Abstract
Neoplastic transformation of mature B cells can be triggered by class-switch recombination of the immunoglobulin gene, which aberrantly targets a protooncogene and promotes translocation. Class-switch recombination is initiated by the B-cell-specific protein activation-induced cytidine deaminase (AID). Using immunohistochemistry with a newly generated monoclonal antibody and quantitative reverse-transcription-polymerase chain reaction (RT-PCR) on microdissected tissue from lymph node, tonsil, and thymus, we demonstrate that AID expression is found in secondary lymphoid organs outside germinal centers and in the thymic medulla at substantial levels. This is accompanied by the presence of circle transcripts, indicating class-switch recombination to be active at these sites. The dominant AID-expressing cell population outside germinal centers displays cytomorphologic properties corresponding to those that define the recently characterized interfollicular large B-cell subset. These findings indicate that interfollicular large B cells and AID-expressing B lymphocytes of the thymic medulla could give rise to mature B-cell malignancies. (Blood. 2006;107:2470-2473)
Introduction
The maturation of a B-cell reaction is achieved by somatic hypermutation (SHM) and class-switch recombination (CSR) of the immunoglobulin gene. Both processes are triggered by activation-induced cytidine deaminase (AID), which promotes the generation of transient DNA double-strand breaks in CSR.1 The vast majority of switching events is carried out faultlessly and facilitates an optimized effector function of a B-cell response. Rarely, however, improper CSR can lead to chromosomal translocations and to the activation of proto-oncogenes.1 Molecular analysis of chromosomal breakpoints revealed that in a variety of B-cell neoplasms, translocations have been caused by aberrant CSR.2 Since CSR critically depends on AID, identifying and characterizing AID-expressing B cells may contribute to the understanding of malignant lymphoma. It is well recognized that centroblasts of the germinal center produce high levels of AID,3 but little knowledge exists on the phenotype and tissue distribution of B cells expressing AID at extrafollicular sites. Here, we demonstrate that outside the germinal center, a major fraction ofAID+ B cells resides in the recently identified subset of interfollicular large B lymphocytes.4
Study design
Generation of an AID-specific monoclonal antibody
BALB/c mice were repeatedly immunized with a C-terminal AID peptide (sequence VDDLRDAFRTLGL) coupled to keyhole limpet hemocyanin. Spleen cells were fused with the mouse myeloma cell line X63-Ag8.653 using standard procedures. Anti-AID hybridoma C12.38 (IgG1) was identified by enzyme-linked immunosorbent assay (ELISA) and cloned twice by limiting dilution. Monoclonal antibody (mAb) was purified by affinity chromatography.
Western blotting
Cell lysates were separated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis under reducing conditions and transferred onto an Immobilon-P membrane (Millipore, Bedford, MA). Bound mAb was labeled with alkaline phosphatase-conjugated goat anti-mouse IgG/Fc (Jackson ImmunoResearch, West Grove, PA) and visualized by NBT-BCIP (Roche, Mannheim, Germany). Mouse mAbs recognizing Simian virus 40 (SV40) T-antigen and beta-actin served as controls.
Tissue and cell lines
Specimens from reactive tonsil, lymph node, and thymus were drawn from our archive of snap-frozen tissues. This material was pseudonymized to comply with the German law for correct usage of archival tissue for clinical research (Deutsches Ärzteblatt 2003; 100 A1632). Approval for these studies was obtained from the University of Ulm ethics board. Tumor cell lines (Nalm6, Reh, P3HR1, Raji, HSB-2, Jurkat) were obtained from ATCC (Manassas, VA). LMH cells were provided by Dr H.-J. Schlicht, Ulm, Germany.
Transfection of LMH cells
The coding region of human AID was amplified by polymerase chain reaction (PCR) from tonsillar cDNA and inserted in-frame into the pCI/cT1-272 vector,5 encoding an hsp73-binding SV40 T-antigen fragment. LMH cells were transiently transfected with the cT272-AID fusion construct (AID/T-antigen) or a mock vector applying the Ca2PO4 method.6
Immunomorphology
Fixed cytospin preparations and cryosections were incubated for 1 hour with anti-AID or CD20 mAb clone L26 (Dako, Hamburg, Germany), respectively. Bound mAb was labeled using the EnVision system (Dako) and visualized by 3-amino-9-ethylcarbazole. Picture acquisition was performed applying an Axiophot microscope (Zeiss, Jena, Germany) equipped with a KY-F75U digital camera (JVC, Friedberg, Germany) and DISKUS software version 4.5 (C.H. Hilgers Technisches Büro, Königswinter, Germany). Images were processed using Adobe Photoshop software version 5.5 (Adobe, Unterschleißheim, Germany).
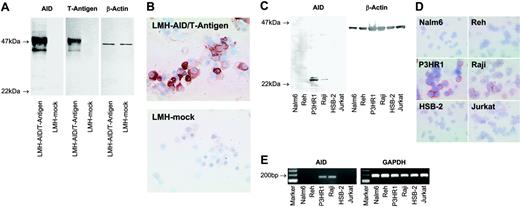
The monoclonal antibody C12.38 specifically recognizes AID. (A) Western blots of lysates from LMH cells transfected with a vector containing the coding sequence of AID and the SV40 T-antigen (LMH-AID/T-antigen) or mock vector (LMH-mock) were probed with mAb recognizing AID (clone C12.38), T-antigen, or β-actin as a loading control. Antibodies against AID and T-antigen both label a 47-kDa AID/T-antigen fusion protein in AID/T-antigen-transfected LMH cells, while mock-transfected cells are not reactive. (B) Cytospins of LMH cells transfected with AID/T-antigen, but not with mock vector stain with anti-AID. A nuclear staining detected in some LMH cells was probably nonspecific, since it was occasionally observed in tissues unlikely to express AID, such as epithelium. (C) Anti-AID mAb detects a 24-kDa band in Western blots from lysates of the human Burkitt lymphoma cell lines P3HR1and Raji, whereas Nalm6, Reh, HSB-2, and Jurkat lysates yield no signal. β-Actin probing was used as a loading control. (D) In cytospin preparations, P3HR1 and Raji cells stain with anti-AID. No labeling is seen with Nalm6, Reh, HSB-2, and Jurkat. (E) RT-PCR reveals AID expression in P3HR1 and Raji cells, but not in Nalm6, Reh, HSB-2, or Jurkat. All cell populations are strongly positive for the housekeeping gene GAPDH. Magnification: 40×/0.75 NA objective (B,D).
The monoclonal antibody C12.38 specifically recognizes AID. (A) Western blots of lysates from LMH cells transfected with a vector containing the coding sequence of AID and the SV40 T-antigen (LMH-AID/T-antigen) or mock vector (LMH-mock) were probed with mAb recognizing AID (clone C12.38), T-antigen, or β-actin as a loading control. Antibodies against AID and T-antigen both label a 47-kDa AID/T-antigen fusion protein in AID/T-antigen-transfected LMH cells, while mock-transfected cells are not reactive. (B) Cytospins of LMH cells transfected with AID/T-antigen, but not with mock vector stain with anti-AID. A nuclear staining detected in some LMH cells was probably nonspecific, since it was occasionally observed in tissues unlikely to express AID, such as epithelium. (C) Anti-AID mAb detects a 24-kDa band in Western blots from lysates of the human Burkitt lymphoma cell lines P3HR1and Raji, whereas Nalm6, Reh, HSB-2, and Jurkat lysates yield no signal. β-Actin probing was used as a loading control. (D) In cytospin preparations, P3HR1 and Raji cells stain with anti-AID. No labeling is seen with Nalm6, Reh, HSB-2, and Jurkat. (E) RT-PCR reveals AID expression in P3HR1 and Raji cells, but not in Nalm6, Reh, HSB-2, or Jurkat. All cell populations are strongly positive for the housekeeping gene GAPDH. Magnification: 40×/0.75 NA objective (B,D).
Double fluorescent immunostaining
Cryosections were incubated with anti-AID and labeled by the EnVision system, followed by signal amplification using Alexa Fluor 546 tyramid (Molecular Probes, Göttingen, Germany). Next, sections were consecutively incubated with CD20, biotin-labeled anti-mouse IgG2a mAb clone R19-5 (BD Biosciences, Heidelberg, Germany), and Alexa Fluor 488-conjugated streptavidin (Molecular Probes). Nuclear counterstaining was performed with DAPI. Slides examined under an Axioskop fluorescence microscope (Zeiss) as described7 using a mercury-vapor light source and an appropriate filter-set. Images were recorded by a C5985 CCD camera (Hamamatsu, Herrsching, Germany) with ISIS3 version 3.02 (Metasystems, Heidelberg, Germany) as an acquisition software, and were processed using Adobe Photoshop.
Microdissection, mRNA isolation, reverse transcription
Microdissection and laser pressure catapulting was performed on hematoxylin-stained frozen tissue sections as described.8 RNA was isolated from microdissected tissue and pelleted cells9 using the PicoPure RNA isolation kit (Arcturus, Mountain View, CA) and Trizol reagent (Invitrogen, Karlsruhe, Germany), respectively, and reverse transcribed as depicted elsewhere.9
Real-time PCR
AID mRNA expression was quantified by real-time reverse-transcription (RT)-PCR9 using GAPDH as a reference gene. Primer sequences were as follows: AIDleft, TGATGAACCGGAGGAAGTTT; AIDright, AGCCGTTCTTATTGCGAAGA; GAPDHleft, GCCAAAAGGGTCATCATCTC; GAPDHright, TGTGGTCATGAGTCCTTCCA.
Detection of circle transcripts
Circle transcripts (CTs) were amplified in a nested RT-PCR with the following primers: Iγ, AGGACACACCAGAGGCTGAC; Cμ, TCTCAGGACTGATGGGAAGC; Iα, CTCTTGGCAGGCAGCCAG; Cγ, CGCTGCTGAGGGAGTAGAGT; Iγint, TGGGAGCA/GC/TGAGGAACAT; Cμint, GAAGCCCCGGGTACTGCT; Iαint, AGGGTGGACCTGCCATGA; Cγint, AGTTCCACGACACCGTCAC. To amplify Iγ-Cμ CT, Iγ plus Cμ (1st round) and Iγint plus Cμint (2nd round) were used. To amplify Iα-Cμ CT, Iα plus Cμ (1st round) and Iαint plus Cμint (2nd round) were used. To amplify Iα-Cγ CT, Iα plus Cγ (1st round) and Iαint plus Cγint (2nd round) were used.
Results and discussion
To identify AID expression in situ on a cellular level, we generated an AID-specific mAb (clone C12.38). In Western blots of lysates from LMH cells overexpressing AID/T-antigen, C12.38 and a mAb against T-antigen labeled the 47-kDa fusion protein, while neither reagent stained control cells transfected with a mock vector (Figure 1A). C12.38 strongly labeled cytospin preparations from AID/T-antigen-expressing but not mock-transfected LMH cells (Figure 1B). Immunoblotting of P3HR1 and Raji Burkitt lymphoma cells with C12.38 yielded a 24-kDa band (Figure 1C), corresponding to the molecular size of AID. No specific band was observed in the immature B-cell lines Nalm6 and Reh, and the T-cell leukemia lines HSB-2 and Jurkat. A similar staining pattern was found by immunocytochemistry (Figure 1D). RT-PCR (Figure 1E) confirmed the AID expression status of the cell lines. Taken together, these results show that C12.38 specifically binds human AID.
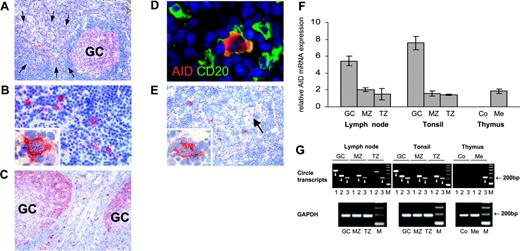
AID expression and class-switch recombination in secondary lymphoid organs and the thymus. AID expression was detected by conventional immunohistochemistry (A-C,E) and, in conjunction with CD20, by double immunofluorescent staining (D). AID is expressed in lymphoid follicles of the lymph node (A) and tonsil (C). In fully developed germinal centers (GCs), AID expression is predominately observed in the dark zone (C). Outside germinal centers, AID+ cells are found scattered throughout the T zone (A,C). The arrows in A indicate an extrafollicular area containing numerous AID+ cells. These AID-expressing cells are large and often display a distinct dendritic cytomorphology (B, and inset in B). Double staining of AID (red) and CD20 (green) shows coexpression of CD20 by AID+ cells (D). AID staining is largely limited to the cytoplasm (inset in B,D). Low numbers of AID+ cells are present in the thymic medulla (E) containing a Hassall corpuscle (arrow). AID+ asteroid cells of the thymic medulla morphologically resemble those in the T zone of the lymph node and tonsil (inset in E). (F) Quantification of AID mRNA by real-time PCR on microdissected tissue shows significant levels of AID expression in the germinal center, mantle zone (MZ), and T zone (TZ) of secondary lymphoid organs, and in the medulla (Me) of the thymus, but not in the thymic cortex (Co). Relative AID mRNA expression is given in arbitrary units as the mean (n = 3) and standard deviation. (G) Iγ-Cμ (1), Iα-Cμ (2), and Iα-Cγ (3) circle transcripts and GAPDH as a housekeeping gene were amplified by nested RT-PCR from microdissected tissue. Circle transcripts are detected in all 3 microcompartments of the lymph node and tonsil, and in the thymic medulla. No circle transcripts can be amplified from the cortical region of the thymus. Specific bands are indicated by arrowheads. The specificity of representative PCR products was confirmed by sequencing (GenBank accession nos. DQ083944-6). All data represent at least 2 independent stainings or measurements. Magnification: 10×/0.3 NA objective (A,C); 40×/0.75 NA objective (B); 63×/1.4 NA objective (D, and panels B and E's insets); and 20×/0.5 NA objective (E).
AID expression and class-switch recombination in secondary lymphoid organs and the thymus. AID expression was detected by conventional immunohistochemistry (A-C,E) and, in conjunction with CD20, by double immunofluorescent staining (D). AID is expressed in lymphoid follicles of the lymph node (A) and tonsil (C). In fully developed germinal centers (GCs), AID expression is predominately observed in the dark zone (C). Outside germinal centers, AID+ cells are found scattered throughout the T zone (A,C). The arrows in A indicate an extrafollicular area containing numerous AID+ cells. These AID-expressing cells are large and often display a distinct dendritic cytomorphology (B, and inset in B). Double staining of AID (red) and CD20 (green) shows coexpression of CD20 by AID+ cells (D). AID staining is largely limited to the cytoplasm (inset in B,D). Low numbers of AID+ cells are present in the thymic medulla (E) containing a Hassall corpuscle (arrow). AID+ asteroid cells of the thymic medulla morphologically resemble those in the T zone of the lymph node and tonsil (inset in E). (F) Quantification of AID mRNA by real-time PCR on microdissected tissue shows significant levels of AID expression in the germinal center, mantle zone (MZ), and T zone (TZ) of secondary lymphoid organs, and in the medulla (Me) of the thymus, but not in the thymic cortex (Co). Relative AID mRNA expression is given in arbitrary units as the mean (n = 3) and standard deviation. (G) Iγ-Cμ (1), Iα-Cμ (2), and Iα-Cγ (3) circle transcripts and GAPDH as a housekeeping gene were amplified by nested RT-PCR from microdissected tissue. Circle transcripts are detected in all 3 microcompartments of the lymph node and tonsil, and in the thymic medulla. No circle transcripts can be amplified from the cortical region of the thymus. Specific bands are indicated by arrowheads. The specificity of representative PCR products was confirmed by sequencing (GenBank accession nos. DQ083944-6). All data represent at least 2 independent stainings or measurements. Magnification: 10×/0.3 NA objective (A,C); 40×/0.75 NA objective (B); 63×/1.4 NA objective (D, and panels B and E's insets); and 20×/0.5 NA objective (E).
Immunohistochemical staining of lymph node (Figure 2A-B) and tonsil (Figure 2C) revealed cytoplasmic AID protein expression in the germinal center of lymphoid follicles. In fully developed germinal centers, AID staining was predominately seen in the dark zone (Figure 2C), consistent with a recent report.10 Outside the germinal center, intensely AID+ cells were scattered throughout the T zone and within the mantle zone (Figure 2A,C). These AID+ cells coexpressed CD20, documenting their B-cell nature (Figure 2D). Extrafollicular AID+ cells were medium sized to large with centrally located nuclei and one or few nucleoli (Figure 2B,D). Frequently, these AID+ B cells formed prominent cytoplasmic extensions, lending them a dendritic cell-like appearance. Immunostaining of adjacent serial sections revealed that approximately 10% to 20% of large CD20+ B cells located in the T zone expressed AID (data not shown). AID+ cells with asteroid morphology were also observed at low numbers in the thymic medulla (Figure 2E) but not in the cortex. The immunohistochemical AID expression pattern was confirmed by quantitative RT-PCR on microdissected tissue (Figure 2F). In the lymph node and tonsil, AID transcripts were most abundant in the germinal center, yet significant expression levels were also found in the mantle zone and in the T zone. In addition, AID mRNA was detected in the thymic medulla, while practically no expression was present in the cortex. AID mRNA expression in the medullary thymus was higher than to be expected in view of the scarce AID+ cells observed by immunostaining. A possible explanation could be a low AID expression by thymic B cells at a level below the detection threshold of immunohistochemistry.
Since AID requires posttranslational modification and interaction with cofactors to become functional,11-13 we determined whether the presence of AID+ cells was accompanied by CSR at extrafollicular sites. Circle transcripts (CTs) are generated during isotype switch recombination from looped-out circular DNA located in between 2 activated switch regions14 and become rapidly degraded. Therefore, their presence indicates recent CSR.15,16 Using nested RT-PCR, we detected Iγ-Cμ, Iα-Cμ, and Iα-Cγ CT (corresponding to IgM→IgG, IgM→IgA, and IgG→IgA switching, respectively) in the germinal centers of the lymph node and tonsil (Figure 2G). CTs were also found in the follicular mantle and the T zone. In the thymus, CTs were detected in the medulla, whereas no CTs were found in the cortex. AID activity thus correlates with gene expression at all microcompartments investigated.
To summarize, we identify a population of strongly AID-expressing B lymphocytes that are located outside the germinal center of secondary lymphoid organs and in the thymic medulla where CSR is active. It is likely that a significant number of extrafollicular AID+ cells comprise B lymphocytes recruited into an extrafollicular antibody response, bypassing the germinal center.17 Some of these cells may also represent B lymphocytes emigrated from the follicle upon binding cognate antigen to receive T-cell help at the B/T boundary.18 The cytomorphology and the distribution pattern of extrafollicular AID+ cells meet the characteristics recently described to define interfollicular large B cells.4 This distinct B lymphocyte subset has been proposed as a nonneoplastic precursor of B-cell malignancies, a notion supported by the data presented here. Clearly, a more detailed characterization of AID+ interfollicular large B cells will be necessary for relating this subset to specific lymphoma entities. Possible candidates might be found in the heterogenous group of diffuse large B-cell lymphoma,4 some of which harbor translocations generated by erroneous CSR.19,20
Prepublished online as Blood First Edition Paper, November 3, 2005; DOI 10.1182/blood-2005-06-2502.
Supported by the Forschungsschwerpunktprogramm des Landes Baden-Württemberg “Mechanismen der Transformation lymphohämatopoietischer Zellen” to P.M. and F.L., and the Deutsche Forschunsgemeinschaft (DFG SMi 505/2-2) to R.S.
G.M. designed research, performed research, and wrote the paper; S.W.P. performed research and analyzed data; B.W. performed research; S.B. performed research; P.R. performed research and wrote the paper; N.F. perfomed research; R.S. contributed vital new reagents; P.M. wrote the paper; O.R. performed research; and F.L. designed research, analyzed data, and wrote the paper.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Our thanks to Elvira Hallauer and Simone Westenfelder for their superb technical assistance.