Abstract
Relapse within the central nervous system (CNS) remains a major challenge for the treatment and cure of childhood ALL. With current intensive treatment, at least one-third of relapses occur within the CNS. Evaluation of CSF is essential to the management of childhood ALL and relies on morphologic examination of cytospin smears. CSF has been re-discovered in the post genomic era as a source of potential protein biomarkers for various diseases. Such proteins may help establish a diagnosis, provide insight into pathogenesis, or identify therapeutic toxicity. In the present study, acellular CSF supernatants from newly diagnosed children with standard risk ALL treated on CCG-1991 were used for protein profiling.
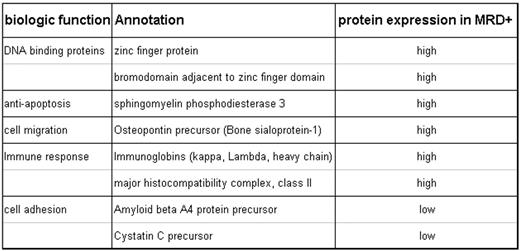
The morphologically negative CSF samples had previously been classified as minimal residual disease (MRD) negative (−) or positive (+) using real-time PCR techniques on centrifuged cell lysates. Three MRD −, 3 MRD +, and 2 morphologically + CSF samples were concentrated and desalted using Microcon filter device. Albumin was removed using a Human Albumin Kit, Albuminomics™. The MRD − and MRD + samples, each containing 15 mcg of protein, were pooled separately and processed for protein identification and quantitation using a multidimensional LC/MS/MS method. Custom software was developed (TandTRAQ) for database searching and peptide quantitation. Protein annotation was primarily derived from ENSEMBL and NCBI sources. Rank order summarization was used to identify putative candidates. In the comparison between MRD + and MRD− samples, we identified 226 unique genome identifiers, with 89 having functional annotation. Of these 89, 49 have high quality protein-level annotation, 35 have mRNA-level annotation, and 5 are predicted (have functional protein evidence). Using a 1.5 fold cut-off, there were 18 up-regulated and 12 down-regulated proteins among the 89 proteins in MRD + compared to MRD − supernatants. Focusing on the 49 with high quality protein-level annotation, 8 proteins show higher-level expression and 2 show lower level expression in MRD + CSF samples (Table). Eight highly expressed proteins are involved in either DNA binding interactions, cell migration, apoptosis, or immune response. Two under-expressed proteins are involved in cell adhesion. The 2 morphologically + CSF samples confirmed 4 over expressed and 1 under-expressed proteins.
This research is the first attempt at protein profiling of CSF from patients with ALL. The proteins identified in this study may further our insight into the pathogenesis of CNS leukemia. Furthermore, future research may find that protein profiling of supernatants is a sensitive way to detect leukemia in CSF and to monitor treatment-related CNS toxicities.
Disclosure: No relevant conflicts of interest to declare.
Author notes
Corresponding author