Abstract
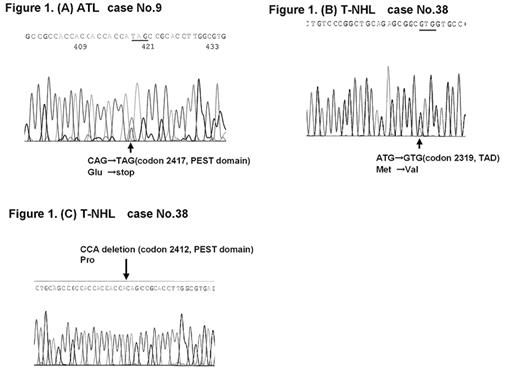
A somatic mutation of the NOTCH1 gene is frequently observed in T-cell acute lymphoblastic leukemia (T-ALL) of immature T-cell leukemia, but its prevalence in mature T-cell leukemia/lymphoma remains unknown. T-cell non-Hodgkin lymphomas (T-NHL) are relatively uncommon malignancies that represent approximately 12% of all lymphomas. The current WHO/EORTC classification recognizes 9 distinct clinicopathologic peripheral T-NHLs. The identification of genetic abnormalities and chromosomal rearrangements has been helpful in recognizing and defining lymphoma entities according to the WHO classification. However, the molecular changes that could play a role in leukemogenesis and progression remain unknown. NOTCH1 gene mutation was analyzed in 53 mature T-cell leukemia/lymphoma in adults, comprising 21 with adult T-cell leukemia (ATL), 25 with T-cell non-Hodgkin lymphoma (T-NHL), seven with T-cell prolymphocytic leukemia (T-PLL). We analyzed the hotspots of the NOTCH1 activating mutation in T-ALL cases located in exons 26 and 27, which encodes the N-terminal and C-terminal region of the heterodimerization (HD-N, HD-C) domain, and in exon 34, which encodes the PEST domain and terminal activation domain (TAD). Mutation detection in these two locations was performed by means of PCR-based denaturing high-performance liquid chromatography (dHPLC) using a WAVE DNA fragment analysis system followed by sequencing. We detected a nonsense mutation, C7249T, resulting in Q2417X, in an ATL patient, and a missense mutation, A6955G, resulting in M2319V, as well as a 3-bp deletion between 7234 and 7236 that resulted in deletion of the proline at codon 2412 in the same allele in a T-NHL, peripheral T-cell lymphoma, unspecified (PTCL-u), patient. NOTCH1 mutation was infrequent in mature T-cell leukemia/lymphoma, but NOTCH1 may be involved in leukemogenesis associated with a variety of T-cell leukemia/lymphoma rather than only with T-ALL.
Disclosure: No relevant conflicts of interest to declare.
Author notes
Corresponding author