWKWyDMOYAHcyh4UYTiz5xl2jnrWxpfZCkMpuCEBzOeZ2-hQlIwJ-Iv3gG39GGJrbQrXLYylNz17PJLHEAvl0JRxMs9RIQIIGmVoglMuYHHpPYkBc5p8yv9ydyJzLnTN4gqBs4HPhxAb1mdvFQ9J7jY-9I4KtKWqGi7v8lXFMEagmKyyu5CnjfO179LuzaUcGDybmu~bHVTC0xMyAz8mgfYgLuQtF167uAy0eleCKm27ZfJcSRP3Hp-l2BiG2vg9xu1s8Xt86XQBGsW~dwT6q4zw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)