Abstract
Tumor necrosis factor (TNF) receptor–associated periodic syndrome (TRAPS) is an autosomal dominant systemic autoinflammatory disease associated with heterozygous mutations in TNF receptor 1 (TNFR1). Here we examined the structural and functional alterations caused by 9 distinct TRAPS-associated TNFR1 mutations in transfected cells and a mouse “knock-in” model of TRAPS. We found that these TNFR1 mutants did not generate soluble versions of the receptor, either through membrane cleavage or in exosomes. Mutant receptors did not bind TNF and failed to function as dominant-negative inhibitors of TNFR1-induced apoptosis. Instead, TRAPS mutant TNFR1 formed abnormal disulfide-linked oligomers that failed to interact with wild-type TNFR1 molecules through the preligand assembly domain (PLAD) that normally governs receptor self-association. TRAPS mutant TNFR1 molecules were retained intracellularly and colocalized with endoplasmic reticulum (ER) markers. The capacity of mutant receptors to spontaneously induce both apoptosis and nuclear factor κB (NF-κB) activity was reduced. In contrast, the R92Q variant of TNFR1 behaved like the wild-type receptor in all of these assays. The inflammatory phenotype of TRAPS may be due to consequences of mutant TNFR1 protein misfolding and ER retention.
Introduction
Tumor necrosis factor (TNF) receptor–associated periodic syndrome (TRAPS; Online Mendelian Inheritance in Man database [OMIM] no. 142680)39 is an autoinflammatory syndrome characterized by episodic fevers, rashes, abdominal pain, and muscle, joint, and systemic inflammation that can lead to secondary amyloidosis. TRAPS is strongly associated with heterozygous mutations in the gene encoding TNF receptor 1 (TNFR1, TNFRSF1A, CD120a, p60-TNFR).1 TNFR1 is a TNF-R superfamily receptor involved in the organization of secondary lymphoid tissue, host responses to microorganisms, and inflammation. The extracellular domain of TNFR1 consists of 4 cysteine-rich domains (CRDs) characterized by structurally important, intramolecular disulfide bonds. A region in the N-terminal CRD (CRD1) called the preligand assembly domain (PLAD) mediates noncovalent homotypic receptor interactions that allow efficient ligand binding and signal transduction.2 Regions in CRD2 and CRD3 interact with trimeric TNF,3 resulting in recruitment of the adapter protein TRADD to the cytoplasmic death domain (DD) of TNFR1.4 TRADD then recruits other molecules to initiate the downstream signaling cascade,5,6 triggering either nuclear factor κB (NF-κB) activation and the production of inflammatory cytokines or caspase cleavage, resulting in apoptosis.7
More than 40 heterozygous mutations in TNFR1 have been reported in TRAPS (http://fmf.igh.cnrs.fr/infevers). All result in alteration or in-frame deletion of amino acids in the extracellular domain, and most are in CRD1 and CRD2. Cysteine mutations are associated with a higher disease penetrance and risk of amyloidosis.8 Distinct from these pathogenic mutations, a polymorphic variant of TNFR1, R92Q, is found in 1% to 2% of the healthy population and associated with a milder form of TRAPS with low penetrance.8 Some TRAPS patients exhibit reduced serum levels of soluble TNFR1 (sTNFR1) and defective activation-induced shedding of TNFR1 from peripheral-blood leukocytes.1,8,9 It has been proposed that this defect plays a role in the exaggerated inflammatory responses seen in TRAPS; this has been referred to as “the shedding hypothesis.” However, TNFR1 shedding defects vary between TRAPS patients and cell types, calling into question the role of this defect in disease pathogenesis. Recent studies have indicated that some TNFR1 mutants are inefficiently expressed on the cell surface,10,11 but the biochemical basis of this trafficking defect, or whether TRAPS mutant receptors interfere with the function or secretion of wild-type receptors, is not known. Here we studied the subcellular localization and function of individually marked wild-type and mutant TNFR1 molecules associated with TRAPS, and found misfolding and abnormal oligomerization of mutant receptors as a key common feature resulting in retention in the endoplasmic reticulum (ER), inability to form soluble receptors, and altered signaling.
Materials and methods
Cells and reagents
HT1080, 293T, and COS-7 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA) and cultured as recommended. Jurkat 4E3 were a gift from Francis Chan (University of Massachusetts Medical School, Worcester). Anti-TNFR1 (H-5; Santa Cruz Biotechnology, Santa Cruz, CA) was used for Western blotting, biotinylated polyclonal anti-TNFR1 (500-3143Bt, Peprotech, Cherry Hill, NJ; and BAF425, R&D Systems, Minneapolis, MN) were used for flow cytometry; and anti-TNFR1 was used for immunporecipitations (mAb 55R-286; BD Pharmingen, San Diego, CA). Other antibodies used were anti–green fluorescent protein (GFP) (Roche, Indianapolis, IN), anti-GFP–HRP (BD Clontech, Palo Alto, CA); Anti-HA (Covance, Princeton, NJ), anti-mouse–PE (Dako Cytomation, Carpinteria, CA), and anti-FLAG (Sigma, St Louis, MO). Plasmids containing 1,4-galactosyltransferase (GalT) or signal recognition particle receptor-β subunit (Srβ) fused to cyan fluorescent protein (CFP) were a gift from Kristina Zaal (NIAMS, NIH).
TNFR1 cloning and mutagenesis
TNFR1 sequences were amplified from cDNAs containing the wild-type or mutant sequences using Pfu Turbo polymerase (Stratagene, La Jolla, CA) as in full-length (amino acid [aa] 1-426) and cytoplasmic domain–truncated (ΔCD, aa 1-211) versions. (cDNAs containing the sequences with the C30R, C43S, T50M, and C52F mutations were a gift from Stefan Siebert [University of Cardiff, Wales].) These were ligated into pECFP-N1 and pEYFP-N1 (BD Clontech) that were modified to contain the TNFR2 signal sequence and HA-tag 3′ to the cloning site, pCDM7 modified to contain a 3C protease cleavage site and the human IgG-Fc domain, and pRGFP2 with or without an N-terminal FLAG tag. Additional mutations were made by Quick Change site-directed mutagenesis (Stratagene). All mutations were confirmed by sequencing.
TNFR1 mutant mice
Details of the production and phenotype of these mice will be reported elsewhere (H.K., A.J.J., and D.L.K., manuscript in preparation). Briefly, a targeting vector was prepared containing nucleotide mutations to produce the T50M mutation in mouse TNFR1. Embryonic stem (ES) cells harboring this mutation were selected and injected into blastocysts. Lines carrying this mutation were crossed to the E1A-cre strain to remove the neomycin selection cassette and backcrossed to C57Bl/6 for at least 5 generations before intercrossing to generate homozygous mutant mice. Presence of the mutant TNFR1 allele was confirmed by resequencing cDNA from the final line. TNFR1 knock-out mice on a C57BL/6 background were obtained from Jackson Labs (Bar Harbor, ME).
Immunoblotting and immunoprecipitations
293T cells were transiently transfected with Fugene 6 (Roche), and harvested after 24 hours. Cell lysis, immunoprecipitations, and Western blotting were performed as previously described.12 To detect soluble versions of TNFR1, cells were cultured in CHO-II-SFM (Invitrogen, Carlsbad, CA). Medium was harvested and centrifuged at 10 000g for 35 minutes to remove cellular debris, and proteins were then acetone precipitated from the supernatant. TNFR1 Fc-fusion expression constructs were transfected into 293T cells and purified using protein-A (Sigma). Fc-fusion proteins (5 μg) or cell lysates containing TNFR1-GFP fusion proteins were mixed with protein-A sepharose beads and supernatant containing TNFα, extensively washed and then run on a 15% polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie blue or blotted for GFP.
Surface plasmon resonance
Experiments were carried out on a BIAcore 3000 instrument (BIAcore, Uppsala, Sweden) using HEPES-buffered saline (HBS-EP) at 25°C. Anti-FLAG monoclonal antibody (diluted in sodium acetate, pH 4.5) was covalently immobilized to a CM-5 chip (BIAcore), according to the manufacturer's guidelines. Soluble TNFα was then captured onto the chip directly from tissue-culture supernatant, which was flowed over until an immobilization level of approximately 1400 resonance units (RU) was generated in all 4 flow cells. The Fc-fusions (TRAPS proteins) were diluted in HBS-EP at a concentration of 50 μg/mL, and a volume of 15 μL was injected into individual flow cells; the first CRD of another TNFR family member, DR5-D1-Fc, was used as a negative control.
Apoptosis assays
To test dominant inhibition by TNFR1 mutants on TNF-induced apoptosis, Jurkat 4E10 cells13 were transfected by electroporation with the indicated ΔCD TNFR1-GFP or DR4-GFP fusion protein expression vectors. Cells were allowed to express proteins and then treated with 100 ng/mL TNFα for 18 hours. Yellow fluorescent protein–positive (YFP+) cells were assayed for cell death by fluorescence-activated cell sorting (FACS) analysis of forward scatter and propidium iodine (PI) uptake. Specific cell death was calculated using the formula 1 – (% live untreated cells/% live treated cells) × 100. To examine spontaneous death induced by transient expression of TNFR1, HT1080 cells were transfected with 0.25 μg of the indicated TNFR1-YFP expression constructs. After 18 hours, cells were harvested, washed once with phosphate-buffered saline (PBS), and then incubated for 15 minutes at room temperature with the mitochondrial potentiometric dye tetramethylrhodamine methyl ester (TMRM).
FRET
Flow cytometric detection of fluorescence resonance energy transfer (FRET) was performed as previously described14,15 using a CYAN cyotflourimeter (Dako Cytomation), and 405/488 nm dual laser excitation. To determine FRET efficiency, the live CFP/YFP double-positive population was gated and the FRET signal was examined in this population. The same CFP/YFP gates were used in all samples within an experiment.
TNFR1 expression testing and localization by flow cytometry
Cells were incubated with anti-TNFR1 or anti–epitope tag antibodies with conventional FACS staining techniques with or without permeablization using cytofix/cytoperm buffers (BD Pharmingen). To test for receptor internalization, transfected cells were incubated with anti-FLAG monoclonal antibody (mAb) and incubated at 37°C for various times until transfer to 0°C to halt endocytosis, followed by treatment with acid (0.25 M glycine-HCl) to remove all cell-bound antibody, permeabilization, and then incubation with anti–mouse PE (Dako Cytomation). Cells were then analyzed by FACS and compared with cells stained without permeabilization.
Confocal microscopy and FRAP
Confocal microscopy was performed at 37°C with an LSM510 confocal microscope (Zeiss, Oberkochen, Germany) in multitrack mode, using a 63 ×/1.4 numeric aperture Plan-apochromat objective. Images were processed using Zeiss LSM510 software version 3.2, and Photoshop version 7.0 (Adobe Systems, San Jose, CA) was used to convert the images to 8 bit digital files and to crop the relevant areas. Fluorescence recovery after photobleaching (FRAP) analysis was done on ER-localized TNFR1 photobleached at 100% laser output for 15 iterations. Fifty images were made at 500-microsecond intervals in the recovery phase. The mean fluorescence of the region of interest was used to calculate t1/2. Fluorescent intensity versus time was plotted against time to determine t1/2 using Prism software (Graphpad, San Diego, CA).
Luciferase assays
HT1080 cells were transfected with 0.1 μg of the indicated TNFR1 expression construct along with 0.2 μg of the NF-κB luciferase reporter construct, 40 ng of renilla luciferase as a transfection efficiency and viability control, and 0.25 μg pcDNA3 to increase the total amount of DNA used. After 24 hours, cells were lysed and luciferase was detected with the dual luciferase reagent system (Promega, Madison, WI). Fold NF-κB induction was determined by dividing renilla-corrected luciferase intensity of each sample by the corrected luciferase intensity of the ΔCD TNFR1 control.
Results
TRAPS-associated mutant TNFR1 are not secreted and do not bind TNF
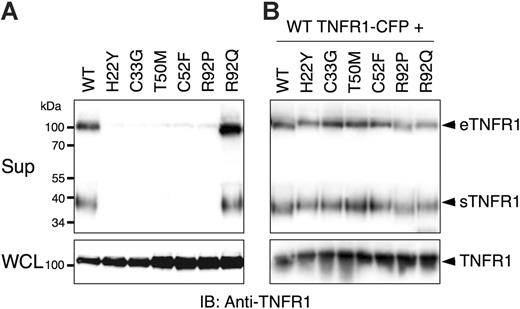
Since TRAPS has been associated with reduced serum levels of soluble receptors, we first investigated whether transfected wild-type or TRAPS-associated mutant TNFR1 molecules could form soluble versions of the receptor. Soluble TNFR1 can be formed by metalloproteinase-induced shedding of surface receptors (sTNFR1), or secretion of full-length receptors in exosome-like vesicles (eTNFR1).16 As expected, cells transfected with wild-type TNFR1-YFP fusion were able to spontaneously produce both forms of soluble TNFR1 (Figure 1A). In contrast, production of soluble TNFR1 by 6 distinct TNFR1 mutants associated with TRAPS was severely inhibited, despite comparable expression in cell lysates. However, the R92Q TNFR1 variant was secreted and cleaved with equal efficiency as the wild-type receptor. Since most TNFR1 mutations in TRAPS patients are heterozygous,1,8,9 we tested whether TRAPS mutant TNFR1 could dominantly interfere with secretion of the wild-type receptor in these assays. Supernatants from cells cotransfected with wild-type and the same group of mutant receptors contained similar amounts of soluble receptors as cells cotransfected with only wild-type TNFR1 (Figure 1B). As a control, we found LAMP-1, which is associated with exosomes,16 in cell-free supernatants but not Fas, another TNFR family member not known to be secreted in exosomes (data not shown). PMA treatment enhanced formation of soluble wild-type TNFR1, but mutant receptor secretion was still severely inhibited (data not shown). These data indicate that TRAPS-associated TNFR1 mutants do not enter the secretory pathway, but also do not appear to interfere with secretion of the wild-type receptor.
Mutations in TNFR1 prevent formation of forming soluble versions of the receptor. 293T cells were transiently transfected with the indicated TNFR1-YFP expression vectors without (A) or with (B) wild-type (WT) TNFR1-CFP. Medium was changed to serum-free medium and the cells were allowed to produce soluble TNFR1. Supernatants (Sup) and whole-cell lysates (WCL) were collected after 24 hours and analyzed by SDS-PAGE and immunoblotting with anti-TNFR1. eTFNR1 denotes exosome-associated full-length TNFR1; sTNFR1, soluble cleaved TNFR1 encoding the extracellular domain only.
Mutations in TNFR1 prevent formation of forming soluble versions of the receptor. 293T cells were transiently transfected with the indicated TNFR1-YFP expression vectors without (A) or with (B) wild-type (WT) TNFR1-CFP. Medium was changed to serum-free medium and the cells were allowed to produce soluble TNFR1. Supernatants (Sup) and whole-cell lysates (WCL) were collected after 24 hours and analyzed by SDS-PAGE and immunoblotting with anti-TNFR1. eTFNR1 denotes exosome-associated full-length TNFR1; sTNFR1, soluble cleaved TNFR1 encoding the extracellular domain only.
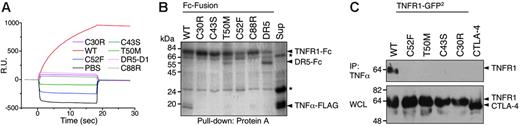
Whether or not they are secreted, mutant TNF receptors in TRAPS should be able to influence TNF-dependent signaling only if they can bind TNF or associate with wild-type receptors. We tested whether TRAPS-associated mutant TNFR1 could bind TNF in a number of in vitro and cell-based assays. Chimeras of 5 distinct TRAPS-associated mutants fused to the Fc portion of human IgG1 were produced and purified. Surface plasmon resonance analysis demonstrated that wild-type TNFR1 strongly bound to TNFα, whereas all 5 mutant receptors failed to show any binding (Figure 2A). In addition, this same panel of mutants failed to pull down TNFα in vitro, whereas the wild-type TNFR1-Fc was able to do so (Figure 2B). FLAG-tagged TNFα could efficiently immunoprecipitate wild-type but not the mutant transfected TNFR1 from cell lysates (Figure 2C). In agreement with this data, TNF could not bind cells transfected with any TRAPS mutant TNFR1 except the R92 functional variant in a FACS-based TNF binding assay (data not shown). Thus, either in vitro or in cells, none of the TRAPS mutant TNFR1 we tested bind TNFα.
Abnormal self-association of TRAPS-associated TNFR1 mutants
One potential consequence of lack of ligand binding by TRAPS mutants could be the formation of mixed complexes with wild-type receptors that alter signaling. For example, non–ligand-binding mutants of Fas in the autoimmune lymphoproliferative syndrome (ALPS) interact with wild-type receptors and thereby inhibit apoptosis.17-19 Both Fas and TNFR1 can form ligand-independent noncovalent oligomers through the PLAD contained in CRD1 of the receptors. Even cytoplasmic domain–deleted (ΔCD) non–ligand-binding extracellular mutants of Fas and TNFR1 can interfere with wild-type receptor signaling through this mechanism.2,15 We therefore tested whether TRAPS-associated TNFR1 mutants could interfere with wild-type TNFR1 signaling. ΔCD wild-type and mutant TNFR1 or DR4-GFP were transfected into a TNFR2-expressing Jurkat cell line that is sensitive to TNFα-induced apoptosis through TNFR1.2 In agreement with previous data, wild-type ΔCD TNFR1 potently interfered with TNFα-induced apoptosis relative to cells transfected with the TRAIL receptor DR4, which does not bind to TNF or associate with TNFR1 (Figure 3A). In contrast, TRAPS mutant TNFR1 failed to interfere with TNFα-induced apoptosis with the exception of the R92Q variant, which again functioned similarly to wild-type truncated TNFR1.
These experiments suggested that TRAPS mutant TNFR1 do not functionally interact with their wild-type counterparts. We have previously used FRET between variants of GFP to demonstrate that TNFR1 and Fas formed specific homotypic complexes through the PLAD.2,15 We constructed CFP and YFP fusions of ΔCD TNFR1 and assayed the ability of a panel of TRAPS mutant receptors to interact with wild-type TNFR1 using FACS-based FRET.14 As expected, the extracellular domain of Fas showed negligible FRET with TNFR1, and wild-type TNFR1 strongly interacted with itself (Figure 3B, top rows; 3C). Receptors harboring TRAPS mutants located both within CRD1, the PLAD (H22Y, C33G, T50M, and C52F), as well as receptors harboring mutations in CRD2 (C88R and R92P), exhibited greatly reduced FRET with wild-type TNFR1 (Figure 3B, left column; 3C, empty bars). Surprisingly, mutant receptors continued to interact with themselves with similar efficiency to that of wild-type receptor self-association (Figure 3B, right column; 3C, filled bars). The R92Q polymorphic variant, however, retained its ability to associate with wild-type TNFR1. These experiments confirm the lack of an interaction between wild-type and mutant TNFR1 molecules and suggest that the mutant receptors may be interacting with themselves through a PLAD-independent mechanism.
TRAPS mutations render TNFR1 unable to bind TNFα. (A) Surface plasmon resonance analysis of TNFα binding. Fc-fusions of wild-type (WT), the indicated mutant TNFR1-Fc, and control DR5-CRD1-Fc were injected onto BIAcore chips coated with immobilized TNFα. Sensorgrams show resonance units (RU) over time. (B) In vitro pull-down of TNFα with Fc-fusion proteins. The indicated purified Fc-fusion proteins were mixed with protein-A Sepharose beads and tissue-culture supernatant containing soluble TNFα-FLAG. After washing, proteins were removed from the Sepharose and run on SDS-PAGE. Gels were stained with Coomassie blue. The final lane is a pull-down of supernatant alone (sup) with anti-FLAG beads to confirm presence of TNFα. Nonspecific bands are indicated by an asterisk. (C) Immunoprecipitation of TNFR1-GFP2 with TNFα-FLAG. Lysate from 293T cells transfected with wild-type or TRAPS mutant ΔCD GFP2-fusion protein expression vectors or a CTLA4-GFP2 control fusion vector were mixed with anti-FLAG Sepharose beads precoated with TNFα-FLAG. After washing, proteins were eluted from the beads and run on SDS-PAGE along with whole-cell lysates (WCL). Immunoblots were performed with anti-GFP–HRP.
TRAPS mutations render TNFR1 unable to bind TNFα. (A) Surface plasmon resonance analysis of TNFα binding. Fc-fusions of wild-type (WT), the indicated mutant TNFR1-Fc, and control DR5-CRD1-Fc were injected onto BIAcore chips coated with immobilized TNFα. Sensorgrams show resonance units (RU) over time. (B) In vitro pull-down of TNFα with Fc-fusion proteins. The indicated purified Fc-fusion proteins were mixed with protein-A Sepharose beads and tissue-culture supernatant containing soluble TNFα-FLAG. After washing, proteins were removed from the Sepharose and run on SDS-PAGE. Gels were stained with Coomassie blue. The final lane is a pull-down of supernatant alone (sup) with anti-FLAG beads to confirm presence of TNFα. Nonspecific bands are indicated by an asterisk. (C) Immunoprecipitation of TNFR1-GFP2 with TNFα-FLAG. Lysate from 293T cells transfected with wild-type or TRAPS mutant ΔCD GFP2-fusion protein expression vectors or a CTLA4-GFP2 control fusion vector were mixed with anti-FLAG Sepharose beads precoated with TNFα-FLAG. After washing, proteins were eluted from the beads and run on SDS-PAGE along with whole-cell lysates (WCL). Immunoblots were performed with anti-GFP–HRP.
Since many of the TRAPS mutations result in the loss of a cysteine, leaving an unpaired cysteine in the TNFR1 extracellular domain, self-association between mutant receptors may result from intramolecular disulfide bonding of the single unpaired cysteines. Alternatively, global unfolding of TRAPS mutant receptors could create multiple free cysteines. We tested these possibilities by cotransfecting CFP- and YFP-tagged TNFR1 containing mutations at both cysteine –33 and –52, which normally form an intramolecular disulfide bond.3 Like the single mutants, the C33G/C52F double-mutant failed to interact with wild-type TNFR1. This mutant was still able to interact with itself by FRET (Figure 3C) and form high-molecular-weight oligomers (data not shown), suggesting that if disulfide bonds underlie the self-association of TRAPS mutants, then cysteines distinct from the mutant residues are also unpaired as a result of more global misfolding.
Mutant TNFR1 do not functionally associate with wild-type TNFR1, but self-associate through intermolecular disulfide bonds. (A) Jurkat 4E3 cells were transfected with the indicated ΔCD TNFR1-YFP expression vectors and treated with TNFα. Cell viability was then determined by forward scatter and PI uptake using flow cytometry. (B-C) FRET analysis of TNFR1 interactions. 293T cells were transfected with TNFR1-CFP and -YFP fusion proteins and analyzed by flow cytometry. In the top set of plots, wild-type (WT) TNFR1-CFP was cotransfected with WT-TNFR1 or Fas-YFP. The dot plot shows the CFP+YFP+ population gating used for FRET analysis in all samples. The bottom set of plots depicts association between CFP-tagged mutant receptors and WT (left) or mutant (right) YFP-tagged receptors. (C) Percentage of FRET-positive cells from all WT and mutant combinations tested, analyzed as in panel B. (D) 293T cells were transiently transfected with ΔCD TNFR1-YFP fusion proteins, and whole-cell lysates were run on a polyacrylamide gel in nonreducing (top) or reducing (bottom) conditions. The proteins were then immunoblotted using anti-TNFR1.
Mutant TNFR1 do not functionally associate with wild-type TNFR1, but self-associate through intermolecular disulfide bonds. (A) Jurkat 4E3 cells were transfected with the indicated ΔCD TNFR1-YFP expression vectors and treated with TNFα. Cell viability was then determined by forward scatter and PI uptake using flow cytometry. (B-C) FRET analysis of TNFR1 interactions. 293T cells were transfected with TNFR1-CFP and -YFP fusion proteins and analyzed by flow cytometry. In the top set of plots, wild-type (WT) TNFR1-CFP was cotransfected with WT-TNFR1 or Fas-YFP. The dot plot shows the CFP+YFP+ population gating used for FRET analysis in all samples. The bottom set of plots depicts association between CFP-tagged mutant receptors and WT (left) or mutant (right) YFP-tagged receptors. (C) Percentage of FRET-positive cells from all WT and mutant combinations tested, analyzed as in panel B. (D) 293T cells were transiently transfected with ΔCD TNFR1-YFP fusion proteins, and whole-cell lysates were run on a polyacrylamide gel in nonreducing (top) or reducing (bottom) conditions. The proteins were then immunoblotted using anti-TNFR1.
To directly test whether mutant receptors form disulfide-linked oligomers, Western blots were performed using lysates of cells transfected with wild-type or TRAPS mutant receptors under reducing and nonreducing conditions. In agreement with previous data, wild-type receptors were primarily monomeric in this assay, which is expected, since PLAD-dependent self-association of TNFR1 occurs through noncovalent interactions.2 However, all of the TRAPS mutant receptors, including both cysteine and noncysteine mutants, with the exception of the R92Q variant, exhibited a high degree of oligomerization: species corresponding to dimers, trimers, and higher-order complexes were visible on nonreducing Western blots (Figure 3D). Addition of DTT reduced these complexes to the size of monomeric TNFR1, although bands for the mutant receptors were considerably sharper, likely due to altered glycosylation. Addition of N-ethyl maleimide, which reacts with the -SH group on unpaired cysteines, to cells before lysis did not block formation of TRAPS mutant receptor oligomers (data not shown), indicating that the disulfide bonds were likely formed in vivo, rather than after cell lysis. Immunoprecipitation experiments showed that mutant receptors could be precipitated with wild-type TNFR1, but these receptors were exclusively found as high-molecular-weight disulfide-linked complexes, indicating that any interactions of wild-type and mutant TNFR1 were not occurring through PLAD-dependent noncovalent interactions (Figure S1A-B, available on the Blood website; see the Supplemental Figures link at the top of the online article).
TRAPS mutants fail to traffic efficiently to the plasma membrane
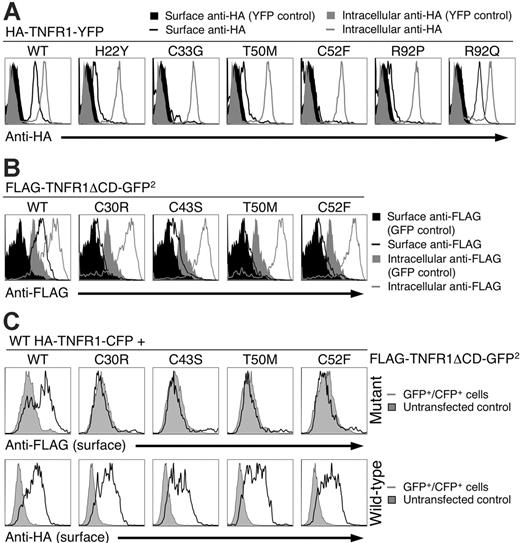
Since incorrectly folded receptors are often either retained in the ER or degraded by the proteosome,20 we examined surface expression of the mutant TNFR1 using a system in which all the receptors were tagged with an N-terminal HA epitope to control for differential binding of antibodies to the mutant receptors, and a C-terminal YFP fusion to allow for direct identification and quantitation of receptor transfection. Surface anti-HA staining was evident on YFP-positive cells transfected with only wild-type and R92Q TNFR1 (Figure 4A, black line). In contrast, cells transfected with the H22Y, C33G, T50M, C52F, and R92P mutant TNFR had either minimal or no surface anti-HA staining, indicating that these mutations severely impaired the ability of the receptors to reach the cell surface. All transfected cells were positive for anti-HA when they were permeabilized before staining, indicating efficient expression of all the TNFR1 constructs (Figure 4A, gray line). Anti-HA did not stain cells transfected with YFP alone either on the surface or intracellularly (Figure 4A, black and gray filled areas, respectively). To determine whether the Golgi retention motif in the TNFR1 cytoplasmic domain21,22 was necessary for the trafficking defect of the mutant TNFR, we analyzed cells transfected with ΔCD TNFR1. As expected, surface levels of wild-type receptors increased, but ΔCD TRAPS mutant TNFR1 still had significantly impaired surface staining, with cysteine-containing mutants most severely affected when surface receptor levels were analyzed with anti-FLAG (Figure 4B) or anti-TNFR1 (Figure S2A), indicating that the extracellular TRAPS mutations impair receptor trafficking independent of cytoplasmic sorting signals.
Mutant TNFR1 do not traffic to the plasma membrane. (A) 293T cells were transiently transfected with the indicated full-length wild-type (WT) or mutant TNFR1-YFP fusion constructs with N-terminal HA tags. Live or fixed and permeabilized cells were then stained with anti-HA followed by anti–mouse PE and analyzed by flow cytometry. The histograms are gated on the YFP+ cells that were live according to forward- and side-scatter characteristics. (B) 293T cells were transfected as with ΔCD FLAG-TNFR1-GFP2 and stained as in panel A with anti-FLAG. The filled gray and black areas in panels A and B represent cells transfected with a YFP expression vector and stained as in panel A. (C) N-terminal HA-tagged WT TNFR1-CFP was cotransfected with wild-type or the indicated mutant N-terminal FLAG-tagged TNFR1-ΔCD-GFP2. Cells were surface stained with anti-HA and anti-FLAG antibodies and analyzed by flow cytometry. Plots shown are gated on the CFP+GFP+ population.
Mutant TNFR1 do not traffic to the plasma membrane. (A) 293T cells were transiently transfected with the indicated full-length wild-type (WT) or mutant TNFR1-YFP fusion constructs with N-terminal HA tags. Live or fixed and permeabilized cells were then stained with anti-HA followed by anti–mouse PE and analyzed by flow cytometry. The histograms are gated on the YFP+ cells that were live according to forward- and side-scatter characteristics. (B) 293T cells were transfected as with ΔCD FLAG-TNFR1-GFP2 and stained as in panel A with anti-FLAG. The filled gray and black areas in panels A and B represent cells transfected with a YFP expression vector and stained as in panel A. (C) N-terminal HA-tagged WT TNFR1-CFP was cotransfected with wild-type or the indicated mutant N-terminal FLAG-tagged TNFR1-ΔCD-GFP2. Cells were surface stained with anti-HA and anti-FLAG antibodies and analyzed by flow cytometry. Plots shown are gated on the CFP+GFP+ population.
To determine if wild-type and mutant receptors could affect each other's surface expression, cells were cotransfected with expression vectors for both wild-type and mutant TNFR1 with different epitope tags so the trafficking of each receptor could be individually followed. Cotransfection of wild-type TNFR1 with mutant receptors failed to relocalize mutant receptors to the cell surface (Figure 4C, top), and induced only a slight downmodulation of the wild-type receptor (Figure 4C, bottom), indicating that for the most part, wild-type and TRAPS mutant receptors traffic independently. We also considered the possibility that the TRAPS mutations could cause rapid internalization of surface-expressed receptors. Internalization of transfected wild-type TNFR1 could be detected within 30 minutes of labeling. However, even after 6 hours of incubation at 37°C, no surface labeling or internalization of C30R TNFR1 could be detected (Figure S2B).
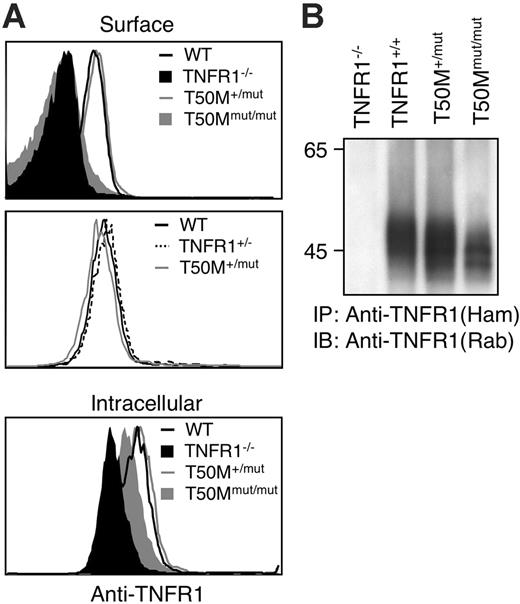
Retention of TRAPS-associated mutant TNFR1 could be a function of receptor overexpression. However, mutant receptors were still not expressed on the cell surface when HT1080 cells were transfected with 20-fold lower quantities of receptor plasmids or in a stable cell line expressing H22Y TNFR1 (Figure S3A,C). Western blot data indicated that the ratio of mutant–wild-type receptors in these experiments was 0.5 to 8.1 (Figure S3B). We also examined TNFR1 surface expression in a line of knock-in mice engineered to express the murine equivalent of the T50M TRAPS mutation from the endogenous TNFR1 locus. In peripheral-blood granulocyte populations, which express the highest level of surface TNFR1, levels of TNFR1 in T50M/+ mice were comparable with both wild-type and heterozygous TNFR1 knock-out mice (Figure 5A), similar to the situation in TRAPS patients, where overall surface TNFR1 expression is not impaired.1,8 In homozygous T50M mice, no surface expression could be detected, despite intracellular staining of TNFR1 in the same cells (Figure 5A). Similar results were found in blood monocytes (data not shown). Immunoprecipitation of TNFR1 also indicated adequate expression of the mutant T50M TNFR1, although the apparent molecular weight was slightly lower (Figure 5B), as was seen with transfected human receptors. These results indicate that altered trafficking of TRAPS mutant TNFR1 occurs with endogenously expressed TNFR1 in mice and in cells expressing lower levels of transfected receptors more closely approximating the case in human TRAPS patient cells.
Retention of TRAPS mutant TNFR1 in the endoplasmic reticulum
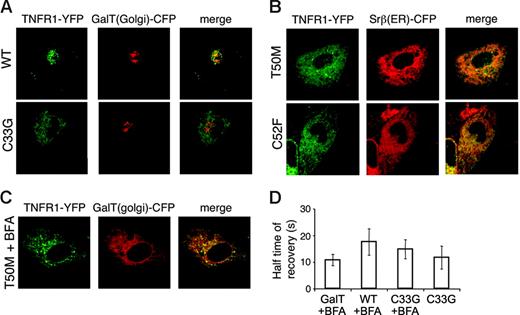
To determine the subcellular localization of TRAPS mutant TNFR1, a variety of cell types transfected with TNFR1-YFP fusion proteins were examined by live cell confocal microscopy. In COS-7 cells, full-length TNFR1 formed cytoplasmic aggregates that were most likely aggresomes, sites of proteasomal degradation for excess proteins,23 since these structures were enlarged and concentrated into perinuclear foci when proteasome inhibitors were added (data not shown). However, when wild-type TNFR1 was transfected into HT1080 cells that lack the SV40 T antigen and express lower levels of receptors, it localized primarily in the Golgi complex, demonstrated by colocalization with the Golgi resident protein GalT24 (Figure 6A, top row). A small proportion of the fluorescent signal was also visible at the plasma membrane. In contrast, mutant TNFR1 exhibited a reticular cytoplasmic localization that did not colocalize with GalT (Figure 6A, bottom row). The pattern of mutant TNFR1 had almost complete overlap with Srβ, an ER resident protein (Figure 6B). Other TNFR1 mutants (H22Y, C33G), including noncysteine and cysteine mutations, localized similarly. Again, like the wild-type receptor, the R92Q variant was localized in the Golgi (data not shown). In Brefeldin A (BFA)–treated cells, Golgi-associated proteins characteristically collapse into perinuclear foci and merge with the ER, whereas ER-associated proteins do not change localization. In accordance with ER localization, mutant TNFR1 failed to relocalize after BFA treatment, whereas the Golgi marker collapsed into structures partially overlapping with the mutant TNFR1 (Figure 6C). In cells cotransfected with wild-type and mutant TNFR1 tagged with different GFP variants, there were no significant alterations in receptor localization compared with single transfected cells; mutant TNFR1 were retained in the ER, while wild-type receptors were predominantly in the Golgi complex (data not shown). This further demonstrates that the mutant and wild-type proteins traffic independently. FRAP experiments showed that TRAPS mutant TNFR1 had similar mobility to WT receptors relocalized to the ER using BFA (Figure 6D). Thus, the mutations in TNFR1 associated with TRAPS do not impair ER exit and transport to the Golgi simply because of reduced diffusion.
Lack of surface TNFR1 in T50M “knock-in” mice. (A) Peripheral-blood neutrophils (GR1+CD11b+) from mice with the indicated genotypes were stained with biotinylated anti-TFNR1 and streptavidin-PE before (surface) or after (intracellular) permeablization. Staining with isotype control biotinylated hamster Ig was equivalent to TNFR1 KO (data not shown). (B) Immunoprecipitation of wild-type and T50M equivalent mutant TNFR1. Splenocytes from mice of the indicated genotypes were lysed and immunoprecipitated with hamster anti–mouse TNFR1, followed by reducing SDS-PAGE and immunoblotting with rabbit anti-TNFR1.
Lack of surface TNFR1 in T50M “knock-in” mice. (A) Peripheral-blood neutrophils (GR1+CD11b+) from mice with the indicated genotypes were stained with biotinylated anti-TFNR1 and streptavidin-PE before (surface) or after (intracellular) permeablization. Staining with isotype control biotinylated hamster Ig was equivalent to TNFR1 KO (data not shown). (B) Immunoprecipitation of wild-type and T50M equivalent mutant TNFR1. Splenocytes from mice of the indicated genotypes were lysed and immunoprecipitated with hamster anti–mouse TNFR1, followed by reducing SDS-PAGE and immunoblotting with rabbit anti-TNFR1.
Retention of TRAPS mutant TNFR1 in the ER. (A-B) HT1080 cells were cotransfected with YFP fusion proteins of full-length wild-type or mutant TNFR1 and CFP fusion proteins of the Golgi marker GalT or the ER marker Srβ. Images were acquired by confocal microscopy. YFP fluorescence is shown in green and CFP fluorescence in red. (C) TRAPS mutant TNFR1 localization is not altered by Brefeldin A. T50M TNFR1-YFP was cotransfected with GalT-CFP as in panel A and cells were treated with Brefeldin A prior to confocal microscopy. (D) FRAP was analyzed in COS-7 cells transfected with wild-type and mutant forms of TNFR1 with the cytoplasmic domain truncated. A small area of the cell was bleached with the laser on high power, and the return of fluorescence to that area over time was monitored. t1/2 ± SD (n = 5) is shown for each condition.
Retention of TRAPS mutant TNFR1 in the ER. (A-B) HT1080 cells were cotransfected with YFP fusion proteins of full-length wild-type or mutant TNFR1 and CFP fusion proteins of the Golgi marker GalT or the ER marker Srβ. Images were acquired by confocal microscopy. YFP fluorescence is shown in green and CFP fluorescence in red. (C) TRAPS mutant TNFR1 localization is not altered by Brefeldin A. T50M TNFR1-YFP was cotransfected with GalT-CFP as in panel A and cells were treated with Brefeldin A prior to confocal microscopy. (D) FRAP was analyzed in COS-7 cells transfected with wild-type and mutant forms of TNFR1 with the cytoplasmic domain truncated. A small area of the cell was bleached with the laser on high power, and the return of fluorescence to that area over time was monitored. t1/2 ± SD (n = 5) is shown for each condition.
Altered signaling by TRAPS mutant TNFR1
Proteins accumulating in the ER can induce ER stress responses, such as the unfolded protein response (UPR), which is characterized by up-regulation of molecular chaperones, or the ER overload response (EOR), which can directly activate NF-κB.25,26 After transfection with either wild-type or mutant TNFR1, HT1080 or 293T cells did not increase BiP/GRP78 expression, the hallmark of the UPR (Figure S4A-B). Also, ΔCD mutant TNFR1 did not induce NF-κB activation more than wild type ΔCD TNFR1, indicating that misfolding of mutant TNFR1 did not activate the EOR (Figure S4C). Thus, TNFR1 does not acutely induce ER stress responses despite ER retention.
Abnormal ligand-independent signaling emanating from intracellularly retained receptors could also contribute to the TRAPS phenotype. Mutant receptors may “hypersignal,” as has been found with dominant active mutations in components of the inflammasome.27,28 Cells transfected with wild-type TNFR1 showed multiple features of apoptosis, including loss of mitochondrial-membrane potential, Annexin V staining, and eventually loss of plasma-membrane integrity (Figure 7A-B, and data not shown). TNFR1-induced cell death was inhibited by the caspase inhibitor zVAD-fmk (Figure 7A). Mutant TNFR1 induced significantly less cell death than wild-type TNFR1, with the exception of the R92Q variant (Figure 7B). Cells transfected with TRAPS mutants also showed a small but significant reduction in NF-κB transactivation by the mutant receptors compared with wild-type TNFR1 (Figure 7C). These data demonstrate that intracellularly retained receptors in TRAPS can signal though the 2 major TNFR1 signaling pathways, but with reduced efficiency.
Mutant TNFR1 do not induce increased spontaneous signaling compared with wild-type TNFR1. (A-B) HT1080 cells were transfected with the indicated full-length TNFR1 or TNFR1-ΔCD or YFP expression vectors and allowed to incubate for 18 hours. Some cells were incubated with 50 μM zVAD-fmk to inhibit caspase activation or 0.1 μM staurosporine (STS) as a positive control for cell death. Cells were then stained with TMRM and analyzed by FACS. (A) Insets indicate percent YFP+TMRM+ (live) cells over YFP+TMRM– (dead) cells. (B) Percentage of cell death was determined by first gating on the YFPmed population and then the TMRM– cells (inset). Error bars indicate SD. (C) HT1080 cells were transfected with the indicated full-length TNFR1 or TNFR1-ΔCD expression constructs along with an NF-κB luciferase reporter and the renilla luciferase (RLTK) as a transfection efficiency control. Cells were also incubated with 50 μM zVAD-fmk to prevent spontaneous cell death. Twenty-four hours later, luciferase expression was examined and NF-κB induction was normalized by comparing it with the renilla luciferase intensity of each sample (±SD). Experiments shown were performed in duplicate, and data are representative of multiple experiments. *P < .05 between WT and mutant TNFR1 constructs by an unpaired Student t test.
Mutant TNFR1 do not induce increased spontaneous signaling compared with wild-type TNFR1. (A-B) HT1080 cells were transfected with the indicated full-length TNFR1 or TNFR1-ΔCD or YFP expression vectors and allowed to incubate for 18 hours. Some cells were incubated with 50 μM zVAD-fmk to inhibit caspase activation or 0.1 μM staurosporine (STS) as a positive control for cell death. Cells were then stained with TMRM and analyzed by FACS. (A) Insets indicate percent YFP+TMRM+ (live) cells over YFP+TMRM– (dead) cells. (B) Percentage of cell death was determined by first gating on the YFPmed population and then the TMRM– cells (inset). Error bars indicate SD. (C) HT1080 cells were transfected with the indicated full-length TNFR1 or TNFR1-ΔCD expression constructs along with an NF-κB luciferase reporter and the renilla luciferase (RLTK) as a transfection efficiency control. Cells were also incubated with 50 μM zVAD-fmk to prevent spontaneous cell death. Twenty-four hours later, luciferase expression was examined and NF-κB induction was normalized by comparing it with the renilla luciferase intensity of each sample (±SD). Experiments shown were performed in duplicate, and data are representative of multiple experiments. *P < .05 between WT and mutant TNFR1 constructs by an unpaired Student t test.
Discussion
Several lines of evidence presented here indicate that the unifying feature of TRAPS mutations is misfolding of the extracellular domain of the receptor leading to retention in the ER. The 9 TRAPS-associated mutations studied here (H22Y, C30S, C30R, C33G, C43S, T50M, C52F, C88R, and R92P) encode both cysteine and noncysteine mutants. Intracellular retention of cysteine mutations appeared to be more complete, indicating that a small percentage of noncysteine mutant TNFR1 molecules may fold correctly. Several findings indicate structural perturbations (beyond the local site of the mutations in TRAPS) that expose multiple reactive cysteine groups in the extracellular domain: (1) Failure of TRAPS mutants to bind TNFα, even when mutations are distant from TNF-binding residues; (2) failure of mutant receptors to associate with wild-type receptors through the PLAD even when mutations are distant from the PLAD; (3) self-association of mutant receptors even when 2 paired cysteines are mutated; and (4) relocalization to the ER, which is a feature of misfolded proteins.29,30 Finally, the finding that the TRAPS T50M mutation engineered into the mouse TNFR1 locus is also retained intracellularly indicates that these features occur in TNF receptors expressed at physiologic levels.
All rare TNFR1 mutations associated with TRAPS share similar relocalization to the ER and failure of surface expression. Interestingly, the R92Q variant TNFR1 behaves like wild-type TNFR1, with apparently normal folding, identical surface expression, TNFα binding, and association with wild-type receptors. Although R92 is in the second CRD, which contains TNF-binding residues, it lies on the opposite face of the receptor from the TNF binding surface. Carriage of the R92Q TNFR1 polymorphism is associated with inflammatory phenotypes other than TRAPS, including increased severity of rheumatoid arthritis and atherosclerotic cardiovascular disease.8,31 Thus, the pathogenesis of TRAPS in patients harboring only the R92Q TNFR1 variant is probably different from that of patients with structurally disrupting TNFR1 mutations.
Unlike ALPS, in which mutant Fas receptors interact with wild-type Fas at the plasma membrane,15 we have found that TRAPS mutant TNFR1 had diminished interactions with wild-type receptors when assayed by FRET. It was previously reported using crosslinking agents that wild-type and TRAPS mutant TNFR1 could associate with each other.32 However, as shown in Figure S1B, coprecipitating receptors were exclusively coaggregated into high-molecular-weight disulfide-linked oligomers, unlike normal receptors that interact noncovalently via the PLAD. These interactions are unlikely to be functionally significant, since the TRAPS mutant TNFR1 could not interfere with TNF-induced apoptosis or secretion of soluble TNFR1 (Figures 1, 3). Furthermore, cotransfected wild-type and mutant TNFR1 trafficked independently of one another, indicating that even if wild-type and mutant receptors interact to some degree, these interactions would be limited by their distinct subcellular localizations.
Other groups have reported that transfected TNFR1 harboring TRAPS-associated mutations have defective surface expression.10,11 In one report,10 overexpressed full-length TNFR1 was found to form cytoplasmic aggresomes in 293T cells. We also observed this phenomenon with both wild-type and mutant TNFR1 expressed at similarly high levels (data not shown). However, in cells that express transfected proteins at lower levels, we found that wild-type TNFR1 is expressed in its characteristic Golgi localization pattern. In these cells the TRAPS mutant TNFR1 clearly overlapped with the ER marker Srβ in live cells. These experiments are in agreement with the lack of surface expression of the mouse T50M TNFR1 mutant in knock-in mice. Therefore, we believe that the ER is likely to be the site of TRAPS mutant TNFR1 relocalization under physiologic conditions.
Misfolding of proteins with ER retention has been associated with a number of genetic diseases, including cystic fibrosis and α1-antitrypsin deficiency associated with the Z allele.30,33,34 These misfolded proteins are thought to drive ER stress signaling pathways that, in some cases, can be proinflammatory.35,36 Very little is known about the consequences of accumulation of misfolded proteins in myeloid and endothelial cells, the cells that express the highest levels of TNFR1. In 293T and HT1080 cells, TNFR1 expression did not acutely provoke either chaperone protein up-regulation characteristic of the UPR, or death-domain–independent NF-κB induction characteristic of the EOR. However, ER stress triggered by mutant TNFR1 may contribute to the exaggerated inflammatory responses of TRAPS patients over longer periods of time in other cell types, or through other signaling pathways. When we examined the direct signaling capability of TRAPS mutant receptors, NF-κB and apoptosis induction was actually reduced, arguing against dominant active TNFR1 signaling as the basis of TRAPS. In this light, it is intriguing that dermal fibroblasts from TRAPS patients were resistant to TNF-induced apoptosis.37
This work has identified a common molecular basis for the altered receptor trafficking of TNFR1 mutants in TRAPS. The common theme of structural perturbation, oligomerization, and altered trafficking of all TNFR1 mutants studied here, combined with the fact that no TRAPS-associated TNFR1 mutations that result in loss of TNFR1 protein expression have been identified in TRAPS, suggests that the disease results from more than simple loss of normal TNF binding or reduced secretion of the mutant receptors. Instead, it seems likely that TRAPS may result from consequences of the abnormally retained TRAPS mutant TNFR1. Therapy with anti-TNF agents for TRAPS may work indirectly to treat the inflammatory symptoms in TRAPS without correcting the underlying molecular abnormality. This may explain the limited effectiveness of anti-TNF agents in some TRAPS patients.38 Therapies directed at inhibiting expression, promoting degradation, or aiding correct folding of the mutant receptors may be additional targets for therapy in TRAPS.
Prepublished online as Blood First Edition Paper, May 9, 2006; DOI 10.1182/blood-2005-11-006783.
Supported by the NIH-University of Oxford Scholars in Biomedical Sciences graduate partnership program (A.A.L.) and the UK Medical Research Council (F.C.K. and G.R.S.). J.R.M. and A.J.J. are Howard Hughes Medical Institute (HHMI)–NIH Research Scholars.
A.A.L. and F.C.K. contributed equally to this work.
A.A.L. and F.C.K. designed, carried out, and analyzed experiments; J.R.M., H.K., A.J.J., and K.M.H. designed and performed experiments; R.M.S, D.L.K., and G.R.S. designed and interpreted experiments; and A.A.L, F.C.K., and R.M.S. wrote the paper. All authors reviewed the paper.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Kristien Zaal and the NIAMS light microscopy facility for excellent technical support and reagents; and Francis Chan, Jennifer Lippincott-Schwartz, David Ron, Amy Lee, Feras Hawari, Stewart Levine, Stephan Siebert, and Paul Brennan for advice and reagents. We would also like to thank Isabela Wieczorek for her assistance.