Mantle cell lymphoma (MCL) is a lymphoproliferative disorder with distinct pathologic and clinical features. Genetically, the vast majority of MCLs carry the t(11;14)(q13;q32). This characteristic chromosomal abnormality leads to deregulation of the CCND1 gene encoding cyclin D1 at 11q13 through juxtaposition with the immunoglobulin heavy chain (IGH) locus at 14q32. By gene expression profiling, 6 cases of MCL that lacked cyclin D1 mRNA expression but had a gene expression signature typical of MCL have recently been described.1 These cyclin D1–negative MCLs exhibited the characteristic morphologic, immunohistochemical, and clinical features of cyclin D1–positive MCL. In line with the lack of cyclin D1 mRNA expression, cyclin D1 protein was absent in these cases. Moreover, the cases did not have the t(11;14) by fluorescence in situ hybridization (FISH) analysis. Interestingly, all 6 cyclin D1–negative MCLs expressed either cyclin D2 or D3, suggesting that these cyclins can substitute functionally for cyclin D1 in MCL.1 Nevertheless, chromosomal aberrations affecting the CCND2 or CCND3 gene loci were not detectable in these cases by interphase FISH.
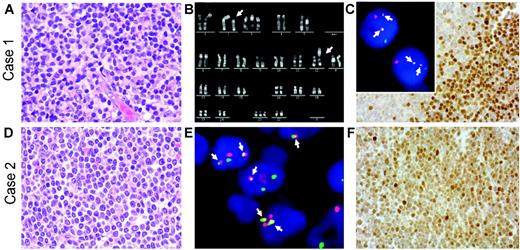
Herein, we present 2 cases of cyclin D1–negative MCL in which cyclin D2 protein expression was associated with a chromosomal translocation juxtaposing the CCND2 gene at chromosomal band 12p13 next to the IGK locus at 2p12. The lymphomas occurred in a 33-year-old woman and a 70-year-old man. Both were diagnosed with stage IV disease with bone marrow involvement. Diagnostic lymph node biopsies showed the typical morphology of MCL with a nodular and diffuse growth pattern (Figure 1A,D). Both lymphomas displayed the immunophenotype typical for MCL (CD5+, CD20+, CD10–, and CD23–), except for lack of cyclin D1 expression. Absence of the t(11;14) or variant CCND1 breaks was proven in both cases by FISH using commercially available probes (LSI IGH/CCND1, LSI CCND1 BAP; Vysis, Downers Grove, IL). Conventional cytogenetic R-banding analysis in the first case revealed the tumor cells to have the karyotype 48,XX,t(2;12)(p12; p13),+3,+21[11] (Figure 1B). Subsequent FISH using recently described probes1,2 showed that the t(2;12) caused breaks in the IGK and CCND2 loci, resulting in IGK-CCND2 fusion (Figure 1C insert). In the second case, metaphase analysis was not possible due to lack of suitable material. Nevertheless, interphase FISH showed breakpoints in the CCND2 and IGK loci, as well as IGK-CCND2 fusion (Figure 1E). Immunohistochemical analyses revealed strong nuclear expression of cyclin D2 protein in the majority of the tumor cells in both cases (Figure 1C,F). According to FISH analysis, there was no evidence for breaks affecting the CCND3, CCNE1, or CCNE2 loci in these cases. Cyclin D1, D3, and E expression was only detectable in a few scattered cells, suggesting expression in a minority of tumor cells or bystander cells in both cases (data not shown).
Our findings indicate that cyclin D2–positive MCL can arise through juxtaposition of the CCND2 and IGK loci. Remarkably, search of the Mitelman Database of Chromosome Aberrations in Cancer3 revealed only one additional lymphoma with the t(2;12)(p12;p13), which was also diagnosed as typical MCL.4 Unfortunately, gene expression profiling was not available in the cases presented here. Nevertheless, the typical morphologic and immunophenotypic features of our cases indicate that rare cyclin D1–negative MCLs exist, in which deregulation of cyclin D2 due to a t(2;12)(p12;p13) substitutes for t(11;14)–driven cyclin D1 expression.
Histologic, immunophenotypic, and cytogenetic findings in 2 cyclin D1–negative/cyclin D2–positive MCLs withIGK-CCND2fusion. (A,D) Hematoxylin and eosin (H&E)–stained lymph node biopsies of cases 1 (A) and 2 (D) showing the typical morphology of MCL. (B) Chromosome R-banding analysis of case 1 revealing the karyotype 48,XX,t(2;12)(p12;p13),+3,+21. (C insert, E) Interphase FISH analysis using double-color, double-fusion probes spanning the IGK (green) and CCND2 (red) loci.1,2 Both cells in case 1 (C insert) and several cells in case 2 (E) show 2 fusion (yellow, arrows) signals indicating IGK-CCND2 juxtaposition in addition to isolated red and green signals indicating intact CCND2 and IGK loci. (C,F) Cyclin D2 nuclear expression in the tumor cells detected by immunohistochemistry using a polyclonal anti–cyclin D2 antibody, as described recently.1 H&E images were visualized under a Zeiss Axioskop 40 microscope (Carl Zeiss, Jena, Germany) equipped with a 40 ×/0.75 objective lens and an RT Slider camera (Diagnostic Instruments, Sterling Heights, MI). Images were processed with MetaVue software (Diagnostic Instruments) and Adobe Photoshop (Adobe Systems, San Jose, CA). FISH images were acquired with a 63 ×/1.40 oil-immersion objective in a Zeiss Axioskop2 fluorescence microscope (Zeiss, Göttingen, Germany) equipped with the appropriate filter sets (AHF, Tübingen, Germany), and were documented and processed using the ISIS imaging system (MetaSystems, Altlussheim, Germany).
Histologic, immunophenotypic, and cytogenetic findings in 2 cyclin D1–negative/cyclin D2–positive MCLs withIGK-CCND2fusion. (A,D) Hematoxylin and eosin (H&E)–stained lymph node biopsies of cases 1 (A) and 2 (D) showing the typical morphology of MCL. (B) Chromosome R-banding analysis of case 1 revealing the karyotype 48,XX,t(2;12)(p12;p13),+3,+21. (C insert, E) Interphase FISH analysis using double-color, double-fusion probes spanning the IGK (green) and CCND2 (red) loci.1,2 Both cells in case 1 (C insert) and several cells in case 2 (E) show 2 fusion (yellow, arrows) signals indicating IGK-CCND2 juxtaposition in addition to isolated red and green signals indicating intact CCND2 and IGK loci. (C,F) Cyclin D2 nuclear expression in the tumor cells detected by immunohistochemistry using a polyclonal anti–cyclin D2 antibody, as described recently.1 H&E images were visualized under a Zeiss Axioskop 40 microscope (Carl Zeiss, Jena, Germany) equipped with a 40 ×/0.75 objective lens and an RT Slider camera (Diagnostic Instruments, Sterling Heights, MI). Images were processed with MetaVue software (Diagnostic Instruments) and Adobe Photoshop (Adobe Systems, San Jose, CA). FISH images were acquired with a 63 ×/1.40 oil-immersion objective in a Zeiss Axioskop2 fluorescence microscope (Zeiss, Göttingen, Germany) equipped with the appropriate filter sets (AHF, Tübingen, Germany), and were documented and processed using the ISIS imaging system (MetaSystems, Altlussheim, Germany).
Supported from grants of the Deutsche Krebshilfe (70-3173-Tr3), the Lymphoma Research Foundation (New York), and the European Union (European MCL Network, LSHC-CT-2004-503351).