Abstract
The expression of tumor-associated antigens (TAAs) might play a critical role in the control of minimal residual disease (MRD) in acute myeloid leukemia (AML), and therefore might be associated with clinical outcome in AML. In a DNA microarray analysis of 116 AML samples, we found a significant correlation between high mRNA levels of G250/CA9 and longer overall survival (P = .022), a similar trend with high mRNA levels of PRAME (P = .103), and a hint for RHAMM/HMMR. In contrast, for other TAAs like WT1, TERT, PRTN3, BCL2, and LAMR1, we found no correlation with clinical outcome. High expression of at least 1 of the 3 TAAs, RHAMM/HMMR, PRAME, or G250/CA9, provided the strongest favorable prognostic effect (P = .005). Specific T-cell responses were detected in 8 (47%) of 17 patients with AML in complete remission for RHAMM/HMMR-R3 peptide, in 7 (70%) of 10 for PRAME-P3 peptide, and in 6 (60%) of 10 for newly characterized G250/CA9-G2 peptide, a significant increased immune response compared with patients with AML patients who had refractory disease (P < .001). Furthermore, we could demonstrate specific lysis of T2 cells presenting these epitope peptides. In conclusion, expression of the TAAs RHAMM/HMMR, PRAME, and G250/CA9 can induce strong antileukemic immune responses, possibly enabling MRD control. Thus, these TAAs represent interesting targets for polyvalent immunotherapeutic approaches in AML.
Introduction
Tumor-associated antigens (TAAs) have been shown to induce specific T-cell immune responses in tumor patients, and thus represent potential target structures for specific immunotherapies.1-4 Recently, a large number of TAAs inducing specific T-cell responses have been identified and functionally characterized in different solid tumors, but several TAAs also seem to be expressed in hematologic malignancies like acute myeloid leukemia (AML).5,6
AML is the most common acute leukemia in adults. With intensive induction therapy, complete remission (CR) rates between 65% and 75% are achieved in younger patients (< 60 years). However, more than 50% of these patients will relapse, leading to an overall survival rate of only 30% to 40% after 5 years. Results are more unfavorable in patients older than 60 years.7,8 Immunotherapeutic approaches targeting TAAs to prevent relapse may represent a promising novel treatment option to improve the outcome of AML patients.
Recently, in patients with AML, specific T-cell responses of cytotoxic T-lymphocytes (CTLs) were detected against the Wilms tumor gene WT1, proteinase 3 (PRTN3), and the receptor for hyaluronic acid–mediated motility (RHAMM; also known as CD168 or HMMR).9-15 For these TAAs, clinical peptide vaccination trials have been initiated including patients with AML.9,16,17 Furthermore, additional TAAs known to induce specific immune responses of CD8+ T cells, such as PRAME, G250/CA9, BCL2, LAMR1, and hTERT, are also frequently expressed in AML.18-21
While immunotherapy eliciting a specific T-cell response to TAAs expressed by leukemic blasts constitutes an interesting option to enhance specific antileukemic effects following chemotherapy or stem cell transplantation, the clinical relevance of TAA expression in the course of AML is largely unknown. The expression of certain TAAs might play an important role in the antileukemic activity of the patient's immune system, by killing residual tumor cells named minimal residual disease (MRD). Therefore, expression of TAAs might be associated with outcome of patients with AML.
Today, novel approaches in genomics, such as DNA microarray technology, have made feasible the simultaneous measurement of thousands of transcripts in a given sample, thereby opening new potential avenues for deciphering transcriptional deregulation in tumorigenesis. Microarray technology has already contributed significantly to our understanding of hematologic malignancies,22-24 in particular providing new insights in the molecular pathology of AML.25-27 In addition, the generated wealth of data provides opportunities for explorative data mining, and for substantiating hypothesis-driven leukemia research.
In the present study, we investigated the influence of the expression levels of leukemia relevant TAAs on the clinical outcome of patients with AML by reanalyzing a previously published large gene expression dataset.25 Interestingly, high expression of the TAAs G250/CA9, RHAMM/HMMR, and PRAME, which we found to be associated with favorable clinical outcome in patients with AML, also induced strong T-cell responses, and therefore represent promising new targets for future monovalent or polyvalent immunotherapeutic approaches.
Materials and methods
Samples
Peripheral blood and bone marrow samples from adult patients with AML were provided by the German-Austrian AML Study Group (AMLSG) with patient informed consent and institutional review board approval from all participating centers. Approval was obtained from the University of Ulm's institutional review board for the reported studies. For cDNA microarray analysis, peripheral blood (n = 53) and bone marrow samples (n = 63) were collected at the time of diagnosis as previously described.25 For functional experiments, peripheral blood samples were collected from patients with AML in CR (n = 20) and from patients with AML who had refractory disease (n = 10). For the antigens G250/CA9 and PRAME, 10 samples from patients with AML were tested, and for the antigen RHAMM/HMMR, 20 samples from patients with AML in CR were examined, 10 in addition to the previously reported cases.9 Furthermore, for all 3 antigens, 10 samples from patients with AML who had refractory disease were analyzed. Peripheral blood sample controls were obtained from 10 healthy volunteers.
Sample preparation
Mononuclear cells (MNCs) were prepared by Ficoll density gradient centrifugation and stored at –80°C for RNA preparation. For the cellular assays, Ficoll-separated MNCs were resuspended with fetal calf serum (FCS) containing 10% DMSO, stored in liquid nitrogen, and thawed for immunologic analyses.
Culture of cell lines
The human cell lines K-562, HL-60, KASUMI-1, KG-1, OCI-AML5, and COS-7 were obtained from the German Collection of Microorganisms and Cell Cultures in Braunschweig, Germany. Cell lines were cultured under standard conditions in RPMI 1640 (Biochrom, Berlin, Germany) containing 10% FCS, 2 mM L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin.
cDNA microarray–based gene expression analysis
To investigate the expression levels of relevant TAAs in adult AML we analyzed a large cDNA microarray–based gene expression dataset which was previously published,25 and is publicly available in the Gene Expression Omnibus database (accession number GSE425).28 Updated clinical follow-up data are listed in Table S1, available on the Blood website (see the Supplemental Table link at the top of the online article).
Fluorescence ratios of well-measured spots were normalized by mean-centering each gene across all arrays within each of 3 array print runs, to minimize potential print run–specific bias.25 For hierarchical clustering, we applied 2-way, average linkage hierarchical clustering29 and visualized results using TreeView version 1.60 (available at http://rana.lbl.gov/EisenSoftware.htm).29 For the correlation with survival data, expression values were dichotomized by the median expression of the respective gene across all AML samples. The terms “high” or “low” TAA expression refer to an expression greater and lower than the median expression across all AML samples, respectively.
Chi-square test, Fisher exact test, Pearson correlation, Student t test, analysis of variance (ANOVA), multivariate proportional hazards analysis, and Kaplan-Meier survival analysis were performed using Excel (Microsoft, Redmond, WA), WinStat (R. Fitch Software, Bad Krozingen, Germany), and the R software package (http://www.r-project.org).
Conventional RT-PCR
We isolated mRNA from stored, frozen MNC pellets by using the mRNA QuickPrep Micro purification kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) according to the manufacturer's recommendations, and assessed RNA quality by gel electrophoresis. mRNA (2.0 μg) was subjected to cDNA synthesis (Superscript II; Gibco BRL, Frederick, MD). Polymerase chain reaction (PCR) for RHAMM/HMMR was performed as previously described.19 For CA9, we used the following primers and conditions: (1) G250/CA9 PCR1: forward primer, 5′ CTA AGC AGC TCC ACA CCC TCT 3′; reverse primer, 5′ TCT CAT CTG CAC AAG GAA CG 3′; 95°C denaturation (1 minute), 60°C annealing (1 minute), 72°C elongation (1 minute), 35 cycles, 1.5 mM MgCl2; and (2) G250/CA9 PCR2: forward primer, 5′ ACT GCT GCT TCT GAT GCC TGT 3′; reverse primer, 5′ AGT TCT GGG AGC GGC GGG A 3′; 95°C denaturation (1 minute), 68°C annealing (2 minutes), 72°C elongation (1 minute), 35 cycles, 1.5 mM MgCl2. For PRAME, reverse transcriptase (RT)–PCR was performed as previously described.19
Real-time RT-PCR
The mRNA expression of RHAMM/HMMR and G250/CA9 was also quantified by real-time RT-PCR using the light cycler SYBR Green I technology according to the manufacturer's protocol (Roche Diagnostics, Mannheim, Germany).19 Expression analysis of RHAMM/HMMR and PRAME was performed as previously described.9,19 Real-time RT-PCR of G250/CA9 expression was performed using the primers of our conventional G250/CA9 PCR 1 approach (“Conventional RT-PCR”) with an initial denaturation at 95°C for 10 minutes, following 40 cycles with 10 seconds at 95°C, 15 seconds at 62°C, and 25 seconds at 72°C. For all quantitative RT-PCR reactions, 0.1 μg mRNA was used for copy number analysis. The amount of mRNA was normalized to the amount of 1 pg of the housekeeping TBP gene.
Peptides used for cellular experiments
To show specific T-cell responses against the antigen RHAMM/HMMR we used the RHAMM/HMMR-derived R3 peptide (ILSLELMKL) in all experiments as described previously.9 For the evaluation of specific T-cell responses against PRAME we tested 4 PRAME-specific peptides in healthy volunteers. In patients with AML, we then used the PRAME-derived P3 peptide (ALYVDSLFFL) and PRAME-derived P1 peptide (VLDGLDVLL), showing favorable results in our healthy volunteer experiments. G250/CA9-derived peptides with human leukocyte antigen (HLA)–A*0201–binding motifs were predicted using different computer algorithms (http://bimas.dcrt.nih.gov/molbio/hla_bind and http://www.uni-tuebingen.de/uni/kxi). Furthermore, peptides were checked for their cleavage pattern by the prediction algorithm for proteasomal cleavage (http://www.uni-tuebingen.de/uni/kxi). Appropriate results were found for the newly characterized G250/CA9-G2 peptide (QLLLSLLLL), which we finally used for the enzyme-linked immunospot (ELISPOT) assays and for specific lysis of G2-pulsed T2 cells (antigen-presenting cells [APCs]) in chromium 51 (Cr-51) release assays. In addition, we tested the other G250/CA9-G1 peptide (HLSTAFARV) that has been recently characterized in patients with renal cell carcinoma.30
Mixed lymphocyte peptide culture
MNCs from patients with AML and healthy volunteers were selected by magnetic beads through a magnetic-activated cell-sorting (MACS) column (Miltenyi Biotec, Bergisch Gladbach, Germany). CD8– APCs were irradiated with 30 Gy and pulsed for 2 hours with either a TAA-derived peptide or a control peptide at a concentration of 10 μg/mL. For all RHAMM/HMMR, G250/CA9, and PRAME peptides, as well as for the control peptides, the same conditions and reagents were used as described previously.9 Results from patients in CR were compared with results detected in samples from patients with AML who had refractory disease.
IFN-γ/granzyme B ELISPOT assays
Interferon-γ (IFN-γ) and granzyme B ELISPOT assays were performed as previously described.9 For IFN-γ ELISPOT assays we used 96-well nitrocellulose plates (Millipore, Schwalbach, Germany) coated with IFN-γ monoclonal antibodies (mAbs; Mabtech, Hamburg, Germany) which were incubated overnight at 4°C and then blocked with 10% human AB serum (German Red Cross, Ulm) for 2 hours at 37°C. Presensitized CD8+ T lymphocytes (1 × 104) and 4 × 104 target cells (peptide-pulsed peripheral blood mononuclear cells [PBMCs]) were added to each well. For the experiments testing different peptides we used a mixture of the peptides. Following overnight incubation in RPMI medium, plates were washed with 1 × phosphate-buffered saline (PBS; supplemented with 0.05% Tween-20), IFN-γ mAbs (0.2 μg/mL) added to each well, and then incubated at room temperature for 2 hours. After washing with streptavidin-alkaline phosphatase (1 μg/mL; Mabtech) for 2 hours, BCIP/NBT (Sigma-Aldrich, Munich, Germany) was used for colorization according to the manufacturer's instructions, and thereafter evaluated by the use of an ELISPOT reader (CTL; Reutlingen, Germany). In accordance, the granzyme B ELISPOT assay was performed according to the manufacturer's instructions (BD, San Diego, CA) as previously described.9 A “positive CTL” reaction was defined as at least 10 spots in case of no background, or more than 2 times the number of background spots in case of low background.
Cr-51 release cytotoxicity assay
T2 cells pulsed with 20 μM G250/CA9-, PRAME-, or RHAMM/HMMR-derived peptides and AML blasts were labeled with Cr-51 for 4 hours as described previously for the antigen RHAMM/HMMR.9 After washing, labeled cells were incubated with CD8+ T cells at effector-target ratios of 2.5:1 to 40:1 for 16 hours at 37°C. Radioactivity in the supernatant from all 96 wells was measured by a gamma counter (PerkinElmer, Boston, MA). The percentage of specific lysis was calculated as [Cr-51 release in the test well – spontaneous Cr-51 release] / [maximum Cr-51 release – spontaneous Cr-51 release].
Results
Expression of TAAs in adult AML
To investigate the expression of TAAs in adult AML, we first reanalyzed our previously published cDNA microarray–based gene expression dataset.25 Interestingly, all investigated genes considered to code for relevant TAAs in AML (TERT, WT1, PRTN3, BCL2, LAMR1, PRAME, RHAMM/HMMR, and G250/CA9) exhibited highly variable expression (ie, ≥ 4-fold from the mean in at least 2 samples) in our large sample set25 encompassing the major cytogenetic AML subgroups (data not shown). As some of the analyzed genes, including G250/CA9, showed low expression levels and had been filtered out in our previous data analysis,25 we performed both conventional and quantitative real-time RT-PCR to confirm our microarray findings for a set of TAAs: RHAMM/HMMR, PRAME, and G250/CA9. Expression of these TAAs was also examined in the leukemia cell lines K-562, HL-60, KASUMI-1, KG-1, and OCI-AML5. For example, K-562 served as a positive control for RHAMM/HMMR as RHAMM/HMMR mRNA expression was found in this cell line. In general, conventional semiquantitative RT-PCR showed a good correlation with microarray findings (data not shown).
In 17 AML samples analyzed by both microarrays and quantitative real-time RT-PCR we observed a good correlation of RHAMM/HMMR mRNA expression levels (Pearson correlation coefficient r = .76). Similar to the RHAMM/HMMR result, we also observed a good correlation for PRAME and G250/CA9 comparing cDNA microarray results with our real-time RT-PCR data (Pearson correlation coefficients r = 0.82 and 0.72, respectively).
Correlation with clinical findings
Correlations of our gene expression data with clinical (age, white blood cell [WBC] counts, blast counts, serum lactate dehydrogenase [LDH] levels, history of a preceding malignancy), cytogenetic (karyotype), and molecular-genetic (FLT3 and MLL aberrations31,32 ) findings are summarized in Table 1. In brief, for TERT and BCL2 we did not find any significant correlation with important clinical characteristics. Higher levels of RHAMM/HMMR and LAMR1 were associated with WBC counts of less than 30 g/L (P = .027 and P = .002, respectively; Student t test), while lower levels of LAMR1 and PRAME were correlated with a history of preceding malignancy (P = .047 and P = .020, respectively; Student t test). We also observed a significant association of PRAME, PRTN3, LAMR1, and G250/CA9 levels with distinct karyotypes (P < .001, P ≤ .001, P = .044, and P = .004, respectively; ANOVA). Cases with a t(8;21), del(7q)/–7, and t(15;17) showed higher levels of PRAME, while cases with complex karyotypes or inv(16) exhibited lower means of PRAME. t(8;21) and inv(16) were associated with higher relative levels of PRTN3, and t(9;11) and del(7q)/–7 with lower levels. On the other hand, for LAMR1 there was a correlation of del(7q)/–7 with higher expression. Interestingly, in G250/CA9, del(7q)/–7 was also associated with higher mRNA levels as were cases with t(9;11), while complex karyotypes and t(15;17) showed a correlation with lower mean G250/CA9 levels. Regarding well-known molecular markers, we only found a significant correlation of higher WT1 levels with FLT3 internal tandem duplications (ITDs; P = .046; Student t test).
Correlation with outcome
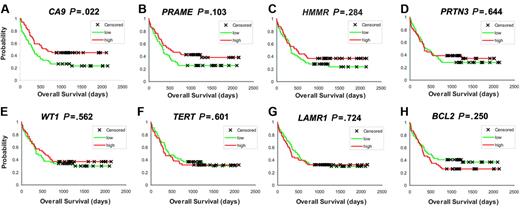
Correlating our findings with outcome data, we observed a statistically significant association of higher G250/CA9 expression with favorable overall survival (P = .022, log-rank test; expression values were dichotomized by the median; Figure 1A). Interestingly, in PRAME and RHAMM/HMMR, 2 additional genes coding for TAAs known to produce potent immune responses,9,20 we discovered similar effects. For PRAME, we saw a trend toward longer overall survival in cases with higher PRAME expression (P = .103; log-rank test), and the Kaplan-Meier plot for RHAMM/HMMR was suggestive for a similar effect (P = .284) (Figure 1B-C). For PRTN3 and WT1, the correlation of expression levels and clinical outcome has been controversially discussed recently.33,34 Our data did not show a correlation with clinical outcome (Figure 1D-E). While there was also no correlation of TERT or LAMR1 expression levels with outcome (Figure 1F-G), higher BCL2 levels suggested poorer overall survival although not statistically significantly (P = .250; log-rank test; Figure 1H).
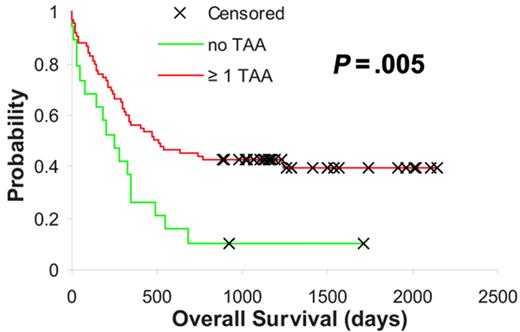
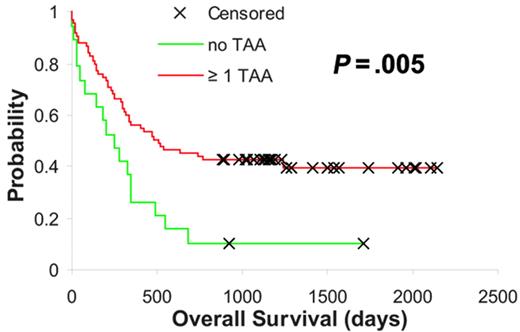
These observations denoted that the expression of certain tumor-specific TAAs was associated with a better outcome following intensive chemotherapy, possibly secondary to yet unknown immunologic mechanisms. Based on this hypothesis we were interested to determine whether the expression of at least 1 of the 3 TAAs in which higher expression correlated with or showed a trend toward better outcome (Figure 1A-C) had an additive influence with regard to improved survival in AML. Thus, we assigned our samples to groups depending on whether they exhibited expression levels of RHAMM/HMMR, PRAME, and G250/CA9 above the median for 0, 1, 2, or all 3 of these TAAs. Interestingly, we observed a statistically significant difference between cases showing “no” TAA expression (count = 0) and the group expressing at least 1 of the 3 TAAs (count ≥ 1), which displayed significantly longer overall survival times (P = .005, log-rank test; Figure 2). These TAA-defined groups (no vs ≥ 1 TAA) were independent of WBC, LDH, a history of secondary AML, cytogenetics, and molecular markers, but there was a significant association between ages older than 60 years and “no TAA” expression (P < .001, Chi-square test; Table 1). Similarly, in multivariate proportional hazards analysis higher expression levels of at least 1 of the 3 TAAs provided significant prognostic information independent of cytogenetics (hazard ratio, 0.546; confidence interval, 0.251-0.841; P = .04). However, in agreement with the univariate analyses the observed survival benefit was not independent of patient age.
TAA expression in AML determined by DNA microarray–based gene expression profiling. (A-H) Kaplan Meier analyses based on G250/CA9, PRAME, RHAMM/HMMR, WT1, TERT, PRTN3, BCL2, and LAMR1 expression levels (P values are indicated; expression values have been dichotomized by the median with higher expression levels color-coded in red and lower levels in green).
TAA expression in AML determined by DNA microarray–based gene expression profiling. (A-H) Kaplan Meier analyses based on G250/CA9, PRAME, RHAMM/HMMR, WT1, TERT, PRTN3, BCL2, and LAMR1 expression levels (P values are indicated; expression values have been dichotomized by the median with higher expression levels color-coded in red and lower levels in green).
AML subgroups based on RHAMM/HMMR, PRAME, and G250/CA9 expression. Kaplan-Meier analysis based on RHAMM/HMMR, PRAME, and G250/CA9 expression level–defined subgroups. Red indicates higher expression of at least 1 of the above-mentioned TAAs, and green indicates low expression levels of all 3 TAAs (P value is indicated).
AML subgroups based on RHAMM/HMMR, PRAME, and G250/CA9 expression. Kaplan-Meier analysis based on RHAMM/HMMR, PRAME, and G250/CA9 expression level–defined subgroups. Red indicates higher expression of at least 1 of the above-mentioned TAAs, and green indicates low expression levels of all 3 TAAs (P value is indicated).
Thereby, our findings support our aforementioned hypothesis, which would also predict stronger specific immune responses in cases with higher TAA expression. Therefore, in a next step we aimed to perform functional analyses to validate our hypothesis.
Specific cellular immune responses to RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides
To characterize the induction of specific T-cell responses in healthy volunteers and patients with AML we tested the RHAMM/HMMR-derived peptide R3, the PRAME-derived peptides P1 and P3, and the G250/CA9-derived peptides G1 and G2 (Table 2). ELISPOT assays for presensitized CD8+ T cells from the peripheral blood were performed for the release of IFN-γ and granzyme B after the presentation of the test peptide presented by T2 cells as described.9
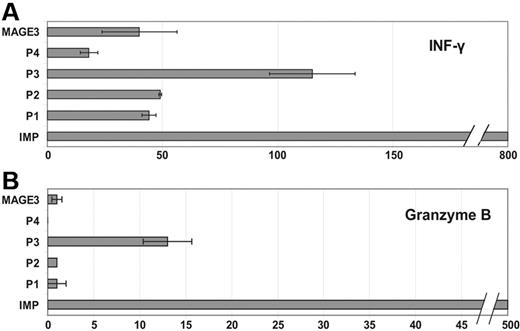
Figure 3 shows representative results for a IFN-γ (Figure 3A) and granzyme B (Figure 3B) release by CD8+ T cells collected from a patient with AML in CR who expressed RHAMM/HMMR, G250/CA9, and PRAME in the leukemic blasts at the time of diagnosis (analyzed by cDNA microarray technology and conventional RT-PCR; Figure 3C). According to the measured gene expression levels, high numbers of reactive CD8+ T cells were counted after stimulation with the RHAMM/HMMR-derived peptides R3 and the G250/CA9-derived peptide G2. Lower numbers of spots were detectable for the PRAME-derived peptide P3. In addition, a high granzyme B release was detected using the influenza matrix protein (IMP) peptide as positive control. As expected, no T-cell reaction was found for the negative control (mere T2 cells without peptide). Cross reactivity was excluded by presensitizing CD8+ T cells with autologous CD8– APCs pulsed with the R3 peptide and thereafter evaluating their granzyme B release upon presentation of an unrelated MAGE-3–derived peptide by T2 cells.
Previously, we had described specific T-cell responses against the RHAMM/HMMR-derived peptide R3, with 39% of AML patients showing CD8+-specific immune responses against the peptide R3.9 In contrast, in healthy volunteers specific immune responses were only found at low frequency for the RHAMM/HMMR-derived peptides.9 Here, we analyzed an additional 10 AML samples, of which 7 were evaluable for specific T-cell responses against the RHAMM/HMMR-derived peptide R3. Taken together, 8 (47%) of 17 AML cases showed specific T-cell responses, which correlated with the respective mRNA expression as previously reported.
For the induction of specific T-cell responses against the PRAME-derived peptides, we examined samples from 10 patients with AML. Positive results in ELISPOT analyses for IFN-γ and granzyme B were observed in 7 (70%) of 10 cases analyzed for the peptide P3 and in 1 (14%) of 7 for the peptide P1, respectively. Figure 4 displays the results of an ELISPOT analysis for IFN-γ and granzyme B for the PRAME-derived peptides P1 and P3 in a patient with AML. While we were able to observe specific T-cell responses against the PRAME peptide P3, no responses were detected against the other PRAME peptide P1 or against the negative control. Cross-reactivity against an irrelevant peptide (MAGE3) was excluded. High IFN-γ and granzyme B release was detected for the IMP peptide serving as a positive control.
Specific immune responses against RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides in a patient with AML who had simultaneous expression of the respective TAAs. ELISPOT analysis for (A) IFN-γ and (B) granzyme B. Strong CD8-mediated immune responses were detected for the IMP peptide used as a positive control and for the RHAMM/HMMR-derived peptide R3 and the G250/CA9-derived peptide G2. Each experiment was carried out in triplicate. A lower intensity was found for the PRAME-derived peptide P3. No T-cell reaction was found for the negative control (T2 cells without peptide). Cross-reactivity was excluded using an irrelevant peptide derived from the TAA MAGE3. These ELISPOT assays were performed using peripheral blood of the patient in CR. (C) DNA microarray–based mRNA expression levels of the genes coding for the respective antigens has been examined at the time of diagnosis in the leukemic blasts: high expression was found for the TAAs RHAMM/HMMR and G250/CA9, while PRAME showed a lower mRNA expression compared with the median expression in 116 patients with AML (for the TAA expression the normalized absolute gene expression values [normalized intensity of the respective feature on the microarry] are depicted).
Specific immune responses against RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides in a patient with AML who had simultaneous expression of the respective TAAs. ELISPOT analysis for (A) IFN-γ and (B) granzyme B. Strong CD8-mediated immune responses were detected for the IMP peptide used as a positive control and for the RHAMM/HMMR-derived peptide R3 and the G250/CA9-derived peptide G2. Each experiment was carried out in triplicate. A lower intensity was found for the PRAME-derived peptide P3. No T-cell reaction was found for the negative control (T2 cells without peptide). Cross-reactivity was excluded using an irrelevant peptide derived from the TAA MAGE3. These ELISPOT assays were performed using peripheral blood of the patient in CR. (C) DNA microarray–based mRNA expression levels of the genes coding for the respective antigens has been examined at the time of diagnosis in the leukemic blasts: high expression was found for the TAAs RHAMM/HMMR and G250/CA9, while PRAME showed a lower mRNA expression compared with the median expression in 116 patients with AML (for the TAA expression the normalized absolute gene expression values [normalized intensity of the respective feature on the microarry] are depicted).
Interestingly, 6 (60%) of 10 of analyzed AML cases exhibited specific T-cell responses against the newly characterized G250/CA9-derived peptide G2. In contrast, only 1 (12.5%) of 8 showed positive results for the peptide G1. In all tested samples, similar results were observed for the ELISPOT assays for IFN-γ and for granzyme B.
We also compared the results of cellular assays from patients in CR to patients with refractory AML. In the latter group, no specific T-cell responses were detected against the RHAMM/HMMR-derived peptide R3 (0 of 10) and the G250/CA9-derived peptide G2 (0 of 10). Specific responses of CD8+ T cells against PRAME-derived peptide P3 were found in only 4 (40%) of 10 of refractory AML patients (Table 2). Therefore, specific T-cell responses of patients with AML who had refractory disease (together, 4 [13%] of 30) were significantly reduced compared with immune responses of patients reaching a CR (21 [57%] of 37; P < .001, Fisher exact test).
RHAMM/HMMR, PRAME, and G250/CA9 mRNA expression in patients with AML exhibiting specific immune responses against the respective antigen-derived peptides
To further investigate our hypothesis, we correlated mRNA expression at the time of diagnosis and immune responses against RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides in the stage of CR of the disease in patients with AML entered within the functional analyses. For this comparative analysis, matched diagnostic mRNA and CR T-cell specimens were available for 25 patients. As expected, specific immune responses significantly correlated with mRNA expression of the respective TAA at the time of diagnosis as determined by conventional RT-PCR, real-time RT-PCR, and/or microarray analysis (P = .03, Fisher exact test; Table 3).
Specific immune responses against peptides derived from PRAME in patients with AML. A high frequency of specific T-cell responses was found against the PRAME-derived peptide P3. This patient with AML showed a strong immune response for the PRAME-P3 peptide in ELISPOT analysis for IFN-γ (A) and granzyme B (B). No immune responses were observed for the other PRAME-derived peptides tested (P1, P2, P4) when compared with the cross-priming experiment against an irrelevant peptide (MAGE3). A strong immune response was detected for the IMP peptide used as a positive control (x-axis: counted spots per 10 000 CD8+ T cells are indicated). Error bars indicate standard deviation.
Specific immune responses against peptides derived from PRAME in patients with AML. A high frequency of specific T-cell responses was found against the PRAME-derived peptide P3. This patient with AML showed a strong immune response for the PRAME-P3 peptide in ELISPOT analysis for IFN-γ (A) and granzyme B (B). No immune responses were observed for the other PRAME-derived peptides tested (P1, P2, P4) when compared with the cross-priming experiment against an irrelevant peptide (MAGE3). A strong immune response was detected for the IMP peptide used as a positive control (x-axis: counted spots per 10 000 CD8+ T cells are indicated). Error bars indicate standard deviation.
Specific cell lysis of RHAMM/HMMR-R3–, PRAME-P3–, and G250/CA9-G2–pulsed T2 cells
We previously described a strong specific lysis of RHAMM/HMMR-R3–pulsed/expressing cells.9 Therefore, we wanted to demonstrate that in addition to RHAMM/HMMR-R3–pulsed cells, both PRAME-P3– and G250/CA9-G2–pulsed T2 cells can also be lysed by CD8+ T cells of patients with AML. Interestingly, using CD8+ T cells of patients with AML, we were able to show specific lysis of peptide-pulsed T2 cells of RHAMM/HMMR-R3–pulsed, PRAME-P3–pulsed, and G250/CA9-G2–pulsed T2 cells (Figure 5A-C).
To determine whether the specific lysis of G250/CA9-G2–pulsed T2 cells was dependent on HLA-ABC molecules, we performed HLA-ABC–blocking experiments. As described earlier for RHAMM/HMMR-R3,9 when blocking HLA-ABC molecules we could not detect a lysis reaction using G250/CA9-G2–primed T cells (Figure 5C). In addition, presensitized CD8+ T cells did not lyse K-562, which lacks HLA-A2 expression, but shows expression of RHAMM/HMMR, PRAME, and G250/CA9 (Figure 5A-C).
Dose-dependent cellular immune responses to RHAMM/HMMR-, PRAME- and G250/CA9-derived peptides
The T-cell reaction to the RHAMM/HMMR-derived peptide R3, the PRAME-derived peptide P3, and the G250/CA9-derived peptide G2 was demonstrated to be dependent on peptide concentration. Using different peptide concentrations we applied ELISPOT assays for granzyme B and IFN-γ to indicate peptide-specific T-cell responses. Based on this approach we were able to define an optimal dose of 10 μg peptide/mL, which was used in consecutive assays. Figure 6 shows a representative dose-dependent reaction against the G250/CA9-derived peptide G2. Similar results were found for PRAME (data not shown). For RHAMM/HMMR, dose-dependent results have been demonstrated already.9
Discussion
Hitherto little has been known about the clinical relevance of TAA expression levels in hematologic malignancies. Recently, DNA microarray technology has become a powerful tool that allows genome-wide gene expression analyses, thereby generating large datasets. This wealth of data opens new potential avenues for hypothesis-driven leukemia research by reanalyzing datasets under different considerations. Thus, we reanalyzed a previously published large AML gene expression dataset,25 which has led to novel insights concerning the clinical relevance of TAA expression in AML.
We focused our analysis on the investigation of the expression levels of TAAs that have been shown both to exhibit the potency to induce specific immune responses in tumors and to be expressed in AML blasts. Therefore, we evaluated the expressions of TERT, WT1, PRTN3, BCL2, LAMR1, PRAME, RHAMM/HMMR, and G250/CA9.9,11,12,18-21 We also validated the DNA microarray-based gene expression data by conventional and quantitative RT-PCR.
Correlation of TAA expression levels with clinical outcome showed that higher levels of TAAs known to be almost exclusively expressed in malignant tissues were positively associated with longer overall survival times. Interestingly, patients whose leukemic cells expressed at least 1 of the TAAs G250/CA9, PRAME, or RHAMM/HMMR had a better clinical outcome than those showing low expression of all 3 TAAs (P = .005, log-rank test). This finding suggests that in AML the expression of distinct tumor-specific TAAs on leukemic blasts might enable the immune system to better eradicate MRD following intensive chemotherapy, thereby leading to decreased relapse rates and longer overall survival times. The fact that an association with longer survival was not observed with the other TAAs (TERT, WT1, PRTN3, and LAMR1) might reflect that different TAAs show different potencies to induce specific T-cell responses and/or that some TAAs are not exclusively expressed on the leukemic blasts. On the other hand, an inverse correlation of survival with BCL2 expression has been previously reported35-37 and might be ascribed to its antiapoptotic function, an effect that might outweigh a potential immunologic benefit.
Interestingly, there was a significant association between older age and “no TAA” expression. As older age is known to be associated with adverse outcome, it might be argued that the examined TAAs merely represent surrogate markers for age. However, none of the markers alone showed a significant association with age, and it is a remarkable observation that in general, leukemic blasts from older patients seem to express less TAAs. This finding might be 1 explanation for the poorer outcome in these cases.
Highly specific lysis of RHAMM/HMMR-R3–, PRAME-P3–, and G250/CA9-G2–pulsed T2 cells in Cr-51 release assays. (A-C) CD8+ T cells stimulated with RHAMM/HMMR-R3 (A), PRAME-P3 (B), and G250/CA9-G2 (C) peptides were able to lyse TAA–peptide pulsed T2 cells and primary AML blasts with expression of the respective TAAs (+). In contrast, primary AML blasts not expressing the respective TAAs (–) and K562 cells expressing all TAAs of interest (RHAMM, PRAME, and G250), but lacking expression of HLA molecules, such as HLA-A2, were not lysed by specific CD8+ T cells. In accordance, T2 cells pulsed with an irrelevant peptide (MAGE3) were not recognized, underlining the TAA specificity of the T-cell immune response (A-C). HLA-ABC–blocked T2 cells pulsed with G250-G2 peptide were not lysed, but pulsed T2 cells treated with isotype control antibody could be recognized by stimulated CD8+ T cells (C).
Highly specific lysis of RHAMM/HMMR-R3–, PRAME-P3–, and G250/CA9-G2–pulsed T2 cells in Cr-51 release assays. (A-C) CD8+ T cells stimulated with RHAMM/HMMR-R3 (A), PRAME-P3 (B), and G250/CA9-G2 (C) peptides were able to lyse TAA–peptide pulsed T2 cells and primary AML blasts with expression of the respective TAAs (+). In contrast, primary AML blasts not expressing the respective TAAs (–) and K562 cells expressing all TAAs of interest (RHAMM, PRAME, and G250), but lacking expression of HLA molecules, such as HLA-A2, were not lysed by specific CD8+ T cells. In accordance, T2 cells pulsed with an irrelevant peptide (MAGE3) were not recognized, underlining the TAA specificity of the T-cell immune response (A-C). HLA-ABC–blocked T2 cells pulsed with G250-G2 peptide were not lysed, but pulsed T2 cells treated with isotype control antibody could be recognized by stimulated CD8+ T cells (C).
Further supporting our findings, expression of PRAME has been shown as an indicator of a good prognosis in childhood AML,38 although this effect might have been secondary to its correlation with favorable cytogenetics such as a translocation t(8;21), which already has been previously described.39 G250/CA9 is expressed in most clear cell forms of renal cell carcinoma (RCC), but not in normal tissues. In accordance with our data in patients with AML, the expression of G250/CA9 has also been shown to be associated with a favorable outcome in RCC.40-42 While some studies have shown a poor clinical outcome of patients with AML expressing WT1 in the leukemic blasts,33,43 others described no correlation of WT1 expression with the clinical outcome,34,44 which is in agreement with our findings.
For RHAMM/HMMR, our results are contrary to the data reported in the literature. In breast cancer and multiple myeloma RHAMM/HMMR expression was found in highly proliferating tumor cells, and there was also an association with the potential to metastasize and with worse clinical outcome.45,46 The connection between increased RHAMM/HMMR levels and poor outcome in these studies might reflect different underlying tumor biology (eg, an impaired lymphatic system in multiple myeloma), which might affect a possible immunologic effect of TAA expression. In AML, RHAMM/HMMR is expressed in 70% of cases, whereas no expression has been detected in normal CD34+ hematopoietic stem cells or in normal tissues (except for testis, placenta, and thymus),19 rendering it a potent immunologic target for the immune system to fight a residual tumor load following chemotherapy in AML. Therefore, RHAMM/HMMR, as well as PRTN3 and WT1, is currently used in peptide vaccination trials for patients with hematologic malignancies,9,10,17 and effective immune responses have been demonstrated.
Dose-dependent cellular immune responses to the G250/CA9-derived peptide G2. A dose-dependent specific immune reaction against G250/CA9-G2–pulsed T2 cells could be detected analyzed by ELISPOT assays for IFN-γ and granzyme B. The optimal dose of 10 μg peptide G250/CA9-G2 per milliliter was used in consecutive assays. For the TAAs PRAME and RHAMM/HMMR,8 similar results were found showing also an optimal peptide dose of 10 μg/mL.
Dose-dependent cellular immune responses to the G250/CA9-derived peptide G2. A dose-dependent specific immune reaction against G250/CA9-G2–pulsed T2 cells could be detected analyzed by ELISPOT assays for IFN-γ and granzyme B. The optimal dose of 10 μg peptide G250/CA9-G2 per milliliter was used in consecutive assays. For the TAAs PRAME and RHAMM/HMMR,8 similar results were found showing also an optimal peptide dose of 10 μg/mL.
To further investigate whether the observed positive clinical effects might result from the induction of specific immune responses, we analyzed the induction of specific T-cell responses by G250/CA9, RHAMM/HMMR, and PRAME. For RHAMM/HMMR and PRAME, HLA-A*0201–restricted immune responses and specific cell lysis of tumor cells have already been described in small AML studies,9,20,47 and an epitope peptide derived from G250/CA9 recognized by specific CD8+ T cells has been described in patients with renal cell carcinoma.30 Here, we reported the induction of specific T-cell responses against the TAA RHAMM/HMMR in a larger cohort of AML patient samples with specific responses in 47% of cases. Moreover, we found specific immune responses against the PRAME-derived peptide P3 in 70% and against the newly characterized G250/CA9-derived peptide G2 in 60% of patients with AML, thereby demonstrating a high frequency of specific T-cell responses against these 3 TAAs in patients with AML. Consistent with an impact on clinical outcome, specific T-cell responses were significantly higher in patients with AML in CR compared with those with refractory disease.
Specific immune responses were found in ELISPOT analysis for IFN-γ and granzyme B, and these could be also confirmed by specific lysis of pulsed T2 cells in Cr-51 release assays. Furthermore, dilution series showed a specific dose-dependent T-cell response against these peptides, which is in concordance with our clinical observations suggesting an immunologic effect based on TAA expression levels. In accordance, we also could demonstrate simultaneous CD8+ T-cell responses against the peptides RHAMM/HMMR-R3, PRAME-P3, and G250/CA9-G2 in individual samples from patients with AML. In the future, the ability to generate these simultaneous immune responses might be helpful in overcoming immunologic tumor-escape mechanisms and to enhance clinical effects of vaccination against tumor cells expressing these TAAs.
In conclusion, our findings are highly suggestive that the expression of some TAAs might play a critical role in immunologic mechanisms, preventing a relapse of AML. Thus, TAAs like RHAMM/HMMR, PRAME, and G250/CA9 represent promising targets for future vaccination trials to further augment immune responses. Based on the observed immune responses and the favorable expression pattern of RHAMM/HMMR in normal tissues, a peptide vaccination trial for patients with hematologic malignancies expressing RHAMM/HMMR is ongoing.9,10,17 Furthermore, the findings of this study suggest that a multipeptide vaccination trial simultaneously targeting several TAAs such as RHAMM/HMMR, PRAME, and G250/CA9 might induce more potent immune responses and might enable us to include a larger cohort of patients with AML in vaccination trials. However, this clearly needs to be addressed within well-designed clinical trials that will help to answer the question of which patients will benefit most from such vaccination protocols with respect to their TAA expression levels.
Authorship
Contribution: J.G. and M.S. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, 8/24/2006; DOI 10.1182/blood-2006-01-023127.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The authors appreciate the excellent technical support of Anita Smaragowska and Marlies Goetz. Furthermore, the authors are indebted to the staff of the Stanford Functional Genomic Facility (SFGF) for providing high-quality cDNA microarrays, and to the staff of the Stanford Microarray Database (SMD) group for providing outstanding database support.
Supported in part by the Deutsche Forschungsgemeinschaft, Bonn, Germany (Forschungsstipendium BU 1339/1 to L.B. and DFG GR 2676/1-1 to J.G. and M.S.) and by a generous grant from the German José Carreras Leukemia Foundation (J.G., M.S., and H.D.).


![Figure 3. Specific immune responses against RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides in a patient with AML who had simultaneous expression of the respective TAAs. ELISPOT analysis for (A) IFN-γ and (B) granzyme B. Strong CD8-mediated immune responses were detected for the IMP peptide used as a positive control and for the RHAMM/HMMR-derived peptide R3 and the G250/CA9-derived peptide G2. Each experiment was carried out in triplicate. A lower intensity was found for the PRAME-derived peptide P3. No T-cell reaction was found for the negative control (T2 cells without peptide). Cross-reactivity was excluded using an irrelevant peptide derived from the TAA MAGE3. These ELISPOT assays were performed using peripheral blood of the patient in CR. (C) DNA microarray–based mRNA expression levels of the genes coding for the respective antigens has been examined at the time of diagnosis in the leukemic blasts: high expression was found for the TAAs RHAMM/HMMR and G250/CA9, while PRAME showed a lower mRNA expression compared with the median expression in 116 patients with AML (for the TAA expression the normalized absolute gene expression values [normalized intensity of the respective feature on the microarry] are depicted).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2006-01-023127/7/m_zh80240605010003.jpeg?Expires=1769177116&Signature=hnBm8jnMSi26J8X1SLlLYo8ZAWHzTp6115VDJY9fd1MKRNlcFaJOCebihm0e~L3m4sCWX-6mxSc3ypJPQKy3~iQHFZ49t8oR~KJujs6AqUby3eKlu8gpbgwEfg63P-s2-7YP~kK0Ri0B9Dat~OwAFkjcXS7UeKAFm~Ooep4ez40uVZsr5I0dQ~alWq5Rm8G48TGbbCaSWzruuJLTwpWOHhhYhSP5dP2hgezTLi8vWn9RdqA0MdZ0V6kn0gqGT0OAMFujX0nORlbNMcvHSKnpDm~CAk0~SqxadDbLZFtnd3~m5AGFfhuRypyStzUphAWr~8ykR~hRTkdQAicZZMf6jw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 3. Specific immune responses against RHAMM/HMMR-, PRAME-, and G250/CA9-derived peptides in a patient with AML who had simultaneous expression of the respective TAAs. ELISPOT analysis for (A) IFN-γ and (B) granzyme B. Strong CD8-mediated immune responses were detected for the IMP peptide used as a positive control and for the RHAMM/HMMR-derived peptide R3 and the G250/CA9-derived peptide G2. Each experiment was carried out in triplicate. A lower intensity was found for the PRAME-derived peptide P3. No T-cell reaction was found for the negative control (T2 cells without peptide). Cross-reactivity was excluded using an irrelevant peptide derived from the TAA MAGE3. These ELISPOT assays were performed using peripheral blood of the patient in CR. (C) DNA microarray–based mRNA expression levels of the genes coding for the respective antigens has been examined at the time of diagnosis in the leukemic blasts: high expression was found for the TAAs RHAMM/HMMR and G250/CA9, while PRAME showed a lower mRNA expression compared with the median expression in 116 patients with AML (for the TAA expression the normalized absolute gene expression values [normalized intensity of the respective feature on the microarry] are depicted).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/13/10.1182_blood-2006-01-023127/7/m_zh80240605010003.jpeg?Expires=1769098607&Signature=pHoahkmR5EtH7jVi3G8H2wb38TvpKw4s36hcc3bEdpt52qKzQXhDzSy1tv~Agmh-0ACQd0FMBuOwPcn9e2d~Dj31Bez4xOpsMM0FYb1VMHM1nFqiEjdbKFR-VgFdT-TKmod8L~h21BVo8zLxy4YhMN16zfir7kknjeMWwjIT71YgaiM9RozxKCl3pDdyvKIMNVhLEx7-YCORXs37gVUFxQsGA0-QudHI~IoCyo6kRM0AgRIgLy5LtX4iz1LZAZoE6hU0pNN~84WSoG0~5mDssErhBLtyo3U51gmn-eiIrJsUP75F~RMOQSdMzM6NMBWemeV3bBbzQLv7eFNN7JZirw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)