Abstract
Aberrant expression of 1 or more transcription factor oncogenes is a critical component of the molecular pathogenesis of human T-cell acute lymphoblastic leukemia (T-ALL); however, oncogenic transcriptional programs downstream of T-ALL oncogenes are mostly unknown. TAL1/SCL is a basic helix-loop-helix (bHLH) transcription factor oncogene aberrantly expressed in 60% of human T-ALLs. We used chromatin immunoprecipitation (ChIP) on chip to identify 71 direct transcriptional targets of TAL1/SCL. Promoters occupied by TAL1 were also frequently bound by the class I bHLH proteins E2A and HEB, suggesting that TAL1/E2A as well as TAL1/HEB heterodimers play a role in transformation of T-cell precursors. Using RNA interference, we demonstrated that TAL1 is required for the maintenance of the leukemic phenotype in Jurkat cells and showed that TAL1 binding can be associated with either repression or activation of genes whose promoters occupied by TAL1, E2A, and HEB. In addition, oligonucleotide microarray analysis of RNA from 47 primary T-ALL samples showed specific expression signatures involving TAL1 targets in TAL1-expressing compared with -nonexpressing human T-ALLs. Our results indicate that TAL1 may act as a bifunctional transcriptional regulator (activator and repressor) at the top of a complex regulatory network that disrupts normal T-cell homeostasis and contributes to leukemogenesis.
Introduction
TAL1/SCL (hereafter referred to as TAL1) is a basic helix-loop-helix (bHLH) transcription factor that is required for normal hematopoiesis,1,2 and whose aberrant expression leads to T-cell acute lymphoblastic leukemia (T-ALL). TAL1 is expressed by the leukemic cells of 60% of patients with T-ALL3,4 as a result of chromosomal translocations or intrachromosomal rearrangements leading to its monoallelic expression, as well as by unknown mechanisms leading to biallelic up-regulation in double-positive thymocytes.5,6 According to the prevailing model of TAL1-induced leukemogenesis, TAL1 acts as a transcriptional repressor through heterodimerization with E2A and HEB, leading to a block of the transcriptional activity of these class-I bHLH factors.7-12 However, transcriptional activation of the RALDH2 gene by TAL1 has also been described suggesting a more complex effect on gene regulation.13 Despite the importance of transcriptional programs downstream of TAL1, the collection of genes directly regulated by TAL1 is mostly unknown. Although TAL1 targets have been reported in the context of early hematopoietic development (KIT),14 red-cell differentiation (GPA and P4.2),15,16 T-cell development (pTA is a likely target of TAL1),17-19 or leukemia (RALDH2),13 none of them has elucidated the regulatory roles that TAL1 plays in the pathogenesis of T-ALL. The identification of a more comprehensive set of genes regulated by TAL1 will lead to improved understanding of the transcriptional role of TAL1 and its regulation circuits that control cell proliferation, differentiation, and apoptosis during T-cell development.
Here, we elucidated the regulatory circuitry regulated by TAL1 in T-ALL using a combination of complementary genome-scale analysis techniques. To identify regions in the genome directly occupied by TAL1 in vivo, we combined chromatin immunoprecipitation and custom-made promoter microarrays (ChIP on chip).20-24 This analysis was combined with TAL1 knockdown by RNA interference (RNAi) and gene-expression profiling in primary samples using oligonucleotide microarrays to analyze the mechanisms of TAL1 transformation on a genomewide scale. Our results support that TAL1 may function both as repressor and as activator of direct target genes whose promoters are also bound by E2A and HEB. We also demonstrate that several of the genes whose promoters are occupied by TAL1 in a T-ALL cell line are also specifically associated with the expression of this transcription factor in human primary leukemias. Our results suggest that transcriptional effects downstream of the aberrant expression of TAL1 in T-cell progenitors are amplified in a complex transcriptional network that results in the disruption of critical mechanisms that control cell homeostasis during thymocyte development.
Materials and methods
Human cell lines
The T-ALL Jurkat cell line clone E6-1 was obtained from the American Type Culture Collection (ATCC; Manassas, VA) and was grown in RPMI media with 10% fetal bovine serum (FBS) in a humidified 5% CO2 atmosphere at 37°C. The EBNA packaging cell line was obtained from ATCC. EBNA cells were grown in Dulbecco modified Eagle medium (DMEM) with 10% FBS in 5% CO2 at 37°C.
RNAi constructs
An RNAi control sequence was obtained from Qiagen (Valencia, CA). The RNAi sequence against TAL1 (nt 879-897) was described in Lazrak et al.25 Both sequences were cloned into the BglII/HindIII sites of the pSuperior (Oligoengine, Seattle, WA) retroviral vector.
Generation of retrovirus
Recombinant retroviruses were produced using the EBNA packaging cell line that was transfected with the pSuperior construct containing the control RNAi sequence or sequence directed against TAL1 along with plasmids encoding the gagpol and VSVg genes. The viral supernatant was collected 48 hours after transfection and filtered through a 0.45-μM filter.
Generation of Jurkat-cell clones with RNAi-induced TAL1 knockdown
Jurkat cells (3 × 106 cells) were infected with 2 mL of viral supernatant in the presence of 9 μg/mL polybrene. Forty-eight hours after infection, the cells were selected in media containing 1 μg/mL puromycin. Oligoclonal cell lines were generated by plating 5 cells/well in 96-well plates. The level of TAL1 knockdown was assessed by Western blot using a mouse monoclonal TAL1-specific antibody BTL73 (generously provided by Dr K. Pulford, Oxford University, United Kingdom).26 To ensure equal loading, the blot was stripped and reprobed with an antibody specific for α-tubulin (Sigma, St Louis, MO).
Total RNA preparation and real-time PCR analysis
Total RNA was extracted using the RNAqueous kit (Ambion, Austin, TX) following the manufacturer's instructions. cDNA was generated using 2 μg RNA and the ThermoScript reverse transcriptase–polymerase chain reaction (RT-PCR) System (Invitrogen, Carlsbad, CA). The cDNA was then analyzed by quantitative PCR using the Sybr Green RT-PCR Core Reagents kit (Applied Biosystems, Foster City, CA). The primer sequences of the oligonucleotides used for RT-PCR are available upon request.
Growth and apoptosis assays
Cell-growth ratios were determined by a colorimetric assay using the Cell Proliferation Kit I (MTT) (Roche, Indianapolis, IN). Briefly, triplicate samples corresponding to 2 clones expressing the control shRNA and 2 clones expressing TAL1 shRNA were plated at a density of 5 × 104 cells/mL. Samples were taken every day and tested using the MTT assay following the manufacturer's protocol. Apoptosis was quantified using the Annexin V: FITC Apoptosis Detection Kit I (BD-Pharmingen, San Diego, CA) following the manufacturer's protocol in 2 Jurkat-cell clones expressing the control shRNA and 2 clones expressing TAL1 shRNA.
Chromatin immunoprecipitation reagents
TAL1 no. 370 antibodies were generously provided by Dr Richard Baer from Columbia University (New York, NY). Typical chromatin immunoprecipitation (ChIP) reactions used 50 μL antisera per experiment, in combination with 100 μL of a protein G magnetic bead suspension obtained from Dynal (Oslo, Norway). Antiserum no. 370 was raised against GST-TAL1,238-331 a fusion protein containing the carboxy-terminal 94 residues of TAL19 ; it recognizes both p42 TAL1 and p22 forms of TAL1.27
E2A was immunoprecipitated for most of the experiments reported here using a rabbit polyclonal antibody28 and verified using a monoclonal antibody G98-271 (BD-Pharmingen) against human E2A that recognizes specifically human E12 and E47, the 2 alternative spliced products of the E2A gene. HEB was immunoprecipitated using Santa Cruz Biotechnology antibody sc-357. Antibody (10 μg) was used per chromatin immunoprecipitation.
ChIP on chip
Complete protocols are available for download in pdf format at http://web.wi.mit.edu/young/TAL1 or http://research.dfci.harvard.edu/looklab/publications/TAL1. The protocol used here was adapted from Odom et al.21 Briefly, cells are fixed with 1% final concentration formaldehyde for 10 to 20 minutes at room temperature, harvested, and rinsed with 1 × phosphate-buffered saline (PBS). The resultant cell pellet is sonicated, and DNA fragments that are crosslinked to a protein of interest are enriched by immunoprecipitation with a factor-specific antibody. After reversal of the crosslinking, the enriched DNA is amplified using ligation-mediated PCR (LM-PCR), and then fluorescently labeled using high-concentration Klenow polymerase and a dNTP-fluorophore. A sample of DNA that has not been enriched by immunoprecipitation is subjected to LM-PCR and labeled with a different fluorophore. Both IP-enriched and unenriched pools of labeled DNA are hybridized to a single DNA microarray containing 13 000 human intergenic regions (for description of microarray design, see http://web.mit.wi.mit.edu/young/TAL1 and http://research.dfci.harvard.edu/looklab/publications/TAL1).
For Jurkat cell line experiments, 5 × 107 to 1 × 108 cells were typically used per chromatin immunoprecipitation. The cells were crosslinked with 1% formaldehyde solution for 10 minutes, rinsed with PBS, and flash-frozen.
ChIP hybridization quality control
The raw data generated from each array experiment were subjected to multiple levels of quality control. First, each scan was examined visually as it was being performed. Samples on microarrays with gross defects (eg, scratches, smeared spots) were repeated. We also determined that no reliable signal was produced from control spots containing Arabidopsis DNA.
Binding site determination and error model
Scanned images were analyzed using GenePix (v3.1 or v4.0; Molecular Devices, Sunnyvale, CA) to obtain background-subtracted intensity values. Each spot was hybridized by both IP-enriched and unenriched DNA, which were labeled with different fluorophores. Consequently, each spot yielded fluorescence intensity information in 2 channels, corresponding to immunoprecipitated DNA and genomic DNA. To account for background hybridization to slides, the median intensity of a set of control blank spots was subtracted for site-specific transcription factors (TAL1). To correct for different amounts of genomic and immunoprecipitated DNA hybridized to the microarray, the median intensity value of the IP-enriched DNA channel was divided by the median of the genomic DNA channel, and this normalization factor was applied to each intensity in the genomic DNA channel. Next, we calculated the log of the ratio of intensity in the IP-enriched channel to intensity in the genomic DNA channel for each intergenic region across the entire set of hybridization experiments. Adjusted intensity values for the IP-enriched channel were calculated from these ratios. A whole-chip error model29,30 was then used to calculate confidence values for each spot on each microarray, and to combine data for the replicates of each experiment to obtain a final average ratio and confidence for each promoter region. Raw promoter array scanning data as well as more detailed information about data interpretation models can be found at http://web.mit.wi.mit.edu/young/TAL1 or http://research.dfci.harvard.edu/looklab/publications/TAL1.
Gene-expression profiling in primary T-ALL samples
Microarray analysis of 45 primary leukemia specimens was performed on diagnostic lymphoblast samples obtained from the Pediatric Oncology Group (POG) tumor bank. All samples were obtained with informed consent and were analyzed under the supervision of the University of New Mexico institutional review board. Gene-expression profiling using oligonucleotide microarrays was performed using Affymetrix U133 arrays (Santa Clara, CA) following standard procedures, and interarray intensity differences were normalized with Dchip (http://Biosun1.harvard.edu/complab/dchip/). Quantitative RT-PCR analysis on T-ALL major oncogenes, including HOX11, HOX11L2, TAL1, LYL1, TAL2 and BHLHB1, was performed as previously described.3 Nearest-neighbor analysis of genes associated with the expression of TAL1 was performed with Genecluster 2.0 (www.broad.mit.edu/cancer/software/genecluster2/gc2).
Results
Identification of TAL1 bound promoters by ChIP on chip
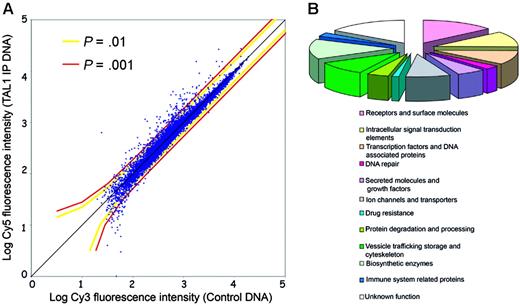
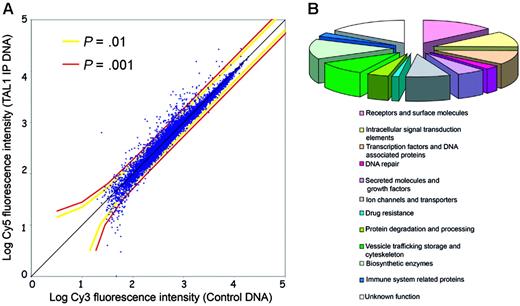
To identify direct target genes regulated by TAL1 in T-ALL, we performed ChIP on chip in Jurkat cells using a polyclonal antibody that recognizes multiple TAL1 isoforms and a custom genomic microarray containing 5′ proximal promoter regions of 13 000 human genes (Hu13K array).21 For this manuscript, the term of direct target includes all promoters that are bound by TAL1, either by direct interaction with the DNA15 or indirectly as a member of a DNA-binding complex, as TAL1 mutants defective in DNA binding are still able to regulate transcription of certain target genes.13 The experiment was performed in triplicate and analyzed to identify promoters that are significantly bound in vivo by TAL1; see Figure 1A, as well as Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). We identified 71 genes whose promoters showed significant binding of TAL1 compared with the chromatin control (P < .001 in triplicate experiments). Classification of these TAL1 direct target genes according to Gene Ontology categories showed that they belong to multiple functional groups (Figures 1B, 2). TAL1 target genes include genes involved in pathways relevant to the disruption of T-cell development and to the pathogenesis of T-ALL, including T-cell differentiation (transcription factor 7 [TCF7]), proliferation (ephrin receptor EphB1 [EPHB1]), and apoptosis (nuclear receptor 4A3 [NR4A3/NOR1] and apoptosis antagonizing transcription factor [AATF/DED]). TAL1-direct targets previously described in T-ALL—RALDH2 and CD4—escaped detection by ChIP on chip on the Hu13K array. Indeed, TAL1 binds to RALDH2 an intronic sequence13 while the binding to CD4 occurs in upstream enhancer regions,8 both not present on the Hu13K promoter array (Table S1).
Identification of TAL1 direct target genes by ChIP on chip in Jurkat cells. (A) Scatterplot representation of a genomic microarray (Hu13K) hybridization with a chromatin immunoprecipitation performed using a TAL1 antibody. The x-axis indicates the log intensity of the fluorescence of the control DNA, labeled with Cy3, while the y-axis represents the log fluorescence intensity of the TAL1 chromatin immunoprecipitates (TAL1-IP) labeled with Cy5. Each dot represents a promoter region from the Hu13K arrays. The colored lines represent the P values calculated by the error model applied to the fluorescence data. (B) Functional classification of the 71 TAL1 targets according to Gene Ontology and/or National Center for Biotechnology Information (NCBI) data for the identified genes. Note the broad spectrum of functional categories of genes regulated by TAL1.
Identification of TAL1 direct target genes by ChIP on chip in Jurkat cells. (A) Scatterplot representation of a genomic microarray (Hu13K) hybridization with a chromatin immunoprecipitation performed using a TAL1 antibody. The x-axis indicates the log intensity of the fluorescence of the control DNA, labeled with Cy3, while the y-axis represents the log fluorescence intensity of the TAL1 chromatin immunoprecipitates (TAL1-IP) labeled with Cy5. Each dot represents a promoter region from the Hu13K arrays. The colored lines represent the P values calculated by the error model applied to the fluorescence data. (B) Functional classification of the 71 TAL1 targets according to Gene Ontology and/or National Center for Biotechnology Information (NCBI) data for the identified genes. Note the broad spectrum of functional categories of genes regulated by TAL1.
We confirmed the ChIP on chip results by performing chromatin immunoprecipitation coupled to quantitative PCR for each of the 71 promoters bound on the arrays. Our results verified the binding of TAL1 to 57 of 71 promoters (80% of the identified targets; Figure 2), which is in agreement with validation rates previously reported for ChIP on chip results using similar platforms.21
Further analysis of the promoters with TAL1 binding by ChIP on chip indicated that approximately 95% (64 of 71) contained E-boxes and that approximately 85% (60 of 71) contained GATA boxes in the proximal promoter sequence. When analyzed specifically for CAGATG, which is the previously described E-box motif preferred by TAL1, 24% (17 of 71) of the TAL1 targets contained this sequence in their promoters. There is a weak enrichment (1.14-fold change) for the CATATG motif in the TAL1 targets compared with all the promoters on the entire Hu13K array, although this does not represent a significant enrichment (1-tailed binomial test P = .318). None of the TAL1-occupied promoters showed a prototypical E-box–GATA motif described by Wadman and colleagues as associated with transcriptional activation by TAL1.31 Only the gene encoding for nicotinic cholinergic receptor alpha 5 (CHRNA5) contained a closely related motif (CAGGTGGGTTTCCGATA) in the proximal promoter region.
A subset of promoters bound by TAL1 in Jurkat cells were randomly selected and assessment of TAL1 binding was performed in MOLT4, a well-characterized T-cell line. This ChIP assay analysis verified TAL1 binding in 3 of 19 promoters tested (RALDH2, AP4B1, and LOC51184), even in the context of a cell line (MOLT4) expressing much lower levels of TAL1 protein than Jurkat cells. This result strongly suggests that at least a fraction of the targets identified by TAL1 in Jurkat cells are likely to be regulated in a broader spectrum of T-cell leukemias.
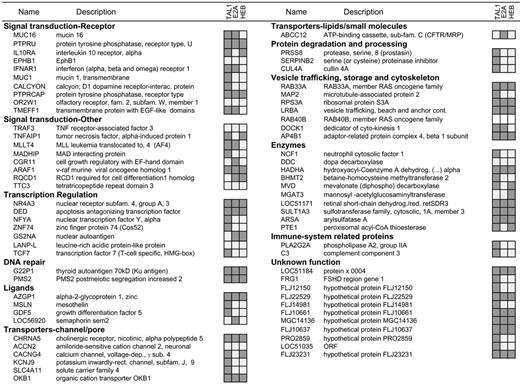
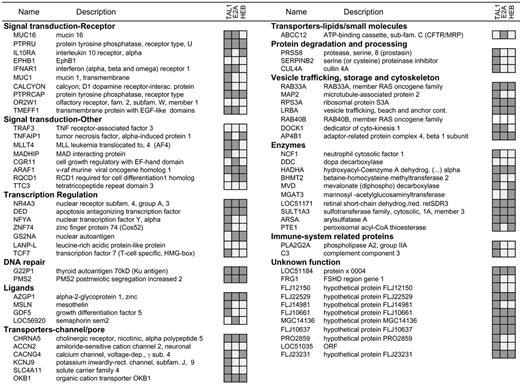
Analysis of TAL1, E2A, and HEB binding to human promoters. Chromatin immunoprecipitation experiments in Jurkat cells were performed with antibodies raised against TAL1, E2A, or HEB. Relative binding to each of the 71 promoters previously identified as TAL1 targets by ChIP on chip was analyzed by gene-specific quantitative PCR. Promoters marked in dark gray were enriched in the chromatin immunoprecipitates compared with those of the control genomic DNA; no enrichment was detected in the immunoprecipitates marked in light gray, while the empty boxes for RAB40B indicates that no suitable primers were identified to perform the quantitative PCR analysis.
Analysis of TAL1, E2A, and HEB binding to human promoters. Chromatin immunoprecipitation experiments in Jurkat cells were performed with antibodies raised against TAL1, E2A, or HEB. Relative binding to each of the 71 promoters previously identified as TAL1 targets by ChIP on chip was analyzed by gene-specific quantitative PCR. Promoters marked in dark gray were enriched in the chromatin immunoprecipitates compared with those of the control genomic DNA; no enrichment was detected in the immunoprecipitates marked in light gray, while the empty boxes for RAB40B indicates that no suitable primers were identified to perform the quantitative PCR analysis.
TAL1 binds to its target promoters in the presence of E2A and HEB
TAL1 is a class II bHLH factor that requires heterodimerization with class I bHLH members such as E2A (E12/E47) and HEB to achieve efficient interaction with E-box (CANNTG) DNA sequences situated in the promoter regions of its target genes.32 To further extend the ChIP on chip results, we analyzed the promoter occupancy of E2A and HEB in this set of TAL1-direct targets in Jurkat T-ALL cells. E2-2, another member of the E-protein family of transcription factors that had been previously described as interacting with TAL1,33 was not expressed at significant levels in Western blot analysis of Jurkat cells (data not shown) and therefore was not tested by ChIP. Chromatin immunoprecipitation using antibodies that recognize both alternative splice forms of E2A (E12 and E47) or HEB, followed by quantitative gene-specific PCR, demonstrated specific association of E2A, HEB, or both E2A and HEB with the promoter regions of 34 (59.6%) of the 57 verified TAL1 targets (Figure 2). These included 5 (8.7%) promoters bound by TAL1 and E2A only, 5 (8.7%) promoters bound by TAL1 and HEB only, and 24 (42.1%) promoters bound by TAL1 complexes containing both E2A and HEB. These data suggest a major role for HEB in the regulation of TAL1-direct targets, a result supported by recent reports indicating accelerated leukemia onset using a TAL1 mouse transgenic model in a HEB+/– background, which is analogous to the effect observed in an E2A+/– background.8 The identification of TAL1 targets that did not bind detectable levels of either E2A or HEB may indicate the existence of as-yetuncharacterized TAL1 transcriptional regulatory complexes in which TAL1 binds to DNA independently of E2A or HEB.
Role of TAL1 in the regulation of its direct targets
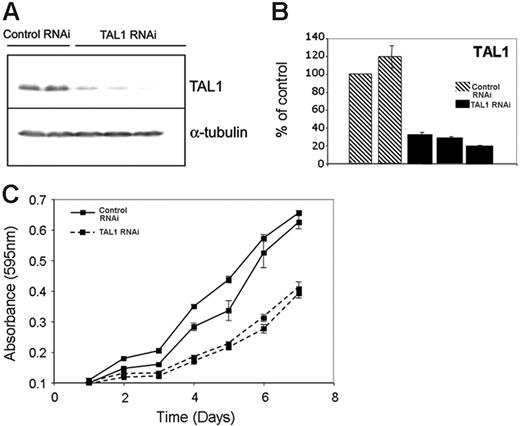
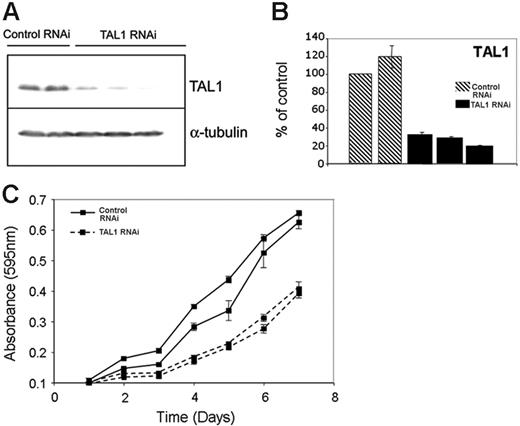
To determine the genes whose proper expression depends, directly or indirectly, on the presence of TAL1 in T-ALL cells, we performed TAL1 knockdown by RNA interference in Jurkat cells. We selected clones of Jurkat cells expressing a short-hairpin RNA (shRNA) against TAL1 and analyzed the expression of the TAL1 protein by Western blotting, choosing clones that showed more than 75% reduction at the protein level for further experiments (Figure 3A). Comparable levels of knockdown were observed for TAL1 RNA, when TAL1 shRNA–expressing clones were compared with controls by quantitative RT-PCR (Figure 3B). Decreased levels of TAL1 expression were accompanied by a diminished proliferative capacity of Jurkat cells, indicating a role for TAL1 in promoting the aberrant growth of T-ALL cells (Figure 3C). Annexin V staining did not show differences in the number of apoptotic cells in the control and TAL1 shRNA–expressing clones, indicating that apoptosis does not seem to play a role in the reduced growth rates shown by TAL1 shRNA–expressing clones (Figure S2).
TAL1 knockdown by shRNA affects growth and gene expression in Jurkat cells. (A) Stable expression of a shRNA against TAL1 in Jurkat cells led to decreased TAL1 protein levels, as measured by Western blotting using a monoclonal antibody against TAL1. The first 2 lanes show results for 2 different clones expressing control shRNAs (Control RNAi), compared with the results of 3 separate clones expressing the TAL1-specfic shRNA in lanes 3-5 (TAL1 RNAi). The membrane was stripped and reprobed with an antibody against α-tubulin to verify equal loading. (B) Quantitative RT-PCR using RNA prepared from the same Jurkat-cell clones shown in panel A that stably express a control shRNA (▧) or the TAL1 shRNA (▪) reveals 70% to 90% knockdown of TAL1 at the RNA level in TAL1 knockdown cells. Error bars indicate standard deviations. (C) TAL1 knockdown affects the growth rate of Jurkat cells. Cells (5 × 104) from clones stably expressing a control or TAL1 shRNAs were plated in RPMI with 5% FBS, and the cell numbers were measured daily using an MTT-based assay.
TAL1 knockdown by shRNA affects growth and gene expression in Jurkat cells. (A) Stable expression of a shRNA against TAL1 in Jurkat cells led to decreased TAL1 protein levels, as measured by Western blotting using a monoclonal antibody against TAL1. The first 2 lanes show results for 2 different clones expressing control shRNAs (Control RNAi), compared with the results of 3 separate clones expressing the TAL1-specfic shRNA in lanes 3-5 (TAL1 RNAi). The membrane was stripped and reprobed with an antibody against α-tubulin to verify equal loading. (B) Quantitative RT-PCR using RNA prepared from the same Jurkat-cell clones shown in panel A that stably express a control shRNA (▧) or the TAL1 shRNA (▪) reveals 70% to 90% knockdown of TAL1 at the RNA level in TAL1 knockdown cells. Error bars indicate standard deviations. (C) TAL1 knockdown affects the growth rate of Jurkat cells. Cells (5 × 104) from clones stably expressing a control or TAL1 shRNAs were plated in RPMI with 5% FBS, and the cell numbers were measured daily using an MTT-based assay.
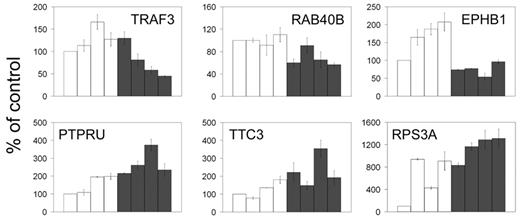
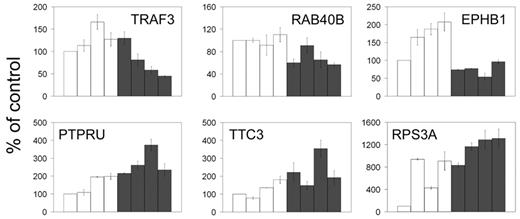
We performed quantitative RT-PCR analysis of 36 of the 71 TAL1-direct target genes in stable clones of Jurkat cells expressing an active TAL1 shRNA. The results demonstrated that TAL1 knockdown resulted in the regulation of only a fraction of the genes identified by ChIP on chip (16%). Specifically, TAL1 down-regulation results in a reduction in the transcription of 3 genes (TRAF3, RAB40B, and EPHB1) and an increase in transcription of 3 (PTPRU, TTC3, and RPS3A) using a Satterthwaite t test at a significance P level less than .05 (Figure 4), indicating a role of TAL1 as a transcriptional activator for TRAF3, RAB40B, and EPHB1, and as a repressor of PTPRU, TTC3, and RPS3A, respectively. There was no significant effect on the expression levels of the remaining 30 TAL1 direct target genes under our experimental conditions. Even though we achieved a 70% to 80% level of TAL1 knockdown, TAL1 is a very highly expressed protein in Jurkat cells, and we cannot exclude the possibility that the remaining TAL1 protein after RNA interference would still be enough to modulate the transcription of some TAL1 target genes in the knockdown cells. It is also possible that many of these targets are not functional in the cell lines we are profiling, yet are active in other tissue types. This is consistent with similar correlations between promoter binding and gene regulation for c-Jun/ATF2, CREB, and the glucocorticoid receptor addressed in several recent publications, demonstrating that promoter occupancy by these regulatory proteins influences 2% to 35% of gene expression.34-37 These findings suggest that although transcription factors may bind to multiple target genes in a given cell type, transcriptional regulation may occur only in those genes whose promoters are simultaneously occupied by a relevant set of tissue-specific transcriptional coregulators.
Expression of TAL1-direct targets in primary samples from human patients with T-ALL
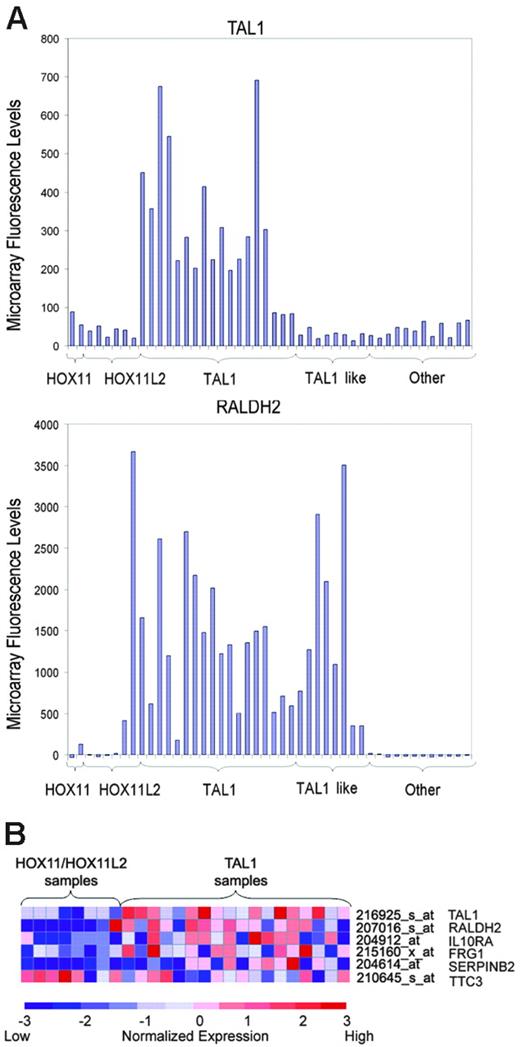
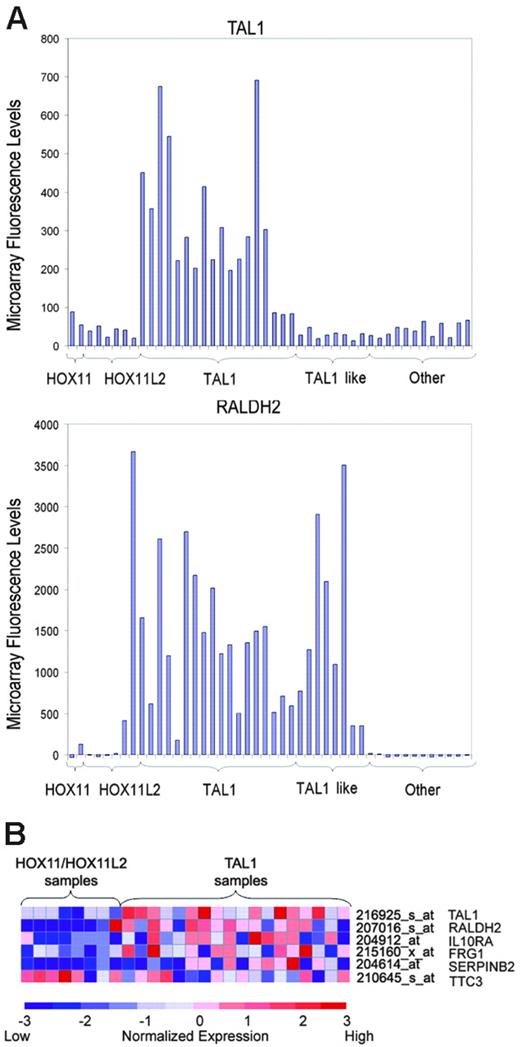
To analyze the association of TAL1 targets identified by ChIP on chip with the aberrant expression of TAL1 in primary tumor lymphoblasts, we performed supervised analysis of microarray gene-expression profiles in a panel of 40 well-characterized T-ALL samples. Patient samples were classified according to the expression of the TAL1, HOX11, HOX11L2, and LYL1 T-cell oncogenes determined by quantitative RT-PCR. In this series, aberrant expression of TAL1 was associated with high levels of RALDH2 expression (Figure 5A), supporting a role of TAL1 in the activation of this direct target gene in T-ALL. Importantly, the presence of high levels of RALDH2 was not restricted to the 16 TAL1-positive samples but was present also in 8 additional patients with T-ALL (Figure 5A). All other T-ALL samples showed lower levels of RALDH2 except for 2 atypical HOX11L2-positive samples. These results suggest that these TAL1-negative RALDH2-high cases—“TAL1-like”—may constitute a distinct group of T-ALL cases that harbor a TAL1-related oncogene(s). This observation is in agreement with our previous gene-expression profiling analysis of T-ALL patients, in which a group of TAL1-related cases were characterized by gene expression signatures highly related to those of TAL1-positive samples.3
TAL1 knockdown reveals regulation of promoters identified by ChIP on chip. Quantitative RT-PCR on RNA prepared from 4 Jurkat-cell clones stably expressing a control shRNA (□) or the TAL1 shRNA (▪) reveals that 3 of the TAL1 target genes are significantly down-regulated by TAL1 knockdown (TRAF3, RAB40B, and EPHB1; top) and 3 are up-regulated (PTPRU, TTC3, and RPS3A; bottom), reflecting genes that would be activated and repressed by TAL1, respectively. Error bars represent standard deviations of triplicate measurements (quantification replicas) normalized to GAPDH levels. The results shown are displayed as a percentage of the mean levels of the control samples. Error bars indicate standard deviations.
TAL1 knockdown reveals regulation of promoters identified by ChIP on chip. Quantitative RT-PCR on RNA prepared from 4 Jurkat-cell clones stably expressing a control shRNA (□) or the TAL1 shRNA (▪) reveals that 3 of the TAL1 target genes are significantly down-regulated by TAL1 knockdown (TRAF3, RAB40B, and EPHB1; top) and 3 are up-regulated (PTPRU, TTC3, and RPS3A; bottom), reflecting genes that would be activated and repressed by TAL1, respectively. Error bars represent standard deviations of triplicate measurements (quantification replicas) normalized to GAPDH levels. The results shown are displayed as a percentage of the mean levels of the control samples. Error bars indicate standard deviations.
Expression profile of TAL1-direct targets in primary T-ALL samples. (A) Microarray fluorescence intensities corresponding to the expression of TAL1 and RALDH2 genes in T-ALL samples. High levels of expression of RALDH2 were detected in all TAL1-positive cases, in agreement with a role of TAL1 in the activation of this direct target gene. Cases were arranged on the basis of the expression of T-ALL oncogenes, including HOX11, HOX11L2, and TAL1. TAL1-like cases denote samples with high levels of RALDH2 similar to those present in TAL1-positive samples, but lacking TAL1 expression. (B) Heat map representing relative expression levels of TAL1 and TAL1 direct target genes in HOX11/HOX11L2-positive cases and TAL1-positive samples. Each column represents 1 of 26 samples positive for HOX11, HOX11L2, or TAL1 by RT-PCR, while each shows the expression pattern of a particular gene identified as a TAL target by ChIP on chip. Relative expression levels are normalized across the samples; levels greater than or less than the mean are shown in shades of red or blue, respectively.
Expression profile of TAL1-direct targets in primary T-ALL samples. (A) Microarray fluorescence intensities corresponding to the expression of TAL1 and RALDH2 genes in T-ALL samples. High levels of expression of RALDH2 were detected in all TAL1-positive cases, in agreement with a role of TAL1 in the activation of this direct target gene. Cases were arranged on the basis of the expression of T-ALL oncogenes, including HOX11, HOX11L2, and TAL1. TAL1-like cases denote samples with high levels of RALDH2 similar to those present in TAL1-positive samples, but lacking TAL1 expression. (B) Heat map representing relative expression levels of TAL1 and TAL1 direct target genes in HOX11/HOX11L2-positive cases and TAL1-positive samples. Each column represents 1 of 26 samples positive for HOX11, HOX11L2, or TAL1 by RT-PCR, while each shows the expression pattern of a particular gene identified as a TAL target by ChIP on chip. Relative expression levels are normalized across the samples; levels greater than or less than the mean are shown in shades of red or blue, respectively.
Combined analysis of ChIP on chip data and gene-expression profiling was used to search for additional genes within the TAL1-direct targets that are associated with the TAL1-positive leukemias. Using a t test nearest neighbor analysis that calculates a combined P value, including the combined significance of the expression data and the ChIP on chip results, we selected 8 TAL1-direct target genes with a combined P value less than .1 (Table 1). Two of these genes—RPS3A and PTPRU—had been analyzed by RT-PCR after TAL1 knockdown by RNA interference and had been found to be repressed by TAL1 (Figure 4), confirming that TAL1 binding to these promoters is associated with changes in gene expression. A more focused analysis designed to avoid the possible confounding effect of the TAL1-like samples focused on the expression signatures of 24 samples containing high levels of expression of HOX11 (n = 2), HOX11L2 (n = 6), and TAL1 (n = 16). This analysis identified 13 TAL1-direct target genes with combined ChIP on chip/t test P values less than .1 (Table 2). Four of these genes, IL10RA, TTC3, FRG1, and SERPINB2, showed marked differences in gene expression between TAL1-positive and HOX11/HOX11L2-positive groups and very significant TAL1 binding in our ChIP on chip analysis, with combined P values less than .01 (Figure 5B). Importantly, 3 of these genes (IL10RA, FRG1, and SERPINB2) showed higher levels of expression in TAL1-positive samples, suggesting that TAL1 may activate their expression, while only 1 (TTC3) showed lower levels of expression in the TAL1 group, and thus is presumably repressed by TAL1 (Figure 5B).
Discussion
We have analyzed the transcriptional effects of TAL1 expression in the context of human T-cell acute leukemia, using TAL1 ChIP on chip analysis coupled with RNA interference to knock down TAL1 expression. The combined analysis of ChIP on chip data and gene-expression profiling in T-ALL samples demonstrates that some of the genes we have identified by ChIP on chip in the Jurkat cell line are also regulated in the primary human leukemias that overexpress TAL1.
Using ChIP on chip, we have identified 71 direct target genes of TAL1 in T-ALL and verified the binding of the transcription factor to 80% of them. Analysis of the promoters bound by TAL1 identified by ChIP on chip indicates that TAL1 frequently binds to its direct target genes as part of regulatory complexes that also contain the class I bHLH factors E2A or HEB. However, in contrast with current models that generally support a role for TAL1 as a transcriptional repressor by interfering with the transcriptional regulation elicited by the tumor suppressor E2A,8,11,12,18,19,38,39 our results suggest that TAL1 inactivation by RNAi results in both activation and repression of multiple direct target genes, in agreement with a more complex transcriptional regulatory role for TAL1 in T-ALL cells. Many of the genes whose promoters are occupied by TAL1 do not seem to be affected by TAL1 down-regulation by RNA interference. Possibly, the level of TAL1 knockdown in our clones is not enough to alter the transcription rates of these promoters, as the knockdown cells still express some TAL1 protein, which may be sufficient to allow basal levels of transcription of specific TAL1 targets. However, an effect on cell growth was observed with the levels of TAL1 knockdown that could be attained, suggesting that a subset of critical TAL1-direct target genes should be measurably regulated. Aberrant expression of TAL1 in T-ALL lymphoblasts results in TAL1 binding to multiple promoter targets, but binding alone may not be sufficient to regulate many of these genes, possibly because lack of tissue-specific transcriptional cofactors. This interpretation is also consistent with the cooperative effects observed between TAL1 and LIM-only domain (LMO) proteins in T-cell transformation.18,40 According to this model, aberrant expression of TAL1 in T-cell progenitors would result in binding to a broad spectrum of direct target promoters, but with limited transcriptional effects. Aberrant expression of LMO proteins in these cells would facilitate the onset of leukemia by reconstituting in T cells transcriptional complexes containing TAL1 and LMO2, which are normally present in hemopoietic progenitors,31,41,42 but still rendering TAL1 inactive for a significant number of promoter targets due to the lack of additional transcriptional cofactors. Consistent with this, low levels of LMO1 mRNA and no expression of LMO2 were detected in Jurkat cells by quantitative PCR.
Gene-expression profiling using oligonucleotide microarrays has previously identified the existence of a group of T-ALL samples with remarkably strong similarities in their pattern of gene expression to TAL1-positive cases. The presence of this group of samples may reflect the activation of TAL1-related transcription factors that would operate on TAL1-target promoters or the activation of mechanistically unrelated oncogenic pathways that only converge with TAL1 in downstream effector pathways. In support of this interpretation, we observed that RALDH2 expression, which can be activated in T-ALL by a TAL1-containing transcriptional complex that binds directly to an alternative intronic promoter sequence, is not strictly limited to TAL1 samples but is also expressed by other T-ALL samples. Thus, TAL1-like cases may harbor an oncogene—presumably an alternative b-HLH transcription factor—which contributes to transformation by controlling the expression of TAL1 direct target genes. Candidate oncogenes to account for the transformation of TAL1-like samples are the LYL1 bHLH transcription factor and the TAL1-related genes TAL2 and BHLHB1, each of which has been activated by chromosomal translocation in T-ALL. However, we failed to detect high levels of expression of these transcription factors in TAL1-like samples in this series (data not shown), suggesting either the existence of additional oncogenic class II bHLH genes or activation of a TAL1-like gene-expression program through an as-yetundefined mechanism.
Overall, our results suggest that the transcriptional effects downstream of the aberrant expression of TAL1 in T-cell progenitors is amplified by a complex oncogenic transcriptional cascade, and illustrate the need to combine microarray gene expression analysis, gene-specific knockdown by RNAi, and ChIP on chip analysis to identify pathways downstream of transcriptional regulators responsible for the malignant transformation of T-cell precursors.
Prepublished online as Blood First Edition Paper, April 18, 2006; DOI 10.1182/blood-2005-08-3482.
Supported by the Lady Tata Memorial Trust (T.P.), a Harvard Hematology Training Grant (J.O.), the Pardee Foundation (A.A.F.), Pollin Research Award (A.A.F.), and National Institutes of Health grants DK070813 (D.T.O.), CA114589-01 (R.S.L.), DK68655 and HG002668 (R.A.Y.), and CA109901 (A.T.L. and R.A.Y.)
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs R. Baer, C. Murre, and K. Pulford for reagents; and Elizabeth Herbolsheimer (Whitehead Institute) and Curtis Glavin (Dana-Farber Cancer Institute) for assistance with the supporting websites.