Abstract
Recurrent reciprocal translocations are present in many hematologic and mesenchymal malignancies. Because significant sequence homology is absent from translocation breakpoint junctions, non-homologous end-joining (NHEJ) pathways of DNA repair are presumed to catalyze their formation. We developed translocation reporters for use in mammalian cells from which NHEJ events can be selected after precise chromosomal breakage. Translocations were efficiently recovered with these reporters using mouse cells, and their breakpoint junctions recapitulated findings from oncogenic translocations. Small deletions and microhomology were present in most junctions; insertions and more complex events also were observed. Thus, our reporters model features of oncogenic rearrangements in human cancer cells. A homologous sequence at a distance from the break site affected the translocation junction without substantially altering translocation frequency. Interestingly, in a direct comparison, the spectrum of translocation breakpoint junctions differed from junctions derived from repair at a single chromosomal break, providing mechanistic insight into translocation formation. (Blood. 2006;107:777-780)
Introduction
Many hematologic and mesenchymal malignancies harbor reciprocal translocations that develop as early and essential events in oncogenesis.1-3 Translocations likely result from contemporaneous DNA double-strand breaks (DSBs) generated by either endogenous (eg, V(D)J recombinase4 ) or exogenous (eg, topoisomerase II poisons5 ) agents. Mammalian cells have multiple pathways to repair DSBs, including mechanisms that involve little or no sequence homology, collectively termed nonhomologous end-joining (NHEJ).6 NHEJ is important for survival in response to clastogens and is critical for the repair of DSBs induced during V(D)J recombination.6,7 Homology-directed repair, an essential pathway in mammalian cells,8 does not appear to be involved in the overwhelming majority of cancer-associated translocations, as breakpoint junctions lack extensive homology,1-3 and translocations involving this pathway are not recovered in model systems.9-12 However, another homologous repair pathway, single-strand annealing (SSA), efficiently generates translocations in model systems.9,10
Given that reciprocal translocations in human cancer cells appear to arise from NHEJ, we developed translocation reporters for use in mammalian cells in which significant sequence homology is absent from the DNA ends.
Study design
DNA manipulations, transfections, and translocation analysis
Targeting vectors, polymerase chain reaction (PCR) conditions, and primers are described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). For translocation experiments, 2 × 107 cells were electroporated with 50 μg pCBASce9,13 or pCAGGS.9,10 Fluorescence in situ hybridization (FISH) was performed as described.9
Pool analysis of repair junctions
Two p5rE cell lines and 2 translocation clones were electroporated with 50
μg pCBASce. After G418 selection, pooled genomic DNA was isolated, digested with I-SceI, and PCR amplified. PCR products were TOPO-cloned (Invitrogen, Carlsbad, CA) and sequenced.
Results and discussion
Reporters to model oncogenic translocations
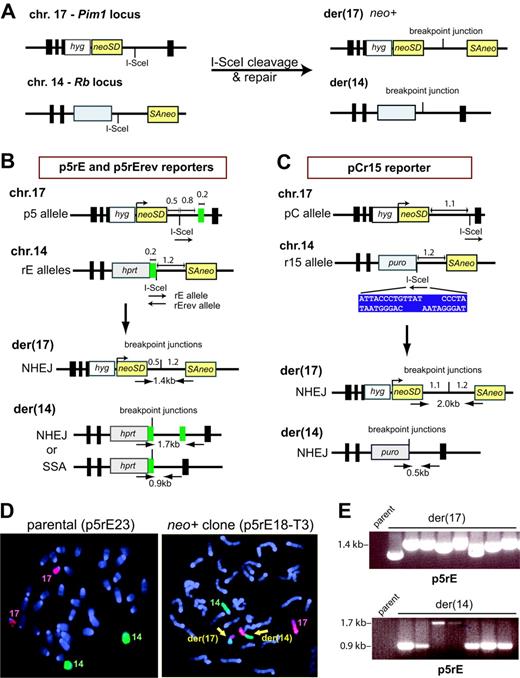
We modified reporters recently developed to study rearrangements at Alu elements9 (Figure 1A). Central to the translocation reporters is a neomycin phosphotransferase gene (neo) split by an intron to form neoSD and SAneo, each of which contains an I-SceI endonuclease cleavage site and is targeted to chromosomes 17 or 14, respectively, in mouse embryonic stem (ES) cells. DSBs generated by I-SceI followed by NHEJ to link neoSD and SAneo generates a neo+ gene on der(17). Repair within intronic sequences allows for the recovery of translocation junctions with a wide array of DNA end modifications.
We constructed 3 translocation reporters based on this design. With p5rE and p5rErev, chromosomal breakage by I-SceI followed by NHEJ with little sequence alteration results in a neo+ gene on der(17) with a 1.7-kb intron (Figure 1B), while formation of der(14) is not under selection. Der(14) can form by NHEJ or SSA, since a 210-bp repeat is located adjacent to the chromosome 14 breakpoint and 0.8 kb from the chromosome 17 breakpoint, allowing for a comparison of these 2 pathways when a repeat is distant from a breakpoint. To test whether compatible overhangs enhance translocation formation, the p5rE reporter contains the I-SceI sites in the same (compatible) orientation, in contrast with the p5rErev reporter, which contains the sites in opposite (incompatible) orientation. Reporter pCr15 contains 2 modifications, an expanded chromosome 17 intron, to accommodate larger deletions, and loss of the repeat, such that der(14) can only form by NHEJ (Figure 1C).
Chromosomal translocations between nonhomologous sequences. (A) Translocation design. A neo gene is split within its intron such that the 5′ portion with the splice donor (neoSD) is targeted to chromosome 17 in mouse ES cells, and the 3′ portion with the splice acceptor (SAneo) is targeted to chromosome 14. DSB induction at the I-SceI sites followed by interchromosomal NHEJ results in a neo+ gene on der(17). Because the breakpoint junction occurs within an intron, a variety of junctions can be recovered. The reciprocal chromosome der(14) also can form but is not under selection. Gray box, selectable marker. (B) Translocation reporters p5rE and p5rErev. The 2 rE alleles differ by whether the I-SceI sites on chromosomes 17 and 14 are in the same (rE) or opposite orientation (rErev). Der(17) arising from NHEJ with minimal sequence alteration reconstructs a neo+ gene with an intron of approximately 1.7 kb; der(14) can arise from NHEJ or SSA at the 210-bp repeat (green box), as shown. In principle, homology-directed repair at the 210-bp repeat could cause translocation formation via a reciprocal exchange. However, consistent with previous results,9-12 homology-directed repair events, which would leave one copy of the repeat on der(17), have not been recovered; hence, these translocation chromosomes are not diagrammed. Key distances are indicated in kb. (C) Translocation reporter pCr15. Der(17) arising from NHEJ with minimal sequence alteration reconstructs a neo+ gene with an intron of approximately 2.3 kb; like der(17), der(14) also can only arise from NHEJ. The I-SceI site is positioned further from the splice donor in the pCr15 reporter than in the p5rE and p5rErev reporters. For the pCr15 reporter, the I-SceI sites are in opposite orientation. The sequence of the I-SceI site upon cleavage is shown (blue box). (D) Fluorescence in situ hybridization using whole mouse chromosome 14-FITC (green) and chromosome 17-Cy3 (red) probes and demonstrating normal chromosomes 14 and 17 in the parental cell line and 2 derivative chromosomes (yellow arrows) in a neo+ clone. (E) Translocation chromosome analysis. PCR primers and sizes of PCR products are indicated on the derivative chromosomes in panel B. A sample of PCR products obtained from p5rE neo+ clones shows der(17) NHEJ junctions of variable length and der(14) NHEJ and SSA products (1.7 and 0.9 kb, respectively). No band is obtained from the parental cell line since the primers are located on different chromosomes.
Chromosomal translocations between nonhomologous sequences. (A) Translocation design. A neo gene is split within its intron such that the 5′ portion with the splice donor (neoSD) is targeted to chromosome 17 in mouse ES cells, and the 3′ portion with the splice acceptor (SAneo) is targeted to chromosome 14. DSB induction at the I-SceI sites followed by interchromosomal NHEJ results in a neo+ gene on der(17). Because the breakpoint junction occurs within an intron, a variety of junctions can be recovered. The reciprocal chromosome der(14) also can form but is not under selection. Gray box, selectable marker. (B) Translocation reporters p5rE and p5rErev. The 2 rE alleles differ by whether the I-SceI sites on chromosomes 17 and 14 are in the same (rE) or opposite orientation (rErev). Der(17) arising from NHEJ with minimal sequence alteration reconstructs a neo+ gene with an intron of approximately 1.7 kb; der(14) can arise from NHEJ or SSA at the 210-bp repeat (green box), as shown. In principle, homology-directed repair at the 210-bp repeat could cause translocation formation via a reciprocal exchange. However, consistent with previous results,9-12 homology-directed repair events, which would leave one copy of the repeat on der(17), have not been recovered; hence, these translocation chromosomes are not diagrammed. Key distances are indicated in kb. (C) Translocation reporter pCr15. Der(17) arising from NHEJ with minimal sequence alteration reconstructs a neo+ gene with an intron of approximately 2.3 kb; like der(17), der(14) also can only arise from NHEJ. The I-SceI site is positioned further from the splice donor in the pCr15 reporter than in the p5rE and p5rErev reporters. For the pCr15 reporter, the I-SceI sites are in opposite orientation. The sequence of the I-SceI site upon cleavage is shown (blue box). (D) Fluorescence in situ hybridization using whole mouse chromosome 14-FITC (green) and chromosome 17-Cy3 (red) probes and demonstrating normal chromosomes 14 and 17 in the parental cell line and 2 derivative chromosomes (yellow arrows) in a neo+ clone. (E) Translocation chromosome analysis. PCR primers and sizes of PCR products are indicated on the derivative chromosomes in panel B. A sample of PCR products obtained from p5rE neo+ clones shows der(17) NHEJ junctions of variable length and der(14) NHEJ and SSA products (1.7 and 0.9 kb, respectively). No band is obtained from the parental cell line since the primers are located on different chromosomes.
Efficient recovery of translocations by NHEJ
After I-SceI expression, neo+ clones were recovered using each reporter at a frequency of 3 to 4 × 10-5, compared with less than 0.2 × 10-7 without I-SceI. Thus, recovery of neo+ clones is not substantially affected by sequence homology in the vicinity of the breakpoints, complimentarity of DNA ends, or a larger segment for deletion. By FISH, 31 of 32 neo+ clones harbored the expected der(17) and der(14) chromosomes (Figure 1D; data not shown), indicating that our reporters faithfully score translocations.
Translocation breakpoint junctions: frequent deletions and microhomology
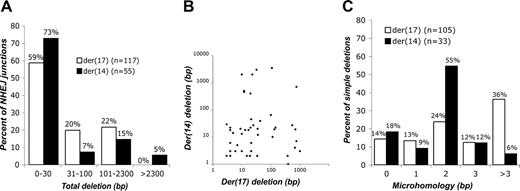
To gain insight into the DSB repair mechanisms leading to translocations, we analyzed 120 neo+ clones (Figure 1E; data not shown). For der(17), 117 neo+ clones gave PCR products; for der(14), 110 neo+ clones gave PCR products, and 7 additional clones had deletions of at least 700 bp as detected by Southern blotting. Of the 234 breakpoint junctions, 172 involved NHEJ and fell into 3 classes: 138 (80%) simple deletions, 20 (12%) insertions, and 4 (2%) complex events, plus 10 unsequenced junctions (Figures S1-S2; data not shown). Although there was some variation in deletion size depending on the particular sequences being joined, the majority of deletions were less than 100 bp for all 3 reporters on both der(17) and der(14) (Figure 2A; Figure S2). Comparing data from a number of malignancies, deletions are observed in a majority of oncogenic translocation junctions as well, with a median deletion length similar to what we observed with our reporters (Table S1). Only 3 der(14) deletions extended more than 2.3 kb, the maximum intron for pCr15, suggesting that our approach recovers most translocations. No correlation was observed between deletion lengths on der(17) and der(14) for an individual neo+ clone (Figure 2B) or between deletion lengths from the centromeric and telomeric ends for an individual derivative chromosome (data not shown). Of the junctions with simple deletions, 117 (85%) exhibited microhomology (identical nucleotides at both ends) (Figure 2C), which is also common in oncogenic translocation junctions (Table S2).
In addition to deletions, insertions and duplications of genomic sequences are observed in a portion of oncogenic translocations (Tables S1-S2). Duplications could result from staggered nicks, followed by filling in of the resulting single-strand overhangs. Duplications of all or a portion of the 4 nucleotide I-SceI overhangs were observed in several neo+ clones (Table S1). Interestingly, although many of the duplications observed in tumors are also only a few bp, duplications reported in several leukemias are more than 100 bp, which would imply a mechanism in which the nicks are staggered at a substantial distance from each other.14,15
Translocation junction analysis. (A) Distribution of deletion lengths in 172 translocation junctions formed by NHEJ for der(17) and der(14) junctions. Junctions are derived from neo+ clones from the 3 reporter cell lines. The der(14) junctions that are not shown here were repaired by SSA. NA indicates not applicable. (B) Scatter plot of translocation deletion lengths in which both derivative chromosome junctions in a neo+ clone arose by NHEJ. No correlation was noted between deletion lengths on der(17) and der(14) (r = -0.085). (C) Distribution of microhomologies in 138 translocation junctions. Only der(17) and der(14) junctions that were repaired by NHEJ without an insertion (ie, simple deletions) are included.
Translocation junction analysis. (A) Distribution of deletion lengths in 172 translocation junctions formed by NHEJ for der(17) and der(14) junctions. Junctions are derived from neo+ clones from the 3 reporter cell lines. The der(14) junctions that are not shown here were repaired by SSA. NA indicates not applicable. (B) Scatter plot of translocation deletion lengths in which both derivative chromosome junctions in a neo+ clone arose by NHEJ. No correlation was noted between deletion lengths on der(17) and der(14) (r = -0.085). (C) Distribution of microhomologies in 138 translocation junctions. Only der(17) and der(14) junctions that were repaired by NHEJ without an insertion (ie, simple deletions) are included.
The median insertion length observed in oncogenic translocation junctions is a few base pair, although longer insertions also have been reported (Table S2). Of 20 insertions we obtained, 12 were less than 10 bp. Four others were more than 100 bp and were derived from a variety of sources (genomic DNA, mitochondrial DNA, I-SceI vector; Figure S1). The complex events contained duplications and inversions of nearby sequences, deletions, and insertions (Figure S1), resembling events reported in some malignancies.14-17
Reporter-specific junctions involving the I-SceI overhangs and sequence repeat also were noted. For p5rE, 24% (13 of 54) of der(17) junctions re-established the I-SceI site, presumably by annealing of the 4 nucleotide, 3′ overhangs. Thus, precise ligation can be used to generate translocation junctions. For p5rE and p5rErev, 65% (62 of 96) of der(14) junctions resulted from SSA compared with 35% (34 of 96) from NHEJ, demonstrating the predominance of SSA when repeats are on opposite sides of the breakpoints, even though one repeat is distant from the I-SceI site. This result suggests that the paucity of oncogenic translocation junctions within repetitive elements is not due to the distance of repetitive elements from breakpoints, but rather to other factors such as sequence divergence, which has recently been shown to substantially reduce Alu-Alu recombination events.9 It remains to be tested whether sequence repeats can influence translocation junction formation when both copies of the repeat are on the same side of the breakpoint, as has been proposed for some translocations.18
Translocation versus intrachromosomal junctions
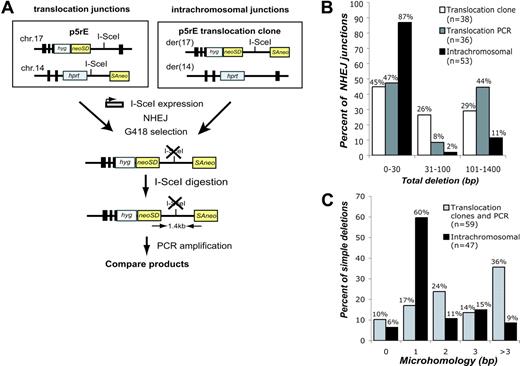
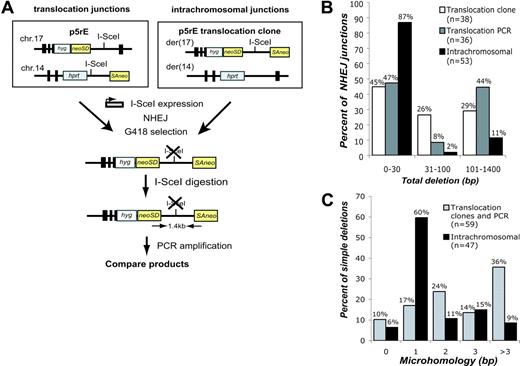
Using similar systems for DSB formation with I-SceI, we estimate that translocations occur approximately 103-fold less frequently in our system than NHEJ involving a single DSB.19,20 Translocations and intrachromosomal repair could occur via the same pathways, or they may arise via distinct pathways. To address this, we directly compared translocation and intrachromosomal junctions derived from DNA ends of the same sequence. To obtain translocation junctions, we expressed I-SceI in p5rE cells, while for intrachromosomal repair, we expressed I-SceI in p5rE translocation clones that had re-established the I-SceI site on der(17) (Figure 3A). For both, PCR was used to isolate breakpoint junctions that had lost the I-SceI site from pooled neo+ cells. Deletion lengths and microhomology obtained with this strategy for translocations did not differ significantly from translocation junctions isolated from p5rE neo+ clones (Figure 3B). The spectrum of translocation junctions differed, however, from the intrachromosomal repair junctions (Figure S2). Overall, only 13% of intrachromosomal repair junctions had deletions more than 30 bp, versus 53% of translocation junctions (P < .001; Figure 3B), and 35% of intrachromosomal junctions had more than 1 bp of microhomology compared to 74% of translocation junctions (P < .001; Figure 3C). Longer deletions and increased microhomology are characteristic of repair junctions from NHEJ-deficient cells (Ku70-/-, Ku80-/-, DNA-PKcs-/-, XRCC4-/-, Lig4-/-),21,22 raising the possibility that translocations occur through NHEJ pathways that are independent of one or more of these factors.
Comparison of translocation and intrachromosomal breakpoint junctions. (A) Strategy to compare translocations and intrachromosomal repair at the same sequence. For translocations, I-SceI was expressed in the p5rE cell line, and for intrachromosomal repair, I-SceI was expressed in 2 p5rE translocation clones that re-established the I-SceI site on der(17) but not on der(14). After transfection, neo+ cells were selected in G418, and genomic DNA was digested with I-SceI prior to PCR amplification with the indicated primers. PCR products were cloned and sequenced. (B) Comparison of deletion lengths at translocation and intrachromosomal breakpoint junctions. The 53 intrachromosomal and 36 of the translocation junctions from p5rE der(17) were isolated by the PCR strategy shown in panel A; 38 translocation junctions from individually isolated p5rE neo+ clones that had not re-established the I-SceI site also are shown for der(17). (C) Comparison of microhomology distributions for translocation and intrachromosomal breakpoint junctions. The translocation junctions are compiled from the PCR strategy shown in panel A and neo+ clonal analysis for p5rE der(17). Only junctions containing simple deletions are included.
Comparison of translocation and intrachromosomal breakpoint junctions. (A) Strategy to compare translocations and intrachromosomal repair at the same sequence. For translocations, I-SceI was expressed in the p5rE cell line, and for intrachromosomal repair, I-SceI was expressed in 2 p5rE translocation clones that re-established the I-SceI site on der(17) but not on der(14). After transfection, neo+ cells were selected in G418, and genomic DNA was digested with I-SceI prior to PCR amplification with the indicated primers. PCR products were cloned and sequenced. (B) Comparison of deletion lengths at translocation and intrachromosomal breakpoint junctions. The 53 intrachromosomal and 36 of the translocation junctions from p5rE der(17) were isolated by the PCR strategy shown in panel A; 38 translocation junctions from individually isolated p5rE neo+ clones that had not re-established the I-SceI site also are shown for der(17). (C) Comparison of microhomology distributions for translocation and intrachromosomal breakpoint junctions. The translocation junctions are compiled from the PCR strategy shown in panel A and neo+ clonal analysis for p5rE der(17). Only junctions containing simple deletions are included.
We have demonstrated that reciprocal translocations between nonhomologous sequences can be recovered in repair-proficient murine cells and that breakpoint junctions recapitulate those found in oncogenic translocations, with frequent deletions and microhomology, as well as insertions and more complex events. Moreover, in a direct comparison, we find that the spectrum of junctions for translocations differs from those derived from repair of a single DSB. Our reporters provide a system for determining the genetic requirements for translocation formation and identifying lineage-specific repair mechanisms in hematopoietic and mesenchymal cells.
Prepublished online as Blood First Edition Paper, September 29, 2005; DOI 10.1182/blood-2005-06-2437.
Supported in part by the Clinical Scholars Biomedical Research Training Program (National Institutes of Health [NIH] CA09512; D.M.W.); the Leukemia and Lymphoma Society (5415-05; D.M.W.); the Dorothy Rodbell Cohen Foundation (B.E.); and NIH GM54688 (M.J.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Margaret Leversha at the MSKCC Molecular Cytogenetics Core Facility for performing the FISH analysis, and members of the Jasin laboratory, especially Felipe Araujo and Jeremy Stark, for helpful discussions.