Abstract
In advanced-phase chronic myeloid leukemia (CML), resistance to imatinib mesylate is associated with point mutations in the BCR-ABL kinase domain. A new generation of potent ABL kinase inhibitors is undergoing clinical evaluation. It is important to generate specific resistance profiles for each of these compounds, which could translate into combinatorial and sequential treatment strategies. Having characterized nilotinib (AMN107) against a large panel of imatinib mesylate–resistant Bcr-Abl mutants, we investigated which mutants might arise under nilotinib therapy using a cell-based resistance screen. In contrast to imatinib mesylate, resistance to nilotinib was associated with a limited spectrum of Bcr-Abl kinase mutations. Among these were mutations affecting the P-loop and T315I. Rarely emerging resistant colonies at a concentration of 400 nM nilotinib exclusively expressed the T315I mutation. With the exception of T315I, all of the mutations that were identified were effectively suppressed when the nilotinib concentration was increased to 2000 nM, which falls within the peak-trough range in plasma levels (3.6-1.7 μM) measured in patients treated with 400 mg twice daily. Our findings suggest that nilotinib might be superior to imatinib mesylate in terms of the development of resistance. However, our study indicates that clinical resistance to nilotinib may be associated with the predominant emergence of T315I.
Introduction
The tyrosine kinase inhibitor imatinib mesylate (Gleevec; Novartis Pharma, Basel, Switzerland) fundamentally changed the standard of care for the treatment of chronic myeloid leukemia (CML). In the International Randomized Interferon versus STI571 Study (IRIS) trial, imatinib mesylate achieved a complete cytogenetic response rate of 84% in patients with newly diagnosed CML after a 42-month follow-up.1 However, in advanced-phase CML and Philadelphia chromosome–positive acute lymphoblastic leukemia (Ph+ ALL), responses to imatinib mesylate may be of a short duration.2-7 Clinical resistance to imatinib mesylate has been attributed to amplification of the BCR-ABL gene,8,9 clonal evolution,9-13 and, most importantly, BCR-ABL gene mutations that obviate binding to the drug target.8,14-19 Consequently, alternative Bcr-Abl kinase inhibitors, such as PD166326, nilotinib (AMN107), or dasatinib (BMS-354825) have been developed. Nilotinib and dasatinib have already entered clinical trials and have displayed promising activity in patients with imatinib mesylate–resistant chronic- and advanced-phase CML20-23 and imatinib mesylate–resistant Ph+ ALL, respectively.21 These compounds are more potent than imatinib mesylate in suppressing Bcr-Abl tyrosine kinase activity and cell growth of Bcr-Abl–transformed cells and are capable of suppressing imatinib mesylate–resistant, mutant forms of Bcr-Abl.24-30 This is in line with structural data, suggesting different binding properties for imatinib mesylate and alternative Abl kinase inhibitors such as PD173955 and nilotinib to the Bcr-Abl kinase domain.30-32 Thus, specific resistance profiles for different therapeutically used Abl kinase inhibitors can be expected to emerge in treated patients, and it may be important to know these resistant profiles upfront to guide therapeutic decisions. To fully characterize nilotinib, we evaluated it against a panel of Ba/F3 cell lines expressing imatinib mesylate–resistant Bcr-Abl. To identify specific resistance mutations in Bcr-Abl which could emerge under therapy with this new agent, we then evaluated nilotinib in the cell-based screening method which we have previously established.33
Materials and methods
Inhibitors
Nilotinib (AMN107) was dissolved in dimethyl sulfoxide (DMSO) to give a 10-mM stock solution which was stored at –20°C. Imatinib mesylate was dissolved in water, and 10-mM stock solutions were stored at –20°C.
Cell culture and DNA constructs
Ba/F3 cells were maintained, transfected, and transformed as described previously.33 Point mutations that were either identified in the nilotinib screen or that are known to be associated with imatinib mesylate resistance in patients with CML or Ph+ ALL were introduced in Mig EGFP Bcr-Abl p18534,35 using the QuickChange mutagenesis kit (Stratagene, Amsterdam, the Netherlands). Mutant p185 was then subcloned as described before.33 Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). Bcr-Abl–specific nested reverse transcription–polymerase chain reaction of the Bcr-Abl kinase domain was performed as described before.33
Resistance screen
Screening for inhibitor-resistant colonies was performed as described previously.33 Briefly, Ba/F3 Mig EGFPp185 wild-type cells were cultured in 96-well plates at a density of 4 × 105 cells per well in the presence of nilotinib at 50, 100, 150, 200, 300, and 400 nM. Culture supernatants were replaced by fresh medium containing inhibitor after 48 hours. Cell colonies that became visible were picked, expanded, and analyzed. Resulting inhibitor-resistant sublines were cultured in the presence of inhibitor at a concentration corresponding to that used during the screen.
Effects of imatinib mesylate and nilotinib on Bcr-Abl autophosphorylation
The phosphorylation status of Bcr-Abl in cell lysates was determined with a capture enzyme-linked immunosorbent assay (ELISA): Ba/F3 cells were seeded at 1 to 2 × 105 cells in 50 μL/well in 96-well round bottom tissue culture plates. Compounds (serial dilutions) were added to the cells and incubated for 1.5 hours (37°C, 5% CO2). Untreated cells were used as controls. After centrifugation, the cell pellets were lysed by addition of 150 μL cold lysis buffer (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM EGTA, 1% NP-40, 2 mM Na3VO4, 1 mM PMSF, 50 μg/mL aprotinin, and 80 μg/mL leupeptin). The lysates (50 μL/well) were transferred to black ELISA plates (Packard HTRF-96 black plates) that had been coated (4°C, overnight) with the capture Ab anti–Abl-SH3 domain Ab 06-466 from Upstate (Charlottesville, VA) (50 ng/well) and blocked (4 hours, 22°C) with PBST, 3% TopBlock (Juro). After 3 to 4 hours of incubation at 4°C, phosphorylation of the captured Bcr-Abl was detected with PY20(AP) diluted in blocking buffer to 0.1 to 0.33 μg/mL. Finally, chemiluminescence was quantified after 45 minutes of incubation with 90 μL per well of the AP substrate (CDPStar RTU with Emerald II from Applied Biosystems [Weiterstadt, Germany] [no. T2388C]) by measuring counts per second (CPS) with a Packard Top Count Microplate Scintillation Counter (Top Count). Compound effects were calculated as the percentage of reduction of the readout for the positive controls. From the dose-response curves, IC50 values were calculated by graphic extrapolation.
Effects of imatinib mesylate and nilotinib on cell growth
Effects on cell viability (and proliferation) were determined by using an MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium)–based method (absorption of formazan at 490 nm; CellTiter 96; Promega, Madison, WI) with measurements taken in triplicates after 24 and 48 hours of culture. Alternatively, the luminescent ATPLite kit (Perkin Elmer Life Sciences; no. 6016947) was used according to the instructions of the suppliers. After 72 hours of incubation in the absence or presence of the drug, light emission (luminescence) was measured with a Packard TopCount. After subtraction of the background signal, compound effects were calculated as the percentage of reduction of the control signal. TGI50 values were determined from the dose-response curves by graphic extrapolation.
Western blot
Ba/F3 cells were cultured for 2.5 hours without and in the presence of inhibitor at the indicated concentrations. Cell lysis, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were done as described previously.36 Abl antibodies were obtained from Pharmingen (8E9) (BD Biosciences, Heidelberg, Germany) and Calbiochem-Novabiochem (Ab3) (Schwalbach, Germany). Antibodies to phosphotyrosine were purchased from Upstate Biotechnology (4G10) (Biozol, Eching, Germany) and Transduction (PY20) (BD Biosciences). Bands were visualized using the enhanced chemoluminescence (ECL) system (Amersham, Braunschweig, Germany).
Results
Activity of nilotinib against known imatinib mesylate–resistant Bcr-Abl mutants
The autophosphorylation activity of the Bcr-Abl fusion protein in cells was quantified with a capture ELISA using a c-Abl–specific capture antibody, together with an enzyme-labeled antiphosphotyrosine antibody and a luminescent substrate. The background tyrosine kinase activity of c-Abl was too low to be detected. The effects of imatinib mesylate and nilotinib on Ba/F3 cell lines expressing wild-type or imatinib mesylate–resistant Bcr-Abl are shown in Table 1. Imatinib mesylate and nilotinib inhibited wild-type Bcr-Abl autophosphorylation with mean IC50 values of 208 and 49 nM, respectively. In confirmation of previous reports,30,37 the Y253H, E255K, and E255V mutants were inhibited with IC50 values between 200 and 800 nM, and the T315I mutant was insensitive to nilotinib at concentrations less than 10 000 nM (data not shown). In addition, a number of less frequently observed imatinib mesylate resistance mutations were potently inhibited by nilotinib with mean IC50 values in the 30 to 200 nM range (Table 1). These effects were highly correlated with those on cell proliferation with nilotinib inhibiting the growth of wild-type Bcr-Abl–expressing Ba/F3 cells with a mean IC50 value of 28 nM (imatinib mesylate, 430 nM) and inhibiting mutant cell lines with mean IC50 values in the 20 to 300 nM range.
Effects of imatinib mesylate and nilotinib on known imatinib mesylate–resistant mutant forms of Bcr-Abl
Bcr-Abl form or construct . | Imatinib mesylate . | . | Nilotinib . | . | ||
|---|---|---|---|---|---|---|
| . | Autophosphorylation . | Proliferation . | Autophosphorylation . | Proliferation . | ||
| Wild-type p185 | 208 (2) | 430 ± 11 (9) | 49 (2) | 28 ± 7 (12) | ||
| M2371 | 399 (2) | 1545 (2) | 34 (2) | 35 (2) | ||
| L248V | 1011 (2) | 2081 (2) | 83 ± 7 (3) | 102 ± 13 (4) | ||
| G250A | 313 (2) | 1269 (2) | 46 (2) | 65 (2) | ||
| G250V | 489 (2) | 624 (2) | 54 (2) | 19 (2) | ||
| E255D | 754 (2) | 1082 (2) | 51 (2) | 24 (2) | ||
| E255R | 1877 (2) | 1567 (2) | 235 (2) | 57 (2) | ||
| E275K | 1038 (2) | 563 (2) | 125 (2) | 29 (2) | ||
| D276G | 1284 (2) | 2486 (2) | 107 (2) | 77 (2) | ||
| E281K | 584 (2) | 1601 (2) | 39 (2) | 49 (2) | ||
| E285N | 919 (2) | 1264 (2) | 188 (2) | 63 (2) | ||
| F311V | 1480 (2) | 3535 (2) | 84 ± 2 (3) | 155 ± 31 (4) | ||
| F317C | 1090 (2) | 694 (2) | 57 (2) | 19 (2) | ||
| F317V | 544 ± 47 (3) | 549 ± 173 (4) | 95 ± 28 (3) | 28 ± 4 (4) | ||
| D325N | 584 (2) | 887 (2) | 69 (2) | 25 (2) | ||
| S348L | 553 (2) | 1370 (2) | 54 (2) | 26 (2) | ||
| E355A | 676 (2) | 1434 (2) | 91 (2) | 38 (2) | ||
| E355G | 601 (2) | 1149 (2) | 67 ± 15 (3) | 47 ± 8 (4) | ||
| F359C | 1130 (2) | 2377 (2) | 201 (2) | 291 (2) | ||
| A380S | 2617 (2) | 3744 (2) | 125 (2) | 179 (2) | ||
| L387F | 530 (2) | 172 (2) | 172 (2) | 39 (2) | ||
| M388L | 517 (2) | 525 (2) | 68 (2) | 20 (2) | ||
Bcr-Abl form or construct . | Imatinib mesylate . | . | Nilotinib . | . | ||
|---|---|---|---|---|---|---|
| . | Autophosphorylation . | Proliferation . | Autophosphorylation . | Proliferation . | ||
| Wild-type p185 | 208 (2) | 430 ± 11 (9) | 49 (2) | 28 ± 7 (12) | ||
| M2371 | 399 (2) | 1545 (2) | 34 (2) | 35 (2) | ||
| L248V | 1011 (2) | 2081 (2) | 83 ± 7 (3) | 102 ± 13 (4) | ||
| G250A | 313 (2) | 1269 (2) | 46 (2) | 65 (2) | ||
| G250V | 489 (2) | 624 (2) | 54 (2) | 19 (2) | ||
| E255D | 754 (2) | 1082 (2) | 51 (2) | 24 (2) | ||
| E255R | 1877 (2) | 1567 (2) | 235 (2) | 57 (2) | ||
| E275K | 1038 (2) | 563 (2) | 125 (2) | 29 (2) | ||
| D276G | 1284 (2) | 2486 (2) | 107 (2) | 77 (2) | ||
| E281K | 584 (2) | 1601 (2) | 39 (2) | 49 (2) | ||
| E285N | 919 (2) | 1264 (2) | 188 (2) | 63 (2) | ||
| F311V | 1480 (2) | 3535 (2) | 84 ± 2 (3) | 155 ± 31 (4) | ||
| F317C | 1090 (2) | 694 (2) | 57 (2) | 19 (2) | ||
| F317V | 544 ± 47 (3) | 549 ± 173 (4) | 95 ± 28 (3) | 28 ± 4 (4) | ||
| D325N | 584 (2) | 887 (2) | 69 (2) | 25 (2) | ||
| S348L | 553 (2) | 1370 (2) | 54 (2) | 26 (2) | ||
| E355A | 676 (2) | 1434 (2) | 91 (2) | 38 (2) | ||
| E355G | 601 (2) | 1149 (2) | 67 ± 15 (3) | 47 ± 8 (4) | ||
| F359C | 1130 (2) | 2377 (2) | 201 (2) | 291 (2) | ||
| A380S | 2617 (2) | 3744 (2) | 125 (2) | 179 (2) | ||
| L387F | 530 (2) | 172 (2) | 172 (2) | 39 (2) | ||
| M388L | 517 (2) | 525 (2) | 68 (2) | 20 (2) | ||
Ba/F3 cells were engineered to express mutant forms of Bcr-Abl p185 which are known to mediate resistance to imatinib mesylate, in extension to frequent clinical isolates already studied.30,37 A cell-capture enzyme linked immunosorbent assay was used to evaluate drug effects on Bcr-Abl kinase activity, and an ATLite assay (Perkin Elmer) was used to evaluate effects on cell growth. Results are expressed as mean IC50 values (nM) ± SEM (number of replicates).
Screening for nilotinib-resistant Ba/F3 clones
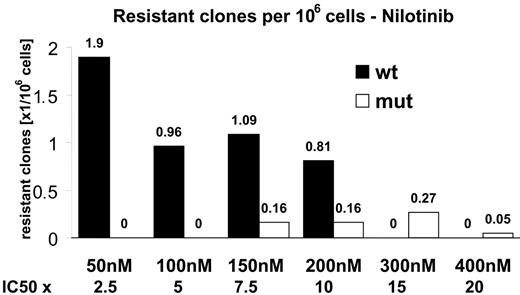
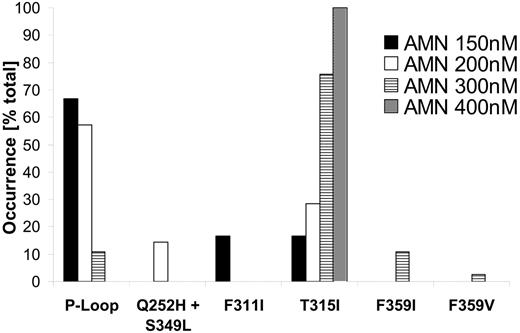
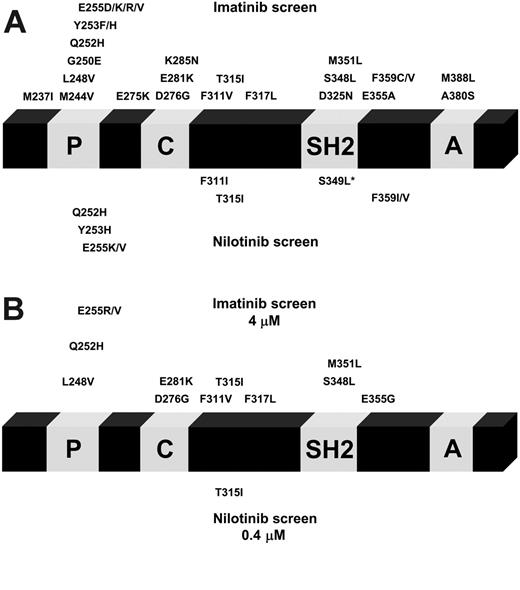
Using a cell-based screening system, we generated a total of 234 Ba/F3 cell clones displaying nilotinib resistance. The frequency of resistant cell clones emerging in the presence of nilotinib at 50 nM, which corresponds to 2.5 times the IC50 value, was 1.9 per million cells (Figure 1), and decreased with increasing nilotinib concentrations to 0.05 per million at 400 nM (20-fold the IC50 value). Thus, an increase of nilotinib concentration by a factor of 8 reduced the frequency of nilotinib-resistant Ba/F3 cell clones by a factor of 38. We then investigated whether the nilotinib-resistant Ba/F3 cell clones harbored Bcr-Abl kinase domain mutations which were reduced in their sensitivity to the drug. Mutant forms of Bcr-Abl were not detected in cell clones surviving 50 nM and 100 nM nilotinib. However, from cell lines growing in the presence of 150, 200, 300, and 400 nM nilotinib, 6 of 48, 6 of 37, 37 of 37, and 2 of 2 resistant clones, respectively, had Bcr-Abl kinase domain mutations. This corresponded to a frequency of mutant clones of 0.16, 0.16, 0.27, and 0.05 per million cells at 150, 200, 300, and 400 nM, respectively (Figure 1). For all concentrations, the P-loop contained 4 of 9 different exchanges identified within the Bcr-Abl kinase domain, including Q252H, Y253H, E255K, and E255V. P-loop mutations predominated at lower nilotinib concentrations (150 and 200 nM; Figure 2). When inhibitor concentrations were increased from 150 to 200 and 300 nM, respectively, we observed a shift from the low-grade resistant P-loop mutant Q252H to more resistant ones (Y253H and E255K/V). At the same time, the ratio of P-loop versus T315I changed in favor of T315I, constituting 88% and 100% of mutations at 300 nM and 400 nM, respectively (Figure 2). T315I represented the most abundant single exchange, which was present in 33 of 51 mutant clones. Less frequently observed mutations at lower concentrations included F311I, a Q252H/S349L double mutant, and F359I (Figure 2). An exchange at F359 to isoleucine and an exchange at S349 have thus far not been described in patients resistant to imatinib mesylate. Compared with the cell-based resistance screen with imatinib mesylate, which we have reported recently,33 nilotinib produced a lower frequency of inhibitor-resistant clones. A frequency of 0.56 per million cells was observed at 4 μM imatinib mesylate (0.1 per million/wild-type and 0.46 per million/mutant),33 as opposed to 0.05 per million (mutant only) at 400 nM nilotinib (Figure 1). In addition, nilotinib produced a limited spectrum of Bcr-Abl kinase domain mutations when compared with imatinib mesylate. For all inhibitor concentrations, the nilotinib screen produced 9 exchanges at 7 different positions, as opposed to 26 exchanges at 21 positions with imatinib mesylate33 (Figure 3A). At 400 nM nilotinib, resistant clones exclusively contained T315I, whereas at 4 μM imatinib mesylate, there were still 12 exchanges at 11 different positions (Figure 3B).
Analysis of nilotinib resistance mutations identified in the screen
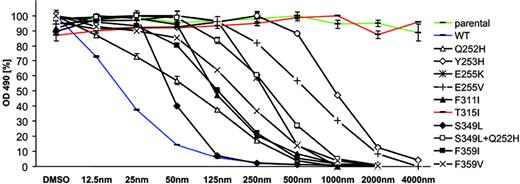
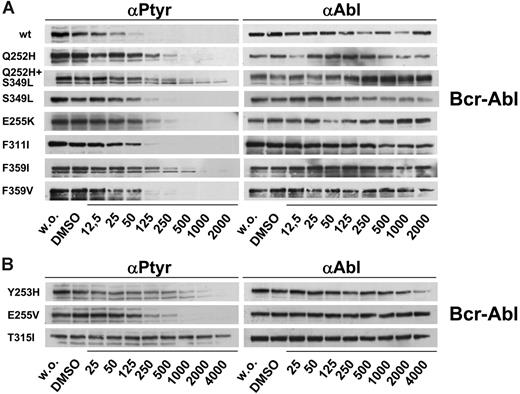
Mutations identified in the nilotinib screen were re-created using site-directed mutagenesis and expressed in Ba/F3 cells. In cell growth assays, dose-response curves indicated that all these mutations mediate resistance to nilotinib (Figure 4; Table 2). P-loop mutations shifted IC90 values from 87 nM for wt Bcr-Abl to concentrations ranging from 383 nM (Q252H) to 2640 nM (Y253H). However, proliferation of clones expressing weak (Q252H, E255K) and also strong P-loop mutations (Y253H, E255V) was effectively inhibited when the nilotinib concentration was increased to 4000 nM (Figure 4). F311I caused a moderate resistance to nilotinib (IC50, 121 nM; IC90, 467 nM). In contrast, in line with previous reports,30,37 T315I caused a complete resistance to nilotinib at concentrations of 4000 nM and above (Figure 4; Table 2). The double mutant Q252H/S349L displayed a cooperative reduction of inhibitor sensitivity compared with both single mutations (Figure 4; Table 2). Both exchanges at F359 were completely inhibited by 500 to 1000 nM nilotinib (Figure 4). In accordance with growth inhibition assays, Bcr-Abl autophosphorylation was suppressed in Ba/F3 cells expressing wild-type Bcr-Abl (Figure 5A). All mutants except T315I displayed complete suppression of Bcr-Abl autophosphorylation at concentrations ranging from 250 nM (F311I, F359V) to 4000 nM (Y253H).
Cellular IC90 values of Bcr-Abl resistance mutations identified in the nilotinib screen
Mutation . | Nilotinib, nM . | Fold, IC90 wt . |
|---|---|---|
| Wild-type p185 | 87 | NA |
| Q252H | 383 | 4.4 |
| Y253H | 2640 | 30.2 |
| E255K | 644 | 7.4 |
| E255V | 1928 | 22.1 |
| F311I | 467 | 5.4 |
| T315I | > 4000 | > 45 |
| S349L | 119 | 1.4 |
| S349L + Q252H | 887 | 10.2 |
| F359I | 433 | 5 |
| F359V | 682 | 7.8 |
Mutation . | Nilotinib, nM . | Fold, IC90 wt . |
|---|---|---|
| Wild-type p185 | 87 | NA |
| Q252H | 383 | 4.4 |
| Y253H | 2640 | 30.2 |
| E255K | 644 | 7.4 |
| E255V | 1928 | 22.1 |
| F311I | 467 | 5.4 |
| T315I | > 4000 | > 45 |
| S349L | 119 | 1.4 |
| S349L + Q252H | 887 | 10.2 |
| F359I | 433 | 5 |
| F359V | 682 | 7.8 |
Ba/F3 cells were transfected with engineered mutant forms of Bcr-Abl and treated as stated in Figure 4. IC90 values and fold increase in comparison to Bcr-Abl wild type-expressing Ba/F3 cells were calculated using the resulting growth curves.
NA indicates not applicable.
Frequency of resistant clones decreases with increasing nilotinib concentrations. Ba/F3 Mig EGFP Bcr-Abl p185 wild-type cells were cultured in 96-well plates in the presence of nilotinib at the indicated concentrations. Resistant colonies were picked and analyzed for the presence of point mutations within the Bcr-Abl kinase domain. Wild-type (□) and mutant (▪) sublines are shown separately. The frequency is shown as resistant clones per million cells at the beginning of culture.
Frequency of resistant clones decreases with increasing nilotinib concentrations. Ba/F3 Mig EGFP Bcr-Abl p185 wild-type cells were cultured in 96-well plates in the presence of nilotinib at the indicated concentrations. Resistant colonies were picked and analyzed for the presence of point mutations within the Bcr-Abl kinase domain. Wild-type (□) and mutant (▪) sublines are shown separately. The frequency is shown as resistant clones per million cells at the beginning of culture.
Occurrence of P-loop mutations and T315I changes inversely with increasing nilotinib concentrations. The relative abundance of different mutations is depicted for each nilotinib concentration that was used in the screen. P-loop mutations included Q252H, Y253H, E255K, and E255V. A Q252H/S349L double mutant is shown separately.
Occurrence of P-loop mutations and T315I changes inversely with increasing nilotinib concentrations. The relative abundance of different mutations is depicted for each nilotinib concentration that was used in the screen. P-loop mutations included Q252H, Y253H, E255K, and E255V. A Q252H/S349L double mutant is shown separately.
Nilotinib produces a limited set of Bcr-Abl kinase domain mutations. The position of identified mutations within the Bcr-Abl kinase domain is shown for imatinib mesylate (above) and nilotinib (below). Point mutations that were identified in imatinib mesylate–resistant clones were described previously and are shown for comparison.33 (A) Twenty-six exchanges affecting 21 different positions were identified in cell clones resistant to 1, 2, or 4 μM imatinib mesylate (above). With nilotinib at 150, 200, 300, or 400 nM, 9 exchanges at 7 positions were found (below). S349L was identified in conjunction with Q252H. *Q252H/S349L double mutant. (B) Resistance mutations identified using imatinib mesylate at 4 μM (above) compared with 400 nM nilotinib (below).
Nilotinib produces a limited set of Bcr-Abl kinase domain mutations. The position of identified mutations within the Bcr-Abl kinase domain is shown for imatinib mesylate (above) and nilotinib (below). Point mutations that were identified in imatinib mesylate–resistant clones were described previously and are shown for comparison.33 (A) Twenty-six exchanges affecting 21 different positions were identified in cell clones resistant to 1, 2, or 4 μM imatinib mesylate (above). With nilotinib at 150, 200, 300, or 400 nM, 9 exchanges at 7 positions were found (below). S349L was identified in conjunction with Q252H. *Q252H/S349L double mutant. (B) Resistance mutations identified using imatinib mesylate at 4 μM (above) compared with 400 nM nilotinib (below).
Discussion
The availability of small molecule kinase inhibitors specifically acting on different oncogeneic tyrosine kinases undoubtedly inaugurated new therapeutic options in a variety of neoplastic disorders. The ABL kinase inhibitor imatinib mesylate induces impressive and durable responses in chronic myeloid leukemia, with a low risk of relapse and only a small percentage of patients being unable to tolerate the drug.1,38,39 However, the development of resistance constituted a major drawback in the treatment of advanced-phase CML. Resistance to imatinib mesylate in CML often is associated with specific point mutations in the BCR-ABL kinase domain that affect binding of imatinib mesylate to its target.8,14-19 Specific mutations mediating imatinib mesylate resistance were also detected in FIP1L1-PDGFRα in patients with hypereosinophilic disorders40,41 and in the C-KIT kinase domain in patients with gastrointestinal stromal tumors.42,43 This phenomenon is not specific for imatinib mesylate, because a resistance mutation in the FLT3 kinase domain has been described in a patient who received PKC412 for acute myeloid leukemia,44 and a T790M mutation in the epidermal growth factor gene (EGFR), which is homologous to the T315I mutation in BCR-ABL, has been reported in patients with lung cancer resistant to the EGFR inhibitor gefitinib (Iressa).45,46
Of all mutations identified, T315I was the only exchange mediating full resistance to nilotinib. Mutations identified in the nilotinib screen were recreated in Bcr-Abl p185 using site-directed mutagenesis and expressed in Ba/F3 cells. After incubation for 24 and 48 hours without and in the presence of nilotinib at the indicated concentrations, proliferation was measured using an MTS-based method. At least 2 independent experiments were performed for each construct. Representative results of 1 experiment after 48 hours of incubation are shown. OD indicates optical density. Values are expressed as mean of triplicates. Bars are ± SE. S349L, although not identified in the screen, was included for comparison with Q252H and the Q252H/S349L double mutant.
Of all mutations identified, T315I was the only exchange mediating full resistance to nilotinib. Mutations identified in the nilotinib screen were recreated in Bcr-Abl p185 using site-directed mutagenesis and expressed in Ba/F3 cells. After incubation for 24 and 48 hours without and in the presence of nilotinib at the indicated concentrations, proliferation was measured using an MTS-based method. At least 2 independent experiments were performed for each construct. Representative results of 1 experiment after 48 hours of incubation are shown. OD indicates optical density. Values are expressed as mean of triplicates. Bars are ± SE. S349L, although not identified in the screen, was included for comparison with Q252H and the Q252H/S349L double mutant.
For imatinib mesylate, cellular IC50 values in BCR-ABL–transformed cell lines are reported to be in the range of 200 to 400 nM.15,30,47,48 In contrast, cellular IC50 values of alternative ABL kinase inhibitors are below 10 nM for the pyridopyrimidine PD166326,25,26 14 nM for the trisubstituted purine analog AP23464,28 20 to 60 nM for the aminopyrimidine nilotinib (AMN107),30 and below 2.5 nM for the disubstituted pyrimidine derivative dasatinib (BMS-354825).29 In addition, these novel compounds are capable of suppressing imatinib mesylate–resistant mutant forms of BCR-ABL, with the exception of T315I.24-30,37 Nilotinib and dasatinib have entered clinical trials and have displayed promising activity in patients with imatinib mesylate–resistant chronic- and advanced-phase CML20-23 and imatinib mesylate–resistant Ph+ ALL, respectively.21 However, data from analyses of resistant patients, especially in the setting of chronic-phase CML, will not be available until a large number of patients have been treated within clinical trials. Because ABL kinase inhibitors based on different chemical scaffolds exhibit distinct modes of binding to the ABL kinase domain,30-32 each inhibitor class or even each single compound may display a distinct profile of amino acid residues critical for binding and inhibition.
Here, we show that nilotinib, in terms of both kinase inhibition as well as Bcr-Abl–dependent cell proliferation, potently inhibits 21 known imatinib mesylate–resistant mutant forms of Bcr-Abl, with IC50 values in the 20 to 300 nM range. As previously reported,30,37 the P-loop mutants Y253H, E255K, and E255V displayed significant resistance to nilotinib with IC50 values in the 200 to 800 nM range, and nilotinib did not inhibit T315I. Because in phase 1 clinical studies in patients with CML, oral administration of nilotinib at 400 mg twice daily provided maximal and trough plasma drug concentrations of 3.6 and 1.7 μM, respectively,20,21 only the T315I BCR-ABL mutant is expected to be insensitive to nilotinib therapy. However, this does not address potential new mutations which might emerge under treatment with nilotinib.
Inhibition of autophosphorylation in Bcr-Abl–mutant, nilotinib-resistant cell lines. Mutations that emerged in the nilotinib screen were reengineered and expressed in Ba/F3 cells. Cells were incubated without and in the presence of nilotinib at the indicated concentrations. Cell lysates were subjected to SDS-PAGE. Blots were probed for phosphotyrosine (left) and Abl (right). (A) Results for resistance mutations displaying cellular IC50 values of less than 500 nM and (B) for mutants causing strong resistance toward nilotinib with cellular IC50 values of 500 nM or above.
Inhibition of autophosphorylation in Bcr-Abl–mutant, nilotinib-resistant cell lines. Mutations that emerged in the nilotinib screen were reengineered and expressed in Ba/F3 cells. Cells were incubated without and in the presence of nilotinib at the indicated concentrations. Cell lysates were subjected to SDS-PAGE. Blots were probed for phosphotyrosine (left) and Abl (right). (A) Results for resistance mutations displaying cellular IC50 values of less than 500 nM and (B) for mutants causing strong resistance toward nilotinib with cellular IC50 values of 500 nM or above.
We have recently reported the results of a cell-based strategy that allows generation of imatinib mesylate–resistant Ba/F3 cell clones in high frequency.33,49 The pattern of mutations identified in these clones corresponded to the pattern observed in patients displaying imatinib mesylate–resistant CML.33 Our findings with nilotinib reported here point to significant advantages over imatinib mesylate that may translate to superior activity of nilotinib in the clinic. When compared with imatinib mesylate, nilotinib produced a lower frequency of resistant cell clones and a reduced spectrum of mutations. The proportion of T315I to P-loop mutations changed in favor of T315I with increasing concentrations of nilotinib. There were only a few clones expressing mutations other than P-loop or T315I. At 400 nM nilotinib, resistant cell clones emerged at a low frequency of 1 per 20 million cells, and these clones exclusively contained T315I. Although bone marrow concentrations are not available, the steady-state trough plasma concentration of 1700 nM following oral administration of 400 mg nilotinib twice daily20,21 is 4 times higher than the maximum concentration of 400 nM used in this screen. In marked contrast, imatinib mesylate at 4000 nM, which approximates to trough concentrations achieved in plasma of treated patients,50 resulted in a frequency of resistant cell clones of 1 per 1.8 million cells.33 These imatinib mesylate–resistant clones still contained a variety of different resistance mutations. Thus, in comparison to imatinib mesylate, nilotinib induced resistant cell clones at a frequency that was by the factor of 10 lower at a 10-fold lower concentration. Consequently, the plasma concentrations of nilotinib achieved in patients treated with 400 mg twice daily might prevent the selection and expansion of resistant disease clones. Moreover, in contrast to imatinib mesylate, clinical resistance to nilotinib is predicted to be associated with the predominant emergence of T315I. Resistance mutations that may arise in the presence of suboptimal nilotinib concentrations include P-loop mutations that mediate a higher degree of nilotinib resistance such as Y253H and E255V. However, because nilotinib plasma levels of up to 3600 nM were measured in phase 1 and 2 studies,20,21 P-loop mutations may emerge less frequently in patients treated with nilotinib, provided that sufficient plasma concentrations are achieved. Also, nilotinib is expected to induce responses in cases in which P-loop mutations cause resistance to imatinib mesylate.
Taken together, our findings indicate that nilotinib should be efficacious in most cases in which BCR-ABL point mutants cause imatinib mesylate resistance and suggest that nilotinib may be superior to imatinib mesylate in terms of the development of resistance. Specific resistance profiles for different ABL kinase inhibitors will guide therapeutic decisions with respect to drug combinations and sequential treatment strategies and will allow the determination of critical plasma concentrations that have to be reached to prevent or overcome specific resistance mutations. This or similar resistance screening approaches can be translated to other neoplastic diseases, whereby rational drug targets are identified and therapeutic kinase inhibitors are available, such as gastrointestinal stromal tumor (GIST), hypereosinophilic syndrome, acute myelogenous leukemia (AML), or lung cancer.
Prepublished online as Blood First Edition Paper, April 13, 2006; DOI 10.1182/blood-2005-12-010132.
Supported by a grant from the BMBF (Federal Ministry of Education and Research) German National Genome Project No. 01-GS-0105 and 01-GS-0447 (J.D. and C.P.).
Two of the authors (P.W.M. and J.M.) are employed by a company (Novartis) whose product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal