Recent studies revisiting the association between plasma thrombin-activatable fibrinolysis inhibitor (TAFI) Ag levels and polymorphisms of the CPB2 gene (coding for TAFI) suggested that TAFI Ag levels were influenced by 2 major quantitative trait nucleotides (QTNs) in European whites. However, the strong linkage disequilibrium (LD) between CPB2 polymorphisms in European whites did not allow one to distinguish which polymorphisms could be the putative QTNs. To get a better insight into the identification of QTNs, a transethnic haplotype analysis contrasting 2 populations of African and European subjects was performed using 13 CPB2 polymorphisms. Results of the haplotype analyses suggested that 3 QTNs had independent effects and explained about 15% of the TAFI variability, consistently in the 2 populations. The lower LD observed in the African population enabled us to identify the 1583T>A SNP located in 3′UTR as one of these QTNs, whereas the -2599C>G and -2345--2344insG SNPs located in the 5′ region might be the 2 other QTNs. A phylogenetic study suggested that these 3 polymorphisms occurred before the period of migration “out of Africa.” Although this transethnic comparison contributed to better map the putative CPB2 QTNs, further studies are required to clarify the role of the promoter region.
Introduction
The thrombin-activatable fibrinolysis inhibitor (TAFI), also known as plasma procarboxypeptidase B,1,2 procarboxypeptidase U,3 or procarboxypeptidase R, is a plasma zygomen that can be activated by the thrombin-thrombomodulin complex into an active enzyme (TAFIa).4-6 TAFIa can then potently attenuate fibrinolysis by removing carboxyterminal lysine residues from partially degraded fibrin during the clot lysis process, which results in a decreased plasminogen binding on fibrin surface and therefore in a decrease of fibrinolytic activity.7,8 Because it plays an important role in the balance between the coagulation and the fibrinolytic system, TAFI has been suspected to constitute a marker for atherothrombotic diseases. Supporting this hypothesis, elevated plasma TAFI concentrations have been reported in patients with deep-vein thrombosis9 and were found more frequently in patients with stable angina pectoris than in a healthy population.10 Plasma TAFI concentrations have also been shown to be higher in patients with type II diabetes mellitus than in healthy controls.11
In healthy individuals, plasma levels of TAFI antigen (TAFI Ag) have been shown to exhibit a large interindividual variability poorly explained by environmental factors.12-14 Plasma levels of TAFI Ag were then suspected to be under a strong genetic influence, a hypothesis supported by several studies showing that polymorphisms of the gene encoding TAFI (named CPB2) were strongly associated with TAFI Ag circulating levels.15,16 The results of a series of combined segregation-linkage analyses strengthened this hypothesis: plasma levels of TAFI Ag were shown to exhibit a strong familial resemblance totally explained by the likely existence of 2 CPB2-linked functional variants.17 These 2 putative variants were postulated to explain more than 70% of the total variability of plasma TAFI Ag levels in healthy adults of European origin.17
The CPB2 gene has been mapped to 13q14.11 and consists of 11 exons spanning approximately 48 kb of genomic DNA.18 A large number of single nucleotide polymorphisms (SNPs) within the CPB2 gene have been described, among which 3 substitutions in the coding sequence,a G to A at nucleotide 505 leading to an Ala to Thr substitution at amino acid 147 (A147T), a C to T at nucleotide 678 leading to a silent mutation (678C>T), and a C to T at nucleotide 1040 leading to a Thr to Ile substitution at amino acid 325 (T325I).19-21 The 2 nonsynonymous polymorphisms were shown to be associated with TAFI Ag levels.15 Moreover, the p.T325I was proved to be functional, the Ile325 allele having increased antifibrinolytic properties in comparison with the Thr325 allele.19 Molecular screening of the 5′ and the 3′ untranslated (3′UTR) regions of CPB2 later identified several SNPs that were all strongly associated with plasma TAFI Ag levels.15,16 However, it was recently demonstrated22,23 that, in some enzyme-linked immunosorbent assays (ELISAs) used for TAFI Ag levels determination, decreased antibody reactivity toward the TAFI Ile325 isoform led to erroneous TAFI Ag levels that were likely to have overestimated TAFI heritability and effects associated with CPB2 polymorphisms in previous studies.15-17 Two subsequent studies readdressed the association between CPB2 polymorphisms and plasma TAFI Ag levels using assays free of isoform-dependent artifact in European white individuals.24,25 Even though the conclusions of both reports were still in agreement with the likely existence of 2 CPB2 polymorphisms underlying TAFI Ag variability, these new analyses suggested that the 2 polymorphisms would explain less than 20% of the total variability of plasma TAFI Ag levels in healthy adults of European origin, compared with the 70% previously estimated.17 However, due to the strong linkage disequilibrium (LD) between all the CPB2 polymorphisms in the European white population, it was not possible to determine which SNPs could be the putative quantitative trait nucleotides (QTNs). In particular, the strong LD between the A147T and the 1583T>A polymorphisms did not allow one to disentangle their effects.24
The lower LD and the greater haplotype diversity observed in African-descent populations26-29 argued that the study of such a population could help to better map the QTNs by identifying informative contrasts between polymorphisms that, in European white populations, could not be discriminated because of strong LD.30,31 In order to get a better insight into the identification of the putative QTNs underlying TAFI Ag variability, we carried out a transethnic association analysis contrasting 2 populations of African and European origins. Since the coding sequence of the CPB2 gene had not been extensively screened, we first performed a molecular screening to identify all common exonic polymorphisms. The detected polymorphisms, as well as those previously identified by the screening of the 5′ and the 3′UTR regions, were studied in relation to plasma TAFI Ag levels by means of haplotype analysis. Finally, a phylogenetic analysis of the CPB2 haplotypes was performed in the 2 populations.
Materials and methods
Study population
The African study sample (170 subjects: 144 men and 26 women) was composed of healthy blood donors aged 19 to 52 years (mean, 29 ± 7 years) recruited in a health care center in Abidjan (Republic of the Ivory Coast). All participants were asked to complete a medical questionnaire and were examined using a standardized epidemiologic protocol. The European study population was composed of 123 healthy men living in the Marseilles area (France). Individuals aged 40 to 60 years (mean, 53 ± 9 years) were recruited on the occasion of a health checkup in a health care center.15 All subjects gave their informed consent to participate in the study and the study was approved by the Centre Hospitalier Universitaire ethics committee. Patients provided informed consent in accordance with the Declaration of Helsinki.
TAFI antigen determination
Blood samples were obtained from the antecubital vein in the morning, after overnight fasting, collected into 3.8% trisodium citrate (0.129 M) (9:1, vol/vol). Platelet-poor plasma was obtained after centrifugation (2500g for 30 minutes at 4°C) and kept frozen below -80°C until analysis. Antigen (Ag) determination of TAFI was performed using the Asserachrom TAFI ELISA (Diagnostica Stago, Asnières, France) according to the manufacturer. This assay is based on 2 monoclonal antibodies raised against TAFI purified from plasma. These antibodies recognize the TAFI 325Thr as well as the TAFI 325Ile isoform, and this assay is free of isoform-dependent artifact as previously described.25,32 Results were expressed as microgram per milliliter.
Detection of new TAFI gene polymorphisms and genotyping
The molecular screening of the entire exonic CPB2 sequence (NM_001872) was performed by comparing 80 chromosomes from 40 European subjects recruited from a systematic screening of a healthy population.15 Genomic DNA was extracted from peripheral blood leukocytes by the salting-out method.33 DNA-sequence variations were identified by polymerase chain reaction (PCR)/single-strand conformational polymorphism (SSCP) and followed by sequencing as previously described.15
The identified SNPs as well as all previously described SNPs of the coding sequence and the 5′ and 3′UTR regions were then genotyped in the present study. Genotyping was performed using polymerase chain reaction (PCR) amplification followed by restriction digestion or using allele-specific PCR.
Statistical analysis
Allele frequencies were estimated by gene counting, and departure from Hardy-Weinberg equilibrium was tested using a χ2 with 1 degree of freedom (df). Allele frequencies were compared between the African and European white populations by a χ2 test with 1 df. Association between each CPB2 polymorphism and TAFI Ag plasma levels was first investigated by use of classical linear model assuming additive allele effects after having tested for the deviation from additivity.
Pairwise LD was estimated using THESIAS software (Tregouet, Paris, France; www.genecanvas.org) based on the SEM algorithm,34 and the extent of disequilibrium was expressed in terms of D′, which is the ratio of the unstandardized coefficient to its maximal/minimal value.35 Haplotype analysis was performed using the THESIAS software, which allows one to simultaneously estimate haplotype frequencies and haplotype effects by comparison with a reference haplotype under the assumption of additive effects of haplotypes on phenotype. The phenotypic mean associated with one dose of each haplotype was reported with its 95% confidence interval. The mean TAFI Ag level of an individual is then the sum of the mean levels associated with his/her 2 haplotypes. A global test of association between haplotypes and the phenotype was performed by a likelihood ratio test (LRT) (χ2 with m - 1 df in the case of m haplotypes). By setting appropriate constraints on parameters, the LRT statistic also allowed us to compare the effects between pairs of haplotypes, in particular those differing by only one nucleotide substitution. Homogeneity of the haplotype effects across populations was tested by introducing corresponding interaction parameters in the haplotypic model. Effects of rare haplotypes (frequency < 3%) were set to 0.
Finally, the evolutionary relatedness of the inferred CPB2 haplotypes was investigated using the maximum parsimony method implemented in the TCS program (Posada, Vigo, Spain).36
Results
Description of CPB2 polymorphisms
Our molecular screening of the coding region in 40 European subjects identified 3 new silent substitutions (a C to T at nucleotide 310 in exon 3B, a T to A at nucleotide 499 in exon 4, and a G to A at nucleotide 663 in exon 6), in addition to the 3 previously described SNPs (678C>T, A147T, and T325I).
All the SNPs of the coding region as well as those previously described in the 5′ and 3′UTR (-2599C>G, -2345--2344insG, -1925T>C, -1690A>G, -1102G>T, -530C>T, -152A>G, -438G>A, 1542C>G, and 1583T>A) were genotyped in the European white and African samples. No carrier of the T499A mutation was detected in either population. The -530C>T, -152A>G, and -1925T>C polymorphisms were in complete association in the African population, while no mutant was detected in European whites. As a consequence, the present analysis included 13 SNPs, 6 in the 5′ region (-2599C>G, -2345--2344insG, -1925T>C, -1690A>G, -1102G>T, -438G>A), 5 in the coding region (310C>T, A147T, 663G>A, 678C>T, T325I), and 2 in the 3′UTR region (1542C>G and 1583T>A).
Allele frequencies and linkage disequilibrium between polymorphisms
Allele frequencies of the 13 SNPs in Africans and European whites are reported in Table 1. All genotype distributions were compatible with Hardy-Weinberg equilibrium in both populations. Most of the SNPs exhibited significant difference in allele frequency between populations.
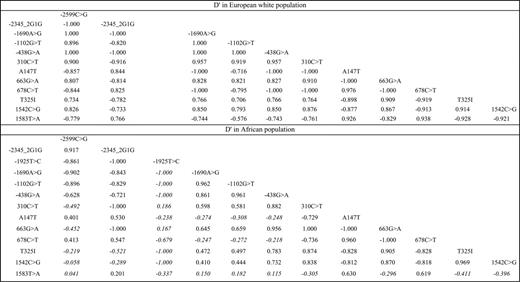
The pattern of LD is reported in Figure 1. In European whites, all SNPs were in strong LD one with each other (the -1690A>G, -1102G>T, and -438G>A SNPs being in complete association). By contrast, SNPs in Africans were distributed into 3 LD blocks, the first one comprising the SNPs of the 5′ region, the second one comprising the SNPs of the coding region and the 1542C>G, and the third one composed of the 1583T>A alone.
Pairwise linkage disequilibrium between CPB2 polymorphisms in D′ in the European white and African populations. All pairwise LDs were significantly different from 0, except those marked in italics.
Pairwise linkage disequilibrium between CPB2 polymorphisms in D′ in the European white and African populations. All pairwise LDs were significantly different from 0, except those marked in italics.
Association of CPB2 polymorphisms with plasma TAFI Ag levels
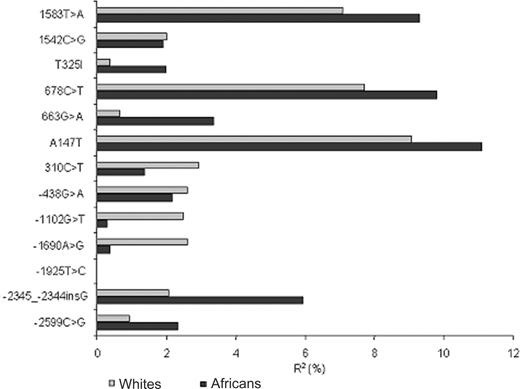
African individuals exhibited lower levels of plasma TAFI Ag than European whites (mean [SE]: 11.23 [2.25] vs 12.37 [2.99] μg/mL; P < 10-3). Figure 2 summarizes the association between CPB2 polymorphisms and plasma levels of TAFI. Full description of association analyses is provided in Tables S1 and S2, available on the Blood website (see the Supplemental Tables link at the top of the online article). In European whites and Africans, 3 SNPs of the coding and 3′UTR regions, A147T, 678T>C, and 1583T>A, were significantly associated with TAFI Ag levels. The percentages of variance explained by these SNPs ranged from 7% to 9% in European whites and from 9% to 11% in Africans. One additional SNP located in the 5′ region, -2345--2344insG, was found associated with TAFI Ag levels only in Africans (R2 = 5.9%, P = .006). In both populations, the SNP most strongly associated with TAFI levels was A147T.
Association of CPB2 haplotypes with plasma TAFI Ag levels
In order to deal with the large number of SNPs, haplotype analyses were first performed separately for the 6 SNPs of the 5′ region on the one hand, and the 7 SNPs of the coding and 3′UTR regions on the other hand.
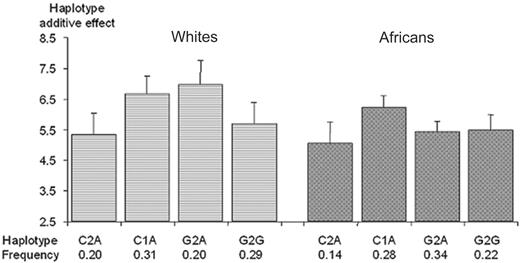
5′ region. In European whites, due to the absence of the -1925T>C SNP and the complete association between the -1690A>G, -1102G>T, and -438G>A SNPs, only 4 haplotypes accounted for the totality of the sampled chromosomes. These 4 haplotypes could be defined by 3 tag SNPs (eg, -2599C>G, -2345--2344insG, and -1690A>G). By contrast, in Africans, due to the lower LD between polymorphisms, the 6 SNPs generated 5 common haplotypes (> 3%) that could be defined by 4 tag SNPs (-2599C>G, -2345--2344insG, -1925T>C, and -1690A>G). The 5 haplotypes generated by these tag SNPs accounted for nearly 95% of the sampled chromosomes. Since in Africans the 2 haplotypes that differed only at position -1925 did not show significant difference in TAFI levels (data not shown), the 4 haplotypes generated by the -2599C>G, -2345--2344insG, and -1690A>G were enough to summarize the association of CPB2 haplotypes with TAFI levels in both populations (Figure 3). These 4 haplotypes explained 8.2% (P = .013) and 6.7% (P = .010) of the variability of plasma TAFI levels in European whites and Africans, respectively. In both populations, the C1A haplotype was associated with a higher mean TAFI level than the C2A haplotype (6.68 vs 5.34 μg/mL [P = .007] and 6.21 vs 5.07 μg/mL [P = .01] in European whites and Africans, respectively), suggesting an increasing effect associated with the -2345 1G allele. In European whites, the comparison of the C2A versus G2A haplotypes indicated an increasing effect of the -2599G allele (5.34 vs 6.96 μg/mL, P = .004), whereas the comparison of the G2A versus G2G haplotypes additionally suggested a decreasing effect of the -1690G allele (6.96 vs 5.69 μg/mL; P = .03). These effects did not reach significance in Africans.
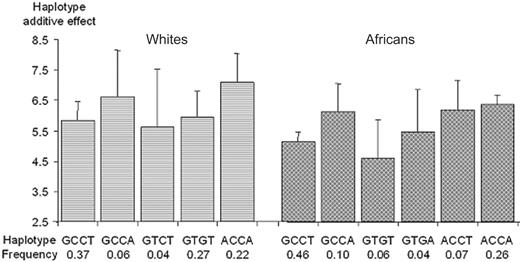
Coding and 3′UTR regions. In European whites, the 7 SNPs generated 5 common haplotypes (frequency, > 3%) accounting for 86% of the sampled chromosomes, whereas in Africans, they generated 6 common haplotypes accounting for 94% of the chromosomes. Although the haplotype structure was not identical in the 2 populations, the common haplotypes could be tagged by the same subset of SNPs consisting of the A147T (G>A), T325I (C>T), 1542C>G, and 1583T>A SNPs (Figure 4). These haplotypes explained 6.7% (P = .11) and 14.6% (P = .001) of the variability of plasma TAFI levels in European whites and Africans, respectively.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the -2599C>G, -2345--2344insG, and -1690A>G polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% CI bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the -2599C>G, -2345--2344insG, and -1690A>G polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% CI bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
In European whites, the 2 haplotypes carrying the +1583A allele were associated with the highest TAFI levels. These 2 haplotypes, only differing at the p.A147T locus, did not show differences in TAFI levels (6.61 vs 7.10 μg/mL, P = .66), suggesting that the effect of the A147T polymorphism observed in univariate analysis was the consequence of its LD with the 1583T>A SNP. No difference in TAFI levels was observed between haplotypes carrying the +1583T allele (χ2 = 0.11 with 2 dfs, P = .94) indicating that, after adjusting for the effect of the g.1583T>A SNP, no other polymorphism was associated with TAFI levels. In Africans, comparisons of the 3 pairs of haplotypes that differed at only the 1583T>A site revealed that the effect of the +1583A allele was homogeneous (χ2 = 1.56 with 2 dfs, P = .46 for the test of homogeneity) across the 3 pairs, the +1583A allele being associated with increased TAFI levels (P < 10-4). No difference in TAFI levels was observed between haplotypes carrying the +1583T allele. In particular, data not shown also indicated that the effect of the 678T>C SNP observed in univariate analysis was also the consequence of its LD with the 1583T>A SNP.
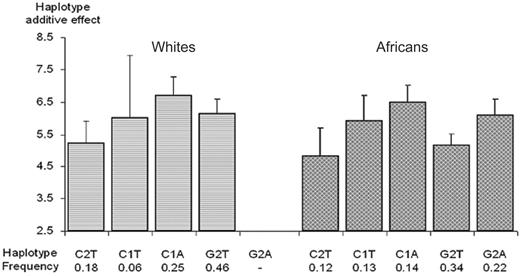
Pooled analysis. In order to cope with the LD between the 5′ and coding/3′ UTR regions, haplotype analysis was performed on the SNPs identified as potentially interesting from the 2 previous subanalyses (ie, the -2599C>G, -2345--2344insG, -1690A>G, and 1583T>A polymorphisms). A first analysis demonstrated that, in both populations, the 2 haplotypes that differed only at position -1690 did not differ in TAFI levels, precluding any effect of this SNP. Therefore, for ease of presentation, Figure 5 illustrates the association between TAFI levels and the main haplotypes generated by the -2599C>G, -2345--2344insG, and 1583T>A SNPs. In both populations, TAFI gene haplotypes explained about 14% of plasma TAFI variability. Except the fact that the G2A haplotype was not present in the European population, the patterns of haplotypic association were very similar in the 2 populations (Figure 5). Haplotype analysis was therefore carried out on the pooled sample of African and European subjects while adjusting for ethnicity, after having checked for the homogeneity of the haplotype effects between populations (test for homogeneity: χ2 = 4.09 with 3 dfs, P = .25). The haplotypes carrying the +1583A allele (C1A and G2A) were associated with higher TAFI levels than their counterparts carrying the +1583T allele (C1T and G2T). The +1583A allele was associated with a population-adjusted average increase of +0.79 (0.28-1.31) μg/mL (P = .003). Besides, the comparison of the C2T versus C1T haplotypes suggested an increasing effect of the -2345 1G allele (+0.93 [0.17-1.69] μg/mL, P = .016), and the comparison of the C2T versus G2T haplotypes additionally suggested an increasing effect of the -2599G allele (+0.70 [0.09-1.32] μg/mL; P = .025). In summary, haplotype analyses indicated that 3 SNPs had independent effects on TAFI levels, -2599C>G, -2345--2344insG, and 1583T>A.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the A147T (G/A), T325I (C/T), 1542C>G, and 1583T>A polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% Ci bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the A147T (G/A), T325I (C/T), 1542C>G, and 1583T>A polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% Ci bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the -2599C>G, -2345--2344insG, and 1583T>A polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% CI bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
Association between plasma TAFI Ag levels and CPB2 haplotypes derived from the -2599C>G, -2345--2344insG, and 1583T>A polymorphisms. Polymorphisms are ordered according to their position on the genomic sequence. Each bar and its 95% CI bracket corresponds to the expected mean of TAFI Ag levels associated with one dose of haplotype under the assumption of additive haplotype effects.
Phylogenetic analysis of CPB2 haplotypes
Phylogenetic analysis was performed to infer the mutational steps among haplotypes. Due to the low to moderate LD between SNPs of the 5′ region and those of the coding/3′UTR region, analyses were performed separately for the 2 regions. Although some haplotypes were population specific, the consensus gene trees were very similar in the 2 populations, supporting the robustness of the results (Figure 6). The reconstructed gene tree for the 5′ region suggested that all haplotypes derived one from another by single nucleotide changes. Haplotypes carrying the different alleles at the -2599 site were divided into 2 distinct clades, whereas the haplotype carrying the 1G allele at the -2345 site derived from a single haplotype. For the coding/3′UTR region, phylogenetic ambiguity was present in the parsimony gene trees reconstructed for both populations, as shown by the loops (Figure 6). However, haplotypes carrying the different alleles at the +1583 site appeared clearly divided into 2 distinct clades.
Consensus parsimony gene trees inferred from CPB2 haplotypes in European whites and Africans. (A) Gene tree constructed from 6 variant sites in the 5′ region (-2599C>G, -2345--2344insG, -1925T>C, -1690A>G, -1102G>T, and -438G>A). (B) Gene tree constructed from 7 variant sites in the coding/3′UTR region (310C>T, A147T [G>A], G633A, 678T>C, T325I [C>T], 1542C>G, and 1583T>A). The relative frequency of each haplotype is indicated by the area of the circle. Haplotypes with a frequency less than 0.02 are not taken into consideration. Full black circles correspond to haplotype not present in the samples. Loops indicate phylogenetic ambiguity due to recombination or to parallel, convergent, or reverse changes. Alleles in underlined bold correspond to the putative QTNs (-2599C>G, -2345--2344insG, and 1583T>A).
Consensus parsimony gene trees inferred from CPB2 haplotypes in European whites and Africans. (A) Gene tree constructed from 6 variant sites in the 5′ region (-2599C>G, -2345--2344insG, -1925T>C, -1690A>G, -1102G>T, and -438G>A). (B) Gene tree constructed from 7 variant sites in the coding/3′UTR region (310C>T, A147T [G>A], G633A, 678T>C, T325I [C>T], 1542C>G, and 1583T>A). The relative frequency of each haplotype is indicated by the area of the circle. Haplotypes with a frequency less than 0.02 are not taken into consideration. Full black circles correspond to haplotype not present in the samples. Loops indicate phylogenetic ambiguity due to recombination or to parallel, convergent, or reverse changes. Alleles in underlined bold correspond to the putative QTNs (-2599C>G, -2345--2344insG, and 1583T>A).
Discussion
Recent studies revisiting the association between plasma TAFI Ag levels and CPB2 polymorphisms have suggested that 2 polymorphisms accounted for about 20% of the plasma TAFI Ag variability in the European white population.24,25 One of these polymorphisms was likely to be located in the promoter region of the TAFI gene, while the second would be located in the coding or 3′ region. However, due to the strong LD between all CPB2 polymorphisms in European whites, it was difficult to distinguish those that have a true functional role from those that are only markers in LD with the CPB2-linked QTNs. One possible strategy to get a better insight into the identification of QTNs is a transethnic comparison in which differences of LD patterns between populations can help to better characterize QTNs, as it was recently shown for angiotensin-1-converting enzyme.31 Indeed, it has been demonstrated that blocks of LDs are smaller in subjects of African origin than in subjects of European origin, which may allow a finer mapping of putative functional polymorphisms.37-39
This report is to our knowledge the first one to investigate the association between CPB2 polymorphisms and plasma TAFI Ag levels in an African population and to compare the results with those obtained in a European white population. All known CPB2 polymorphisms as well as 3 new silent coding mutations, 310C>T, T499A, and 663G>A, were investigated by means of haplotype analysis. Haplotype analysis suggested that among the 3 polymorphisms in the coding and 3′UTR regions associated with plasma TAFI levels in univariate analysis in both populations, the 1583T>A polymorphism was the only one independently associated with the phenotype. While this polymorphism was in relatively strong LD with the SNPs located in the 5′ region in the European white population, the absence of LD in the African population enabled us to clearly demonstrate that the increasing effect of the +1583A allele was independent of any other polymorphism. In addition, 2 SNPs in the 5′ region were found associated with TAFI levels. In both populations, the -2599G and -2345 1G alleles were associated with increased TAFI levels, the effect of these SNPs being independent from each other and from the 1583T>A SNP.
The present data confirmed that CPB2-linked QTNs located in the 5′ and 3′UTR regions independently influenced plasma TAFI levels.24,25 The proportion of TAFI variability attributable to CPB2 haplotypes25 was remarkably similar (around 15%) in the 2 populations despite differences in allele frequencies and environmental backgrounds. The phylogenetic trees were also quite similar between the 2 populations despite the existence of population-specific haplotypes. The fact that the 3 putative QTNs occurred on the same branches of the trees in both populations is in favor of mutations dating from the period before the migration “out of Africa.”
As already suggested from our previous study,25 the 3′UTR QTN might be the 1583T>A, a hypothesis that is consistent with a recent work showing that haplotypes containing the +1583T allele result in a transcript with decreased stability.40 Unexpectedly, the A147T polymorphism did not appear to have its own effect in haplotype analysis, although it exhibited the strongest association with TAFI levels in univariate analysis. This is probably explained by the fact that it was in LD with the 3 putative QTNs and cumulated their effects in univariate analysis. Of importance, as already observed,25 the p.T325I polymorphism was no longer associated with TAFI levels when using the new method free of artifact. Our analysis additionally suggested the existence of 2 QTNs in the 5′ region, -2599C>G and -2345 2G/1G, instead of only one as suggested by our previous studies.24,25 The -2345 2G/1G SNP was, however, not investigated in the first study,24,25 while in the second study, the strong LD in the 5′ region might have hampered a correct identification of the genetic effects.25
While the greater diversity of the CPB2 gene in African subjects helped to identify the 1583T>A SNP as one of the QTLs underlying TAFI levels, the strong LD observed in the 5′ region did not allow us to get a complete insight into the identification of the putative QTNs located in this region. Since our molecular screening was performed only on chromosomes from European subjects, it cannot be ruled out that we missed some SNPs specific to the African population that could have contributed to better map the QTNs. Our screening strategy had about 100% power of detecting any mutation with frequency of 0.05 or more and 56% of 0.01 or more. We cannot exclude that some rare mutations have been missed. Similarly, the search for additional CPB2 SNPs into a public database, such as that of the HAPMAP project (www.hapmap.org), reveals that except several intronic SNPs, only one synonymous SNP located in the 3′UTR region with frequency less than 3.8% was not studied in our analysis. However, our sample size would have been too relatively low to be able to demonstrate any low/moderate effect of this SNP. Had this SNP had a strong effect on TAFI Ag levels, our screening strategy based on comparing 40 DNAs from individuals with low TAFI levels to 40 DNAs from individuals with high TAFI levels15 would have likely detected this SNP. Despite a poor influence of environment on TAFI levels,12,24 we also cannot exclude an interaction between genotype and environment on determination of TAFI plasma levels.
Because of the LD among the 13 SNPs studied, a standard Bonferroni correction for multiple testing could not be applied since it would have been too conservative. Using the method proposed by Li and Ji,41 the number of independent components underlying the LD structure of the set of SNPs was estimated to be approximately 6. After correcting for this number, which corresponds to consider significant any P value of .008 or less, the effect of the 1583T>A SNP remained significant, whereas that of the 2 5′ QTNs was no longer significant. Larger studies conducted in populations of different origins are needed to refine the role of the 5′ region.
In conclusion, this transethnic study helped to better map the CPB2-linked QTNs responsible for the genetic control of TAFI Ag levels. Data from Africans and Europeans converged to identify the 1583T>A polymorphism as one of the QTNs, whereas larger epidemiologic studies and functional experiments are required to better clarify the contribution of the promoter region.
Prepublished online as Blood First Edition Paper, May 16, 2006; DOI 10.1182/blood-2006-01-008094.
C.F. and D.-A.T. contributed equally to the work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 6. Consensus parsimony gene trees inferred from CPB2 haplotypes in European whites and Africans. (A) Gene tree constructed from 6 variant sites in the 5′ region (-2599C>G, -2345--2344insG, -1925T>C, -1690A>G, -1102G>T, and -438G>A). (B) Gene tree constructed from 7 variant sites in the coding/3′UTR region (310C>T, A147T [G>A], G633A, 678T>C, T325I [C>T], 1542C>G, and 1583T>A). The relative frequency of each haplotype is indicated by the area of the circle. Haplotypes with a frequency less than 0.02 are not taken into consideration. Full black circles correspond to haplotype not present in the samples. Loops indicate phylogenetic ambiguity due to recombination or to parallel, convergent, or reverse changes. Alleles in underlined bold correspond to the putative QTNs (-2599C>G, -2345--2344insG, and 1583T>A).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/5/10.1182_blood-2006-01-008094/2/m_zh80170600260006.jpeg?Expires=1769197304&Signature=bt1G4Z8FRe2qS5AvDWRnNjaTcgO7ushLwhTeSqyXgkdB10J9IXY261M7Ua4E3mj3~qUIPUUvDIXVHlVpGI3vnXoLHucQu9FgehONuIqKVHNME8tDTZQ9Wi8Cd7ZNGAJPR4XQI0uQIruAHjQLFghpYJu3opOkdIsio~fro9akgUPEGfRhc8K3uLwygbTZwrroD5tIz87Fq0~VbwommPwPUIzGQNqQAYD9-LILAg9bn8TnjW48jZeCrUZxciXB6U60-crovalqw8zYFzNLTJJVzEoGuVgJbjoUE9GQN79uTuoYTjLTUNvfyJk0XrnFuMsIcYmht7C-l8WLP2aexma8gg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
