Intrinsic sensitivity of newly diagnosed chronic myeloid leukemia (CML) patients to imatinib (IC50imatinib) correlates with molecular response. IC50imatinib is defined as the in vitro concentration of drug required to reduce phosphorylation of the adaptor protein Crkl by 50%. We now show that interpatient variability in IC50imatinib is mainly due to differences in the efficiency of imatinib intracellular uptake and retention (IUR). In 25 untreated CML patients, the IC50imatinib strongly correlated (R2 = –0.484, P = .014 at 2 μM imatinib) with the IUR of [14C]imatinib. The addition of prazosin, a potent inhibitor of OCT-1 cellular transporter, reduced the IUR and eliminated interpatient variability. IC50 values for the more potent BCR-ABL inhibitor nilotinib (AMN107) did not correlate with IC50imatinib (R2 =–0.0561, P > .05). There was also no correlation between IC50nilotinib and the IUR for [14C]nilotinib (R2 = 0.457, P > .05). Prazosin had no effect on nilotinib IUR, suggesting that influx of nilotinib is not mediated by OCT-1. In conclusion, whereas OCT-1–mediated influx may be a key determinant of molecular response to imatinib, it is unlikely to impact on cellular uptake and patient response to nilotinib. Determining interpatient and interdrug differences in cellular uptake and retention could allow individual optimization of kinase inhibitor therapy.
Introduction
Chronic myeloid leukemia (CML) is a hematologic malignancy characterized by the presence of the Philadelphia chromosome resulting from a balanced, reciprocal translocation between chromosomes 9 and 22. At the molecular level, this translocation results in the formation of the BCR-ABL fusion gene that translates to a 210-kDa BCR-ABL protein.1-3 This protein possesses a constitutively activated tyrosine kinase domain, the activity of which is critical to the pathogenesis of CML.4,5
Imatinib mesylate (Glivec, formerly STI571; Novartis Pharmaceuticals, Basel, Switzerland) is a compound based upon the 2-phenylaminopyrimidine skeleton that potently inhibits the ABL tyrosine kinase activity of c-ABL, BCR-ABL, and Tel/ABL. In addition, imatinib inhibits the tyrosine kinase activity of the PDGF receptors6 ARG7 and c-Kit6,8 and the macrophage colony-stimulating factor receptor c-fms9 . The majority of newly diagnosed CML patients respond excellently to imatinib, with 95% achieving complete hematologic response and more than 80%, a complete cytogenetic remission.10 However, despite this, there remains a group of patients that demonstrates suboptimal response to imatinib therapy, defined as failure to achieve a major cytogenetic response (MCyR) at 6 months and/or a complete cytogenetic response (CCyR) at 12 months.11 These patients have a significantly higher risk of disease progression.12
We have recently demonstrated that sensitivity to imatinib-induced kinase inhibition in vitro is predictive of molecular outcome in previously untreated chronic-phase CML patients.13 In order to assess imatinib sensitivity, we have used the in vitro phosphorylation status of the adaptor protein Crkl (CT10 regulator of kinase-like), a known downstream substrate of BCR-ABL,14,15 to calculate the IC50 for imatinib. In this assay, we have demonstrated that a significantly higher proportion of patients with a low IC50imatinib (high intrinsic sensitivity to imatinib-induced kinase inhibition) achieve major molecular responses (3 log reduction in BCR-ABL as determined by quantitative reverse-transcriptase–polymerase chain reaction [RQ-PCR]) by 12 months when compared with those patients with high IC50imatinib.
This demonstrates that the molecular response of chronic-phase (CP) CML patients to imatinib can be predicted prior to drug exposure, and indicates that CP CML patients have varied intrinsic sensitivity to imatinib-induced kinase inhibition. While the in vitro kinase inhibition assay (IC50 assay) was shown to be a reliable predictor of response, the reason for heterogeneity in IC50imatinib values remains unclear. Imatinib-insensitive point mutations in BCR-ABL and BCR-ABL amplification are implicated in the development of secondary resistance; however, these have been excluded as causes of high IC50imatinib values in CML patients at presentation.13
Drug transporters play a key role in uptake and elimination in vivo via the gastrointestinal tract (GIT) and at the cellular level are important determinants of intracellular drug concentration. Drug efflux proteins have been extensively studied, and their overexpression has been frequently implicated as a cause of resistance, in several diseases.16 The best documented efflux pump is the p-glycoprotein (ABCB1 or MDR1), a full-length ATP-binding cassette transporter superfamily member17,18 that is associated with multidrug resistance (MDR).19,20 Recent studies have concluded that imatinib is a substrate for ABCB1,21-26 although this is not without conjecture.27,28 Furthermore, the interaction of imatinib with ABCG2 (BCRP, breast cancer resistance protein) has been reported,29-33 although there is no consensus as to whether imatinib is a substrate for this protein, or simply an inhibitor.34 There is also some evidence that other proteins such as ABCC3 and MVP35 may also be involved in imatinib efflux. The mechanism for the efflux protein involvement in resistance is not well understood but mutations and polymorphisms do occur in ABCB1.36-38 MDR is most commonly an acquired phenomenon, arising following drug exposure.
While playing a critical role in drug transport, influx proteins are less well documented in the area of multidrug resistance than the efflux proteins, particularly in the hematopoietic system. The major influx proteins belong to the largest superfamily of transporters, the solute carrier (SLC) superfamily.39 In humans, the SLC22 subfamily consists of 12 members encompassing the polyspecific organic cation transporters (OCTs). The OCT proteins are associated with the transport of endogenous and exogenous substrates, which exist as cations at physiologic pH, across the plasma membrane in either direction.40,41 It is this family, in combination with the organic anion transporters (OATs), that is involved in the absorption, distribution, and elimination of drugs in vivo.42
Recently, it was reported that imatinib was most likely transported into cells via the OCT-1 influx protein,24 a solute carrier that mediates only uptake and not efflux. In a small number of CML cell lines and primary CML samples collected before and during imatinib therapy, this study demonstrated varying levels of OCT-1 mRNA. A more recent study supported these findings,43 leading to the conclusion that influx proteins may play a role in the varying responses observed in some patients to imatinib monotherapy.
In this current study, we have examined the role of transport proteins in mediating the intrinsic sensitivity to imatinib-induced kinase inhibition observed in our in vitro sensitivity (IC50imatinib) assay. Furthermore, we have assessed intrinsic sensitivity to the novel, more potent and selective kinase inhibitor nilotinib (AMN107; Novartis Pharmaceuticals)44-47 and compared this with the intrinsic sensitivity to imatinib in a group of CML patients at presentation. We have also explored the contribution of common influx and efflux proteins to the in vitro intracellular concentration of nilotinib. As nilotinib proceeds through phase 1 and 2 clinical studies, both as a single agent and in combination with imatinib, these experiments will provide critical evidence relating to drug transport, sensitivity, and drug-drug interaction, which will assist in the development of rational treatment strategies.
Patients, materials, and methods
Patient population and blood samples
Blood was collected from chronic-phase CML patients at presentation to the Royal Adelaide Hospital hematology clinic. All samples were collected with informed consent, in accordance with the Declaration of Helsinki, and, importantly, all samples were collected before commencement of imatinib therapy. Approval for the study was obtained from the institutional review boards of the Institute of Medical and Veterinary Science (Adelaide, Australia) and the Royal Adelaide Hospital.
Mononuclear cells (MNCs) were isolated from blood using Lymphoprep (Axis Shield, Oslo, Norway) density gradient centrifugation. Experiments were performed on either cryopreserved or fresh cells, on the day of isolation or after overnight storage in Hanks balanced salt solution (HBSS: Ca++ and Mg++ free; JRH Biosciences, Lenexa, KS) medium supplemented with 5% fetal bovine serum (FBS; Trace Biosciences, Sydney, Australia).
Cell lines
Human K562 and CCRF-CEM cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The ABCB1-overexpressing variant of CCRF-CEM, CEM/VBL100, was kindly provided by Professor Leonie Ashman (University of Newcastle, Newcastle, Australia).
Kinase inhibitors
The 2-phenylaminopyrimidine derivatives imatinib mesylate (formerly STI571) and nilotinib, together with [14C]-imatinib and [14C]-nilotinib, were kindly provided by Novartis Pharmaceuticals. Stock solutions of imatinib were prepared at 1 mM and 10 mM in distilled water, sterile filtered, and stored at 4°C. Stock solutions of nilotinib were prepared at 10 mM in dimethylsulfoxide (DMSO; Sigma Aldrich, Mannheim, Germany) sterile filtered and stored at 4°C.
Drugs and inhibitors
The OCT inhibitors prazosin (Sigma Aldrich), progesterone (Sigma Aldrich), and procainamide (Sigma Aldrich) were used at 100 μM, 10 μM, and 10 mM, respectively. The ABCB1 inhibitor PSC833 (kindly provided by Novartis Pharmaceuticals) and the ABCG2 inhibitor fumitremorgin C (FTC; Alexis, San Diego, CA) were both used at 10 μM.
Western blot analysis and determination of IC50 values
Western blot for p-Crkl was performed essentially as previously described.13 Briefly, patient cells were incubated for 2 hours48,49 with varying concentrations of imatinib or nilotinib. Concentrations used for imatinib ranged from 0 μM to 50 μM, and for nilotinib from 0 μM to 10 μM. Following incubation, cells were washed with cold PBS and lysed in Laemmli buffer50 by boiling for 12 minutes. Cell lysates were clarified by microfugation and stored at –20°C. Protein lysates were resolved on an SDS/10% polyacrylamide gel and electrophoretically transferred to nitrocellulose PVDF (Amersham Biosciences, Poole, United Kingdom). Following blocking, the membrane was probed with anti-Crkl antibody (Santa Cruz, Santa Cruz, CA), detected with ECF substrate (Amersham Pharmacia, Piscataway, NJ), and analyzed by Fluor Imager analysis (Molecular Dynamics, Sunnyvale, CA). Signals were quantified using Image Quant software (Molecular Dynamics), and the ratio of phosphorylated Crkl (p-Crkl) to nonphosphorylated Crkl was determined using Image Quant analysis. IC50 values were determined as the in vitro concentration of drug required to reduce phosphorylation of the adaptor protein Crkl (p-Crkl) by 50%.
Radiolabeled drug uptake (IUR assay)
Radiolabeled drug uptake studies were performed in triplicate, under culture conditions mimicking those of the IC50 assay.13 In all assays, [14C]-labeled drug was admixed to the desired concentration with unlabeled drug.24 Essentially the assay was performed as per Zhang et al51 under the aforementioned culture conditions. In brief, 200 000 cells were incubated for 2 hours at 37°C in the presence of varying concentrations of imatinib or nilotinib ranging from 0 to 2 μM. Isotopes were resuspended to 1 mg/mL and specific activities were 3.3 MBq for [14C]-imatinib and 3.31 MBq for [14C]-nilotinib. After incubation, the cellular and aqueous phases were separated, and incorporation was determined using a Top Count Microplate Beta Scintillation counter (Canberra Packard, Meriden, CT) following the addition of Microscint20 (Perkin Elmer, Boston, MA) scintillation fluid.
Expression of efflux proteins
Expression levels of ABCB1 and ABCG2 were assessed using flow cytometric analysis. Phycoerythrin (PE)–conjugated anti-ABCB1 antibody, UIC2 (Immunotech, Marseilles, France), and anti-ABCG2 (R & D Systems, Minneapolis, MN) were used for analysis. Staining was performed on ice for 40 minutes and analysis was performed on an Epics XL-MCL Flow Cytometer (Beckman Coulter, Miami, FL). Results were analyzed relative to isotype-matched controls. Detection limits of this assay were determined using limiting dilution of the ABCB1-expressing cell CEM-VBL100 and ABCB1-negative parental line CCRF-CEM. Using this approach, the assay was sensitive to the 1% level.
Data analysis
The Mann-Whitney rank sum or t test was used to define differences between groups, and correlation was performed using Spearman rank order. The Fisher exact test was used to determine the P value corresponding to the exact probability distribution.
Results
Correlation between IC50imatinib and intracellular concentration of imatinib
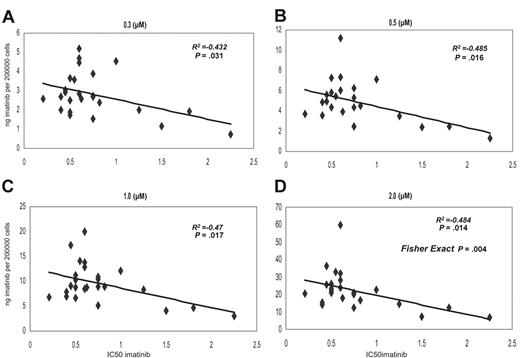
We have compared the IC50imatinib and the intracellular concentration of [14C]-imatinib achieved in 25 samples collected from presentation chronic-phase CML patients prior to imatinib therapy. The IC50imatinib ranged from 0.2 to 2.25 μM, with a median of 0.6 μM in this patient cohort. The intracellular uptake and retention (IUR) of imatinib achieved in vitro was measured following the addition of 0, 0.3, 0.5, 1, and 2 μM imatinib and a 2-hour incubation at 37°C. There was a significant correlation between IC50imatinib and IUR at 0.3, 0.5, 1, and 2 μM concentrations (P = .03, P = .016, P = .017, and P = .014, respectively) (Figure 1A-D). Furthermore, when the data at 2 μM was divided into low and high IC50imatinib and low and high IUR about the median for both groups, the Chi square statistic (Fisher exact test) demonstrated a significant relationship (P = .004) between the 2 variables (Figure 1D, 2.0 μM).
Grouping the data into low and high IC50imatinib about the median revealed a significant difference in intracellular concentration achieved in the 2 groups at 0.5-, 1.0-, and 2.0-μM concentrations, confirming the association between IC50imatinib and IUR (Figure 2A).
Dot plot demonstrating the correlation between the IC50imatinib and IUR for imatinib. Concentrations were as follows: (A) 0.3 μM, (B) 0.5 μM, (C) 1.0 μM, and (D) 2.0 μM.
Dot plot demonstrating the correlation between the IC50imatinib and IUR for imatinib. Concentrations were as follows: (A) 0.3 μM, (B) 0.5 μM, (C) 1.0 μM, and (D) 2.0 μM.
Correlation between IUR imatinib and molecular outcome
There was a significant correlation between IUR imatinib and molecular outcome (as determined by real-time RQ-PCR for BCR-ABL) at 3 months in 19 patients where molecular data were available (R2 = 0.58, P = .041). Furthermore, restricting analysis to the cohort of 11 patients with 12-month molecular follow-up, there was a significant correlation at both 3 and 12 months (R2 = 0.793, P = .003 and R2 = 0.643, P = .042, respectively), indicating that the IUR is predictive of molecular outcome.
Correlation between IC50imatinib and IC50nilotinib
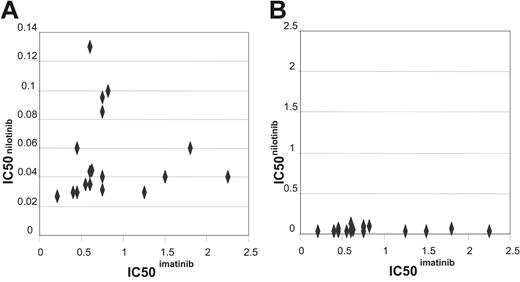
Comparative IC50 assays for both imatinib and nilotinib were performed on cells collected from a cohort of 17 patients (Figure 3A). Assays were performed concurrently on the same sample. The lack of correlation between the IC50 values (R2 = –0.0561, P = .831) suggests that the 2 values are unrelated. The IC50 values for nilotinib were on average 14-fold lower than those observed for imatinib, in keeping with published data on cell lines,46,47 and confirming that nilotinib is a more potent inhibitor of BCR-ABL kinase activity than imatinib in patient samples.
Comparing the interpatient variability with respect to IC50s for imatinib and nilotinib revealed the mean IC50imatinib was 0.774 with a standard deviation of 0.472, in comparison with IC50nilotinib where the mean was 0.05 with a standard deviation of 0.02. This indicates that there is less interpatient variability in IC50 with the new more potent inhibitor (Figure 3B). Furthermore, all patients had IC50nilotinib well below the median plasma concentration of 2 μM as reported in the phase 1 trial where a maximum tolerated dose was not yet achieved.45 In contrast, several patients had IC50imatinib that fell outside the normal range of imatinib concentration that is clinically achieved.52
Correlation of IC50nilotinib and intracellular concentration (IUR) of nilotinib achieved
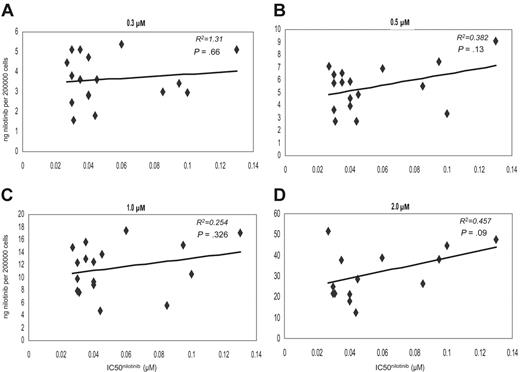
There was no correlation between the IC50nilotinib and the IUR of nilotinib achieved in vitro, at any of the tested concentrations following 2-hour incubation at 37°C (R2 =–0.561, P = .831) (Figure 4A-D). All comparative assays were performed concurrently on the same sample. Furthermore, grouping the data into low and high IC50nilotinib about the median revealed no significant difference in intracellular concentration achieved in the 2 groups at 0.3-, 0.5-, 1.0-, and 2.0-μM concentrations (P = .958, P = .616, P = .526, and P = .472, respectively), further confirming the poor association between IC50nilotinib and IUR (Figure 2B). This indicates that unlike imatinib, interpatient variability in nilotinib IUR cannot explain the observed variability in IC50nilotinib in this patient cohort.
Graph demonstrating the significant difference in IUR between the low and high IC50imatinib groups. (A) For imatinib, median IUR shown in parentheses. (B) Graph demonstrating the similarity in IUR for nilotinib, between the low and high IC50nilotinib groups. Median IUR shown in parentheses. (C) Effect of the OCT-1 inhibitor prazosin on IUR for low and high IC50imatinib groups. (D) Effect of the OCT-1 inhibitor prazosin on IUR for low and high IC50niltoinib groups. Error bars indicate one standard deviation (SD).
Graph demonstrating the significant difference in IUR between the low and high IC50imatinib groups. (A) For imatinib, median IUR shown in parentheses. (B) Graph demonstrating the similarity in IUR for nilotinib, between the low and high IC50nilotinib groups. Median IUR shown in parentheses. (C) Effect of the OCT-1 inhibitor prazosin on IUR for low and high IC50imatinib groups. (D) Effect of the OCT-1 inhibitor prazosin on IUR for low and high IC50niltoinib groups. Error bars indicate one standard deviation (SD).
Effect of the OCT-1 inhibitor prazosin
Prazosin is a potent inhibitor of OCT-1 and to a lesser extent OCT-3 (comparative IC50 values for OCT-1, OCT-2, and OCT-3: 1.8, > 100, and 13 μM, respectively).53 The effect of prazosin on imatinib and nilotinib uptake was assessed in cells from 25 and 22 patients, respectively. The addition of prazosin to patient cells significantly reduced IUR of imatinib at all imatinib concentrations (P = < .001). The impact of prazosin was more significant in the low IC50imatinib group (P < .001) than the high IC50 group where intracellular concentrations were already lower (P = .009) (Figure 2C). There was no significant difference between the 2 groups in IUR achieved following prazosin treatment (P = .93), indicating that prazosin removes the difference in IUR between the low and high IC50 groups. This suggests that OCT-1–mediated influx is the probable underlying mechanism of variable IC50imatinib.
Dot plot comparing the IC50imatinib and IC50nilotinib. (A) Dot plot comparing the IC50imatinib and IC50nilotinib graphed as appropriate for the values achieved. (B) Dot plot comparing the IC50imatinib and IC50nilotinib graphed on the same scale to demonstrate the difference in interpatient variation between the 2 drugs.
Dot plot comparing the IC50imatinib and IC50nilotinib. (A) Dot plot comparing the IC50imatinib and IC50nilotinib graphed as appropriate for the values achieved. (B) Dot plot comparing the IC50imatinib and IC50nilotinib graphed on the same scale to demonstrate the difference in interpatient variation between the 2 drugs.
In contrast, there was no statistical difference between the IUR for nilotinib in patient cells assayed in the presence or absence of prazosin at any of the tested concentrations (P > .05) (Figure 2D).
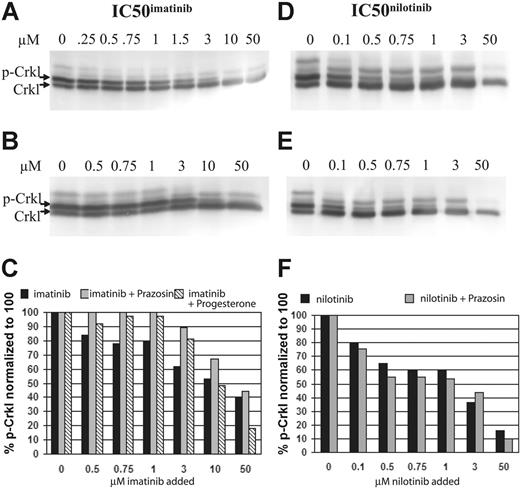
In a related series of experiments, prazosin (100 μM) was added to the IC50imatinib assay of the BCR-ABL–positive cell line K562. In these experiments, it was demonstrated that the addition of prazosin markedly increased the IC50imatinib (Figure 5A-B). Similarly, progesterone, a weaker OCT-1 inhibitor (IC50 3.1 μM)53 was substituted for prazosin in a second series of experiments, and while the increase in percent of p-Crkl was less marked, and became limiting at 10 μM, the effect of imatinib blockade was still demonstrable (Figure 5C). In contrast, when the same experiment using prazosin was performed with nilotinib, there was no significant difference between either the IC50nilotinib or the percent of p-Crkl at the concentration of drug assayed (Figure 5D-F).
Effect of procainamide on IUR
The effect of procainamide was examined on the IUR for both imatinib and nilotinib in 10 patients. Procainamide is a potent inhibitor of OCT-2, a lesser inhibitor of OCT-1, and does not inhibit OCT-3 (comparative IC50 values for OCT-1, OCT-2, and OCT-3: 90, 50, and 738 μM, respectively).41 Treatment of cells with procainamide resulted in a reduction of IUR in the 10 patients analyzed, although the effect failed to reach statistical significance. When the patients were divided into low and high IC50imatinib, the addition of procainamide resulted in a statistically significant reduction in IUR in those patients with a low IC50imatinib, however, there was no significant difference in IUR in the high IC50imatinib group (Table 1).
Like prazosin, the addition of procainamide to the nilotinib assay resulted in no significant difference in IUR in the 10 patients. Furthermore, there was also no significant difference when the data were grouped into low and high IC50 cohorts, indicating that nilotinib is not a substrate for either OCT-1, OCT-2, or OCT-3 (Table 1).
Dot plots showing the correlation between the IC50 and IUR nilotinib. Concentrations were as follows: (A) 0.3 μM, (B) 0.5 μM, (C) 1.0 μM, and (D) 2.0 μM.
Dot plots showing the correlation between the IC50 and IUR nilotinib. Concentrations were as follows: (A) 0.3 μM, (B) 0.5 μM, (C) 1.0 μM, and (D) 2.0 μM.
Effect of temperature on IUR imatinib and nilotinib
It has been previously shown that imatinib uptake is temperature dependent,24 suggesting the process is primarily one of active transport. We have shown a significant difference between assays performed at 4°C and 37°C in the IUR of imatinib at 2 μM (P = .038), confirming an active uptake component, but unlike the findings of Thomas et al24 not at any lower concentration tested (P > .05 at 0.3, 0.5, and 1 μM). In contrast, there was no significant difference in IUR between 4°C and 37°C for nilotinib (P > .05 at 0.3, 0.5, 1, and 2 μM). While this finding suggests that the uptake of nilotinib is largely passive, an active component cannot be conclusively excluded.
Western blots and corresponding graphs. (A) Standard IC50imatinib assay. (B) IC50imatinib assay performed with the addition of 100 μM prazosin. (C) Combined graphic representation of panels A-B. (D) Standard IC50nilotinib assay. (E) IC50nilotinib assay performed with the addition of 100 μM prazosin. (F) Combined graphic representation of panels D-E.
Western blots and corresponding graphs. (A) Standard IC50imatinib assay. (B) IC50imatinib assay performed with the addition of 100 μM prazosin. (C) Combined graphic representation of panels A-B. (D) Standard IC50nilotinib assay. (E) IC50nilotinib assay performed with the addition of 100 μM prazosin. (F) Combined graphic representation of panels D-E.
The implications of ABCB1 on the IUR of either drug achieved in the 14-C assay: cell line analysis
ABCB1 has been implicated as an imatinib transporter.21-25 To assess the contribution of this transporter to IUR, assays were performed in the presence and absence of a known ABCB1 inhibitor. PSC833 is a nonimmunosuppressive analog of cyclosporine, which is a potent inhibitor of ABCB1.54,55 Expression of ABCB1 in CEM-VBL100 was demonstrated by flow cytometry. Using this line and parental CEM (non-ABCB1 expressing) cells, we show that the IUR of imatinib is reduced in VBL cells when compared with CEM (CEM 32, VBL100 10 ng/200 000 cells). The addition of PSC833 resulted in a reversal of the reduced IUR in VBL100 cells (25 ng/200 000 cells), confirming the involvement of ABCB1.
In contrast, there was no difference between the IUR of nilotinib in CEM when compared with VBL100 cell lines (CEM 35, VBL100 38 ng/200 000 cells), suggesting that nilotinib may not be a substrate for ABCB1. In addition, PSC833 resulted in a decreased IUR of [14C]-nilotinib in all assays (18 ng/200 000 cells). While this finding is unexpected, it further indicates that in contrast to imatinib, nilotinib is unlikely to be a substrate for ABCB1.
The role of efflux on the IUR of either drug achieved in the 14-C assay: primary cells
In addition to ABCB1, ABCG2 has been proposed as a transporter of imatinib.29-33 MNCs from 10 patients were analyzed immediately prior to the IUR assay for expression of ABCB1 and ABCG2 using flow cytometry. No measurable expression of either protein, above that of isotype-matched controls, was seen in any patient (median ABCB1: 0.3%, range: 0-1.9%; median ABCG2: 0.1%, range: 0%-2.5%). Using limiting-dilution experiments of positive cell lines, we have demonstrated sensitivity of this assay to the 1% level. This suggests that in this cohort, expression of these efflux proteins is not a feature of blood MNCs from CML patients at presentation prior to imatinib treatment.
Using [14C]-imatinib, the IUR has been examined in the presence and absence of the ABCB1 inhibitor PSC833 and the ABCG2 inhibitor FTC. In keeping with the flow cytometry results, there was no significant difference in the IUR in the presence or absence of either inhibitor when compared with the standard assay (P > .05) (Table 1). Identical IUR assays were then performed on the same 10 patients using [14C]-nilotinib. While there was no significant difference in IUR when FTC was used as an inhibitor, when PSC833 was included there was, as observed unexpectedly in the cell lines, a statistically significant reduction in the IUR in all patients (P < .05) (Table 1).
Discussion
Imatinib mesylate is the current standard of care for chronic-phase CML patients. Results from clinical trials such as IRIS,10 where 80% of patients achieved complete cytogenetic remission, have mandated this practice. While this response is significantly higher than that achieved with previous therapies, implicit in this is that up to 20% of patients have suboptimal response to imatinib. The reason for this varied response in chronic-phase patients remains unclear; however, we have recently demonstrated that intrinsic sensitivity to kinase inhibition at the time of presentation is a predictor of outcome.13
In support of our previous findings, a recent publication has also demonstrated that the imatinib-induced reduction in the total phosphotyrosine levels in CD34+ cells collected from CML patients at presentation is also predictive of outcome.56 These in vitro studies indicate that the variation in response observed in the clinic is a result of varying sensitivities to imatinib-induced BCR-ABL kinase inhibition, and that the extent of kinase inhibition achieved is a major determinant of imatinib response. The underlying reason for this variation in sensitivity to kinase inhibition between patients is therefore the focus of these current studies.
Using [14C]-imatinib, and identical assay conditions to that used for the IC50imatinib assay, we now demonstrate significant correlation between the level of kinase inhibition achieved in vitro and the level of uptake and retention (IUR) of imatinib achieved. This finding suggests that interpatient variation in in vitro sensitivity is mainly mediated by variations in the uptake and retention of imatinib over a 2-hour period. This is a significant finding implicating influx and efflux processes in determining intrinsic imatinib sensitivity.
Previous studies suggest that imatinib transport may be mediated via the OCT proteins, which are members of the solute carrier (SLC) superfamily.24,43 To investigate the contribution of the OCT proteins, assays were performed in the presence of the known inhibitors, prazosin and procainamide. The pattern of inhibition observed in these studies suggests that OCT-1 is the key influx mechanism for imatinib, in keeping with previous reports.24,43 The addition of both prazosin and procainamide removed the difference in IUR measured in the low and high IC50imatinib groups, suggesting, importantly, that OCT-1 mediates the difference between low and high IC50imatinib and, by inference, the difference in sensitivity to imatinib-induced BCR-ABL kinase inhibition.
Furthermore, the addition of prazosin, at concentrations far in excess of the IC50 for the drug,53 did not completely inhibit the intracellular uptake of imatinib. The ability of inhibitors to normalize imatinib uptake between patients suggests that in addition to the OCT-1–mediated mechanisms that demonstrate interpatient variability, there is an underlying uptake of drug that is unrelated to the OCTs and varies little between patients. This secondary uptake mechanism is unlikely to contribute to the variability in intrinsic sensitivity of individual patients. While we would speculate that this mechanism may indeed be passive, an additional active pathway cannot be excluded.
In previous studies, Thomas et al24 demonstrated varying levels of OCT-1 mRNA expression in 4 chronic-phase and 2 acceleratedphase CML patients, but this was not linked to clinical outcome and functional uptake assays were not performed. Crossman et al43 demonstrated a significant difference in the mRNA expression level of this protein between cytogenetic responders (CyRs) and nonresponders (CyNRs). Our current study differs from the study by Crossman et al43 as in their study most patients were assessed after an extensive period of prior therapy. Furthermore, in our study all 25 patients achieved CCyR by 12 months of therapy, hence all patients would fall into the CCyR group of Crossman et al. We have studied mRNA expression levels using RQ-PCR in 19 patients (data not shown) where mRNA was available for analysis. In these patients, we found a trend between the level of expression of OCT-1 and the IUR of imatinib, but in this small patient cohort this trend failed to reach statistical significance (R2 = 0.427, P = .074). We are currently examining a larger patient cohort, and also monitoring changes in OCT-1 mRNA expression throughout the imatinib treatment course. As the influx activity of OCT-1 represents a composite of protein amount and function,41,57 it is probable that an assay measuring the actual influx activity of OCT-1 may be more informative than a measure of the level of OCT-1 mRNA.
Multidrug resistance mediated by efflux pumps is implicated as a cause of chemotherapy failure in many leukemias.16 To investigate the contribution of the major efflux pumps, ABCB1 and ABCG2, on IUR, patients were screened for antigen expression using flow cytometry. In addition, the IUR imatinib assay was performed in the presence and absence of PSC833 (inhibitor of ABCB1) and FTC (inhibitor of ABCG2). Flow analysis performed in the ABCB1-expressing cell line CEM-VBL100 confirmed expression of the efflux pump. IUR assays demonstrate that imatinib is a substrate for ABCB1 and validate the ability of this assay to detect efflux in expressing lines. In patient samples, no discernable antigen expression above that observed in isotype-matched controls was observed before therapy. Furthermore, there was no significant difference in IUR imatinib in the presence of either inhibitor. These findings indicate that unlike the influx pumps, drug efflux is unlikely to contribute to the variability in IC50imatinib observed in this cohort. Taken together, these data further suggest that patient intrinsic sensitivity to kinase inhibition as demonstrated in the IC50imatinib assay is primarily a consequence of the contribution of OCT-1–mediated uptake.
Nilotinib is a novel, potent inhibitor of ABL kinase that is structurally related to imatinib and was rationally designed around the crystal structure of the imatinib-ABL interaction. Nilotinib has a demonstrated potency 20 to 30 times that of imatinib44-47,58,59 in cell line analysis. In keeping with this finding, we demonstrate that the IC50nilotinib is significantly lower than the IC50imatinib in the 17 patient samples tested. Despite the structural similarity in these compounds, there is no correlation between the IC50 for imatinib and nilotinib in this patient cohort. Furthermore, we have demonstrated less interpatient variability with this drug in comparison with that observed with imatinib. In a clinical context, it is significant that blood levels generally achieved are far higher than the IC50nilotinib, indicating that, unlike imatinib, achieving sufficient kinase inhibition with nilotinib to augment a sustained response is likely to be more uniformly achievable.
In direct contrast to imatinib, the IC50 for nilotinib does not appear to be related to the IUR for the drug, and there is no significant difference between the IUR in the low and high IC50nilotinib cohorts. There is no significant difference in IUR when the inhibitors prazosin and procainamide are used, indicating that neither OCT-1, OCT-2, nor OCT-3 is implicated in nilotinib influx.
Experiments with the ABCB1-expressing cell line, CEM-VBL100, and the nonexpressing CCRF-CEM parental line demonstrate no difference in [14C]-nilotinib uptake between the 2 lines, suggesting that nilotinib is not a substrate for this efflux pump. Of interest, the addition of the ABCB1 inhibitor PSC833 resulted in a uniform reduction in IUR, which reached statistical significance in the patient group overall, and also in the high IC50nilotinib cohort. This finding is unexpected and remains unexplained, but it suggests that nilotinib transport is not totally passive and that it might involve some active transport mechanism. Speculatively, there are several possible explanations for this observation: First, PSC833 may directly compete with nilotinib for uptake, resulting in less of the drug being taken into the cell. Alternatively, PSC833 might down-regulate or otherwise impair the nilotinib uptake mechanism. Third, PSC833 might complex with nilotinib, to result in either reduced influx or, as PSC833 is a known substrate of ABCB1,60 in facilitated transport of nilotinib out of the cell via this efflux pump. Further experimentation designed to address these hypotheses is ongoing.
We have demonstrated that the IUR for imatinib is a critical determinant of the IC50imatinib, which we have previously shown to be predictive of molecular outcome to 12 months in CP CML patients treated with imatinib. We now demonstrate that in a cohort of patients, IUR imatinib, like IC50imatinib, is predictive of molecular response to 12 months. Follow-up studies on a larger patient cohort are currently being undertaken. Furthermore, we now confirm previous suggestions24,43 that imatinib is a substrate for the organic cation transporter OCT-1, within which polymorphisms have been shown to affect substrate translocation.61 Using an overexpressing cell line, we show that imatinib is also a substrate for ABCB1, but demonstrate that this is not a significant contributor to IUR in imatinib-naive patients. In contrast, in this series of patients, we have shown that nilotinib is between 5 and 50 times more potent than imatinib, and that there is no correlation between the IC50imatinib and the IC50nilotinib. This is a particularly important finding in the clinical setting, indicating that a predicted poor response to imatinib does not correlate with the prediction of a poor response to nilotinib. Furthermore, the IC50nilotinib does not correlate with IUR for nilotinib, and nilotinib is not a substrate for the major OCT proteins or ABCB1.
Although some weak bases and uncharged compounds are transported by OCTs, the majority of substrates translocated by these transporters are organic cations.43 Consequently, it is likely that imatinib is recognized by OCT-1 in the cationic form, resulting from the high degree of protonation of the basic N-methylpiperazine group at physiologic pH. In contrast, nilotinib lacks this basic group and, in place, possesses an N-arylimidazole that is relatively weakly basic. Therefore, nilotinib exists in the cationic state to a far lesser extent than does imatinib, which greatly reduces its likelihood to be recognized by OCTs as a substrate for influx into cells.
In conclusion, differential expression or function of OCT-1 appears to be a significant determinant of patient response to imatinib mesylate. Whether these differences are expression or functionally related has yet to be determined; however, the outcome of either of these effects is clearly demonstrable in the rapid screening IUR and IC50 assays. In contrast, nilotinib displays marked differences in influx and efflux patterns, which does not involve OCT proteins. Based on our findings in this patient cohort, it is likely that response to nilotinib in CML patients at presentation will be more uniform than those responses observed with imatinib. These findings may also be relevant to nilotinib activity against c-Kit– and PDGF-mediated diseases. In these cases, potency of imatinib and nilotinib is similar,46 and more uniform uptake of nilotinib may provide a critical advantage over imatinib.
Prepublished online as Blood First Edition Paper, April 4, 2006; DOI 10.1182/blood-2005-11-4687.
Supported in part by a grant from the Cancer Council of Australia, with support from Novartis Australia.
Two of the authors (S.R.Q. and P.W.M.) are employed by a company (Novartis Pharma Ltd) whose product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ms Stephanie Zrim for maintaining all cell lines used in these studies and Ms Amity Venables for performing preliminary fluorescence-activated cell sorter (FACS) analysis. We thank Dr Susan Branford and Ms Chani Fields for provision of OCT-1 primers for mRNA analysis, and Dr Peter Diamond for his considerable expertise and support in performing RT-PCR assays. We would also like to thank Dr Kevin Lynch and Novartis Australia for their continued support.