Abstract
The precise mechanism of rituximab plus cyclophosphamide/doxorubicin/vincristine/prednisone (R-CHOP) therapy in diffuse large B-cell lymphoma (DLBCL) is not fully elucidated. Besides overcoming bcl-2-mediated chemoresistance, antibody-dependent cellular cytotoxicity (ADCC), which is activated by effector cells via immunoglobulin G (IgG) fragment C receptors (FcRs), was also proposed as a mechanism of rituximab. The current study evaluated the impact of FcR polymorphism on the response to R-CHOP therapy for DLBCL with the basis that FcR polymorphism can affect rituximab's affinity for ADCC effector cells. The FCGR3A and FCGR2A gene polymorphisms were determined in DLBCL patients receiving R-CHOP (n = 113) compared with CHOP therapy (n = 85). The FCGR3A valine (V) allele was significantly correlated with a higher complete response rate to R-CHOP compared with the phenylalanine (F) allele (88% in V/V vs 79% in V/F vs 50% in F/F; P = .002), while no difference was found between FCGR2A polymorphisms. In addition, V/V allele was associated with faster achievement of response than other alleles. The impact of the FCGR3A gene polymorphism on response rate was not noted in the CHOP group. In terms of overall or event-free survival, no difference was found according to FCGR3A or FCGR2A alleles. The FCGR3A single nucleotide polymorphism (SNP) is predictive of response to R-CHOP, but does not correlate with survival in patients with DLBCL.
Introduction
The introduction of rituximab plus cyclophosphamide/doxorubicin/vincristine/prednisone (R-CHOP) chemotherapy has significantly improved the treatment outcome of diffuse large B-cell lymphoma (DLBCL).1 However, not all patients with DLBCL respond to R-CHOP chemotherapy. Although rituximab has been shown to enhance antibody-dependent cellular cytotoxicity (ADCC) in follicular lymphoma (FL),2 the mechanism of action of R-CHOP in DLBCL has not been clearly defined. Mounier et al3 have suggested that rituximab possesses synergistic activity with CHOP that can overcome bcl-2-mediated chemoresistance. However, rituximab also appears to have an anti-DLBCL effect by itself. A phase 1 study of rituximab therapy in combination with interleukin-12 (IL-12) in B-cell non-Hodgkin lymphoma (NHL) revealed that the overall response rate was 64% (7 of 11 patients) with 5 complete remissions (CRs) in CD20-positive DLBCL patients who were previously treated with anthracycline-containing chemotherapy.4 These observations would suggest that a different mechanism of action rather than overcoming bcl-2-mediated chemoresistance may be involved in the effect of single-agent rituximab, and that it may be independent of CHOP.
While the precise mechanism of action of rituximab has not been fully elucidated, several theories have been proposed, including ADCC, complement-dependent cytotoxicity (CDC), and induction of apoptosis.5 Rituximab is a chimeric monoclonal antibody (mAb) against the CD20 antigen. It has a fragment ab (Fab) domain that binds to the CD20 antigen on B lymphocytes and a fragment c (Fc) domain that recruits immune effector cells to mediate B-cell lysis through ADCC.6
Several types of FcR have been discovered, including FcγRI, FcγRIIa/IIb/IIc, and FcγIIIa/IIIb.5 FcγRIIIa and FcγRIIa are activating receptors due to the presence of an immunoreceptor tyrosine-based activation motif either in the accessory signaling γ chain (FcγRIIIa) or cytoplasmic domain (FcγRIIa). FcγRIIIa is expressed on macrophages, natural killer (NK) cells, and some dendritic cells, while FcγRIIa is expressed on macrophages, neutrophils, dendritic cells, and some mast cells. Previous studies that have examined single nucleotide polymorphisms (SNPs) of Fc receptor (FcR) genes demonstrated that FCGR3A gene SNPs are associated with response to rituximab in patients with FL.7 This is in contrast to patients with chronic lymphocytic leukemia,8 and these differences in response to rituximab are correlated with binding affinity of FcR with ADCC effector cells, suggesting a more important role of ADCC in lymphoma.9-11 It is known that the FCGR3A of valine (V) allele has a higher affinity to human immunoglobulin G (IgG) than the phenylalanine (F) allele, and that cells bearing the FCGR3A V allele mediate ADCC more effectively.12,13
In this study, we evaluated the impact of FCGR3A or FCGR2A gene SNPs on the outcomes of primary R-CHOP therapy in 113 patients with DLBCL, including treatment response and overall and event-free survivals, compared with its contribution on clinical outcomes of CHOP therapy in 85 patients with DLBCL.
Patients, materials, and methods
Patient characteristics and treatment protocol
A total of 113 patients who received R-CHOP chemotherapy between April 2002 and March 2005 as a frontline regimen for CD20-positive DLBCL were included in this retrospective study from 6 hospitals in Republic of Korea (Kyungpook National University Hospital, Chonnam National University Hwasun Hospital, Korean Cancer Center Hospital, Dongsan Medical Center, Pusan National University Hospital, and Yeungnam Medical Center). A control group was created from 85 patients who received CHOP therapy as primary chemotherapy for CD20-positive DLBCL between October 1991 and September 2003 who had available paraffin-embedded tissue samples and who had provided informed consent for future research. This study was approved by the institutional research board at the Kyungpook National University Hospital (Daegu, Korea).
The baseline characteristics of the patients are summarized in Table 1. Overall, among 113 patients (median age, 60 years; 64 men, 49 women), 45 (40%) patients had stages 3 and 4 disease and 37 (33%) patients had intermediate-to-high or high International Prognostic Index (IPI) scores. R-CHOP followed by involved-field radiation was given to 37 (33%) patients, while R-CHOP alone was given to 76 (67%) patients. A median of 6 cycles of R-CHOP therapy was given (range, 1-8 cycles). The comparative information between the R-CHOP group and the CHOP group is shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
R-CHOP chemotherapy was administered as follows: one course of chemotherapy consisted of an intravenous infusion of cyclophosphamide 750 mg/m2, adriamycin 50 mg/m2, vincristine 2 mg, and oral administration of 100 mg prednisone on days 1 to 5 (CHOP), which was repeated every 3 weeks. Rituximab 375 mg/m2 was infused over 4 to 6 hours at day 1 before CHOP chemotherapy began. Patients with stage 1 or 2 disease typically received 4 courses of chemotherapy followed by involved-field radiotherapy (30-40 Gy), while patients with advanced stage disease received 6 to 8 courses of chemotherapy followed by radiotherapy to bulky sites. Maintenance treatment with rituximab after completion of R-CHOP therapy was not administered. Of the 113 patients, 5 received high-dose chemotherapy with autologous stem cell rescue due to relapse after response (n = 3) or initially high IPI score (n = 2). The response to R-CHOP therapy was evaluated after completion of 2 to 3 courses of R-CHOP chemotherapy and 1 month after completion of all planned cycles of R-CHOP, then every 4 months.
FCGR3A/FCGR2A gene polymorphism study
Two single-nucleotide polymorphisms of FcR genes were evaluated in the current study involving the FCGR3A and FCGR2A genes as previously described7,14 : peripheral blood or bone marrow samples derived from patients were taken before treatment with informed consent. Genomic DNA was extracted from the blood samples using the Wizard genomic DNA purification kit (Promega, Madison, WI). For the control group, paraffin-embedded tissue samples were used for DNA purification and only FCGR3A genotyping had been performed.
Two FcR gene polymorphisms were detected based on polymerase chain reactions (PCRs) using primers amplifying a short fragment of DNA containing the polymorphic sites. The first PCR primers for the FCGR3A gene SNP at locus 158 were 5′-ATA TTT ACA GAA TGG CAC AGG-3′ and 5′-GAC TTG GTA CCC AGG TTG AA-3′, while the second PCR primers for the FCGR3A gene at locus 158 were 5′-ATC AGA TTC GAT CCT ACT TCT GCA GGG GGC AT-3′ and 5′-ACG TGC TGA GCT TGA GTG ATG GTG ATG TTC AC-3′, generating a 94-bp fragment. The PCR primers for the FCGR2A gene at locus 131 were 5′-GGA AAA TCC CAG AAA TTC TCG C-3′ and 5′-CAA CAG CCT GAC TAC CTA TTA CGC GGG-3′. The amplifications were performed on a GeneAmp PCR System 9600 (Applied Biosystems, Foster City, CA) in a 20 μL reaction volume containing 100 ng genomic DNA, 10 pmol of each primer, and Q water (Merck, Darmstadt, Germany) treated with 0.1% DEPC (Amresco, Solon, OH) using AccuPower PCR Premix (Bioneer, Daejeon, Korea). The PCR program for first-step amplification for the FCGR3A gene at locus 158 consisted of 35 cycles at 95°C for 60 seconds, 57°C for 90 seconds, 72°C for 90 seconds, and a final elongation step at 72°C for 8 minutes; second-step amplification for the FCGR3A gene at locus 158 consisted of 35 cycles at 95°C for 60 seconds, 64°C for 60 seconds, 72°C for 60 seconds, and a final elongation step at 72°C for 9.5 minutes. The PCR program for the FCGR2A gene at locus 131 consisted of 35 cycles at 94°C for 15 seconds, 55°C for 30 seconds, 72°C for 40 seconds, and a final elongation step at 72°C for 7 minutes. The PCR products were checked on a 2% agarose gel and then subjected to a restriction fragment-length polymorphism (RFLP) analysis.
To distinguish the SNPs, the restriction enzyme NlaIII (New England BioLabs, Beverly, MA) was used for the FCGR3A gene at locus 158, and BstUI (New England BioLabs) for the FCGR2A gene at locus 131. Each 5 μL of the PCR products was digested for 2 hours with 5 U NlaIII (FCGR3A gene at locus 158, at 60°C), and BstUI (FCGR2A gene at locus 131, at 37°C). The digested products were separated on a 2% low-melting-temperature agarose gel (Cambrex Bio Science Rockland, Rockland, ME) and stained with ethidium bromide. For the FCGR3A gene at locus 158, the V allele was identified as 2 digested bands at 61 bp and 33 bp, while the F allele was identified as one undigested band at 94 bp. For the FCGR2A gene at locus 131, the histidine (H) allele was identified as a 337-bp DNA band, while the arginine (R) allele was identified as a 316-bp and 21-bp band. Then, selected PCR-amplified DNA samples were examined by DNA sequencing to confirm the genotyping results.
Definitions
Clinical responses to R-CHOP chemotherapy were determined by physical examination and computed tomography before starting subsequent radiation therapy once patients were planned to receive involved-field radiation. The responses were scored according to International Working Group criteria.15 Overall survival (OS) was measured from day 1 of the first cycle of R-CHOP until the date of death or last follow-up. Event-free survival (EFS) was calculated from day 1 of the first cycle of R-CHOP until treatment failure (disease progression, recurrence, or death of any cause). To exclude potential effects of high-dose chemotherapy with autologous stem cell rescue on survival, patients were considered as censoring cases at the time of transplantation. The time to response to R-CHOP therapy was calculated as an interval between day 1 of the first cycle of R-CHOP and the day response was confirmed, including complete and partial responses.
Statistical analysis
Clinical data were analyzed according to information available as of November 2005. The primary objective of the current study was to correlate FCGR3A/FCGR2A SNPs with the response to R-CHOP therapy. The secondary objectives were to correlate FCGR3A/FCGR2A SNPs with survival outcomes including OS and EFS, and with the time to response to R-CHOP therapy. In addition, the correlation between FCGR3A SNP and FCGR2A SNP was also analyzed in the current study.
The clinical characteristics and treatment outcomes of the patients were compared using Chi-square, Fisher exact, or Mann-Whitney U tests according to the FCGR3A/FCGR2A SNPs. The genetic association between FCGR3A and FCGR2A gene SNPs was determined using the Chi-square test. The clinical characteristics and the response to primary chemotherapy were compared according to the FCGR3A/FCGR2A SNPs in the R-CHOP group and CHOP group, respectively. Logistic regression analysis was conducted to determine the predictive value of the SNPs on response to R-CHOP for 105 patients who had both FCGR3A and FCGR2A SNP data available. Variables included for the logistic regression were stage (stages 1 and 2 vs 3 and 4), IPI score (0-2 vs 3-5), age (< 60 years vs ≥ 60 years), performance status (Eastern Cooperative Oncology Group [ECOG] 0,1 vs ≥ 2), lactate dehydrogenase level (normal vs beyond elevated), extranodal involvement (≤ 1 site vs ≥ 2 sites), and FCGR3A (V/V allele vs non-V/V allele) and FCGR2A (H/H allele vs non-H/H allele) alleles. Odds ratio (OR) and 95% confidence intervals (CIs) for relative risks were also calculated by logistic regression analysis.
Survival estimates were calculated using the Kaplan-Meier method. Differences in OS, EFS, and the time to response to R-CHOP therapy according to the FCGR3A/FCGR2A SNPs were compared using log-rank tests or a Wilcoxon test (for the time to response to R-CHOP therapy). For the multivariate survival analyses using the Cox proportional hazard model to define the prognostic factors for OS, EFS, and the time to response to R-CHOP therapy, a backward conditional procedure was conducted until the P value for the likelihood ratio test was above .05 with the following variables: stage (stages 1, 2 vs 3, 4), IPI score (0-2 vs 3-5), age (< 60 years vs ≥ 60 years), performance status (ECOG 0 or 1 vs ≥ 2), lactate dehydrogenase level (normal vs beyond normal range), extranodal involvement (≤ 1 site vs ≥ 2 sites), and FCGR3A (V/V allele vs non-V/V allele) and FCGR2A (H/H allele vs non-H/H allele) allele. The hazard ratio (HR) and 95% CI were also estimated. A cut-off P value of .05 was adopted for all the statistical analyses. The statistical data were obtained using an SPSS software package, version 11.5 (SPSS, Chicago, IL). Multivariate analyses were performed on 105 of 113 patients; FcγRIIa allele data was unavailable for 8 patients.
Results
Frequency of FCGR3A and FCGR2A alleles
In the R-CHOP group, the distribution of the VV, VF, and FF alleles of FCGR3A was 47%, 48%, and 5%, respectively, while the distribution of HH, HR, and RR alleles in FCGR3A was 56%, 37%, and 7%, respectively. No significant correlation between FCGR3A and FCGR2A alleles was demonstrated (P = .142 by χ-square test). The distribution of VV, VF, and FF alleles of FCGR3A in the CHOP group was 57%, 31%, and 12%, respectively (Table S1).
Patient characteristics according to FCGR3A and FCGR2A allele status
The patients' characteristics are summarized in Table 1. In brief, no differences in patient and disease characteristics were observed more frequently according to FCGR3A and FCGR2A alleles, except for extranodal DLBCL involvement, which was more common with the FCGR2A H/H allele. In addition, except for sex discrepancy, no differences in patient and disease characteristics were observed more frequently according to FCGR3A and FCGR2A alleles in the CHOP group (Table S1).
Response to frontline R-CHOP therapy according to FCGR3A and FCGR2A alleles
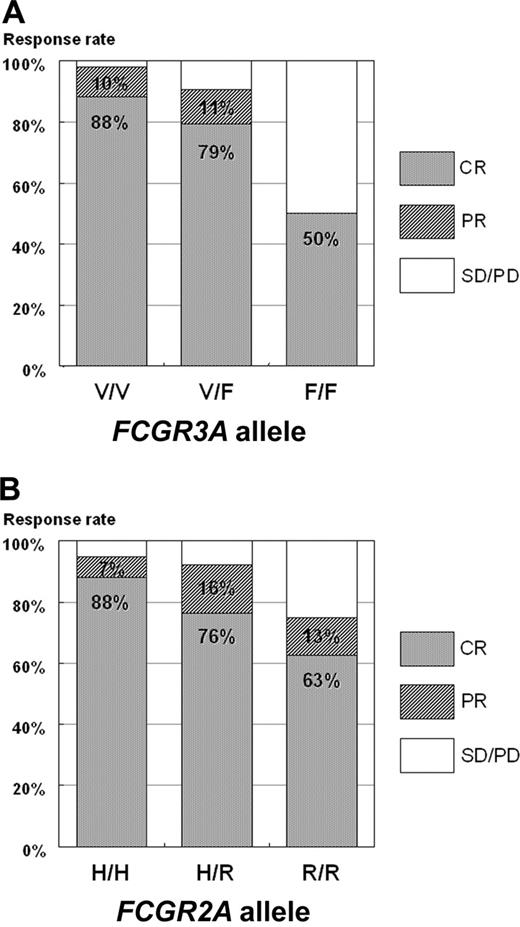
Of 110 patients evaluable for response to chemotherapy, the overall response rate (ORR) was 92% (101 of 110 patients) with a CR of 82% (90 of 110 patients), and a partial response (PR) rate of 10% (11 of 110 patients). As shown in Table 1 and Figure 1, a higher CR rate was observed in the FCGR3A V/V allele (88%) compared with the FCGR3A V/F (79%) or F/F allele (50%; P = .002). The difference in ORR was also significant in favor of the FCGR3A V/V allele (ORR of 98% vs FCGR3A V/F [90%] or F/F allele [50%]; P < .001). A statistically significant difference in response rate was not observed according to FCGR2A allele even though the trend of better response rate was observed in FCGR2A H/H allele (95% ORR vs FCGR2A H/R [92%] or R/R allele [75%]; P = .137).
Response to frontline R-CHOP therapy according to FCGR3A and FCGR2A alleles. (A) P = .002. P = .001 when CP and PR are compared with DC and PD. (B) P = .178.
Response to frontline R-CHOP therapy according to FCGR3A and FCGR2A alleles. (A) P = .002. P = .001 when CP and PR are compared with DC and PD. (B) P = .178.
When comparing the response to frontline chemotherapy between the R-CHOP and CHOP groups, it was significantly different in favor of the R-CHOP group with respect to an achievement of OR (91% vs 70%; P < .001) or CR (81% vs 53%, P < .001; Figure S1A). Meanwhile, the impact of the FCGR3A V/V allele on OS or CR rates was not noted in the CHOP group as shown in Table S1 and Figure S1B.
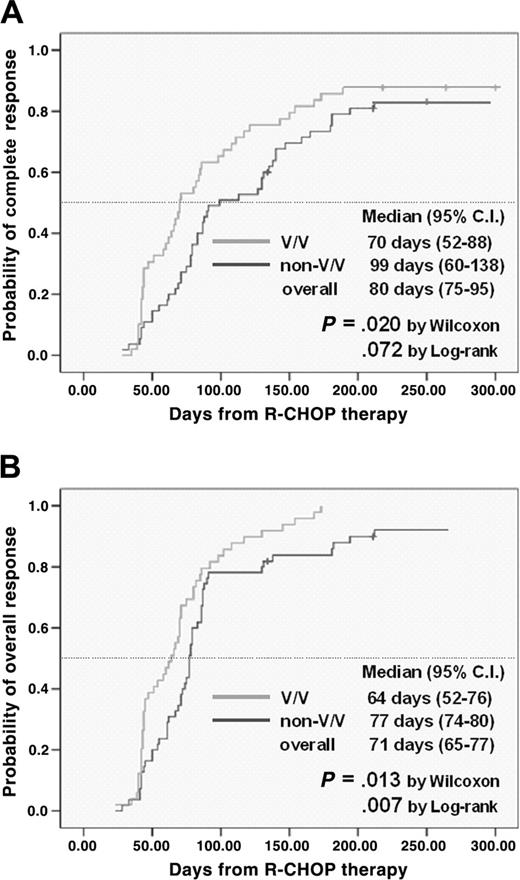
When analyzing the time to CR with R-CHOP (Figure 2A), we found that the group with the FCGR3A V/V allele had a more rapid time to CR (median, 70 days; 95% CI, 52-88 days) compared with non-V/V allele (median, 99 days, 95% CI 60-138 days; P = .020 by Wilcoxon test). A similar trend was also observed in terms of the time to achievement to OR (Figure 2B) with a more rapid response in the group with the FCGR3A V/V allele (median, 64 days; 95% CI 52-76 days) versus non-V/V allele (median, 77 days, 95% CI 74-80 days; P = .013 by Wilcoxon test). However, a significant influence of the FCGR2A allele was not observed on the time to achievement of CR (P = .515) or OR (P = .789).
Survival analysis according to FCGR3A and FCGR2A allele
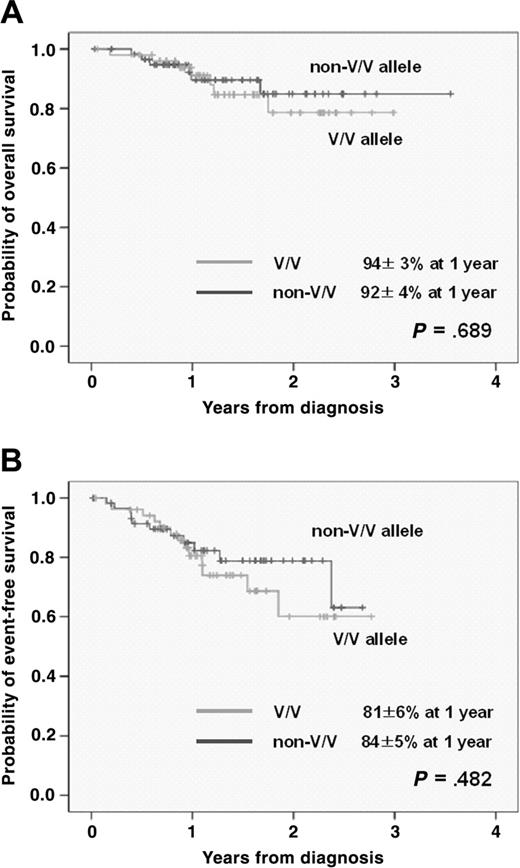
With a median follow-up duration of 420 days (range, 12-1297 days), 22 (19%) patients had relapsed or progressed, with 13 (12%) deaths. The 1- and 2-year OS rate was 93% ± 3% and 82% ± 5%, while 1- and 2-year EFS rates were 83% ± 4% and 70% ± 6%, respectively. When comparing OS and EFS according to FCGR3A and FCGR2A alleles, neither the FCGR3A nor the FCGR2A allele had any impact on OS or EFS (Figure 3).
Multivariate analysis
The results of the multivariate analysis are summarized in Tables 2, 3, 4. When analyzing predictive factors for response to R-CHOP therapy, the FCGR3A V/V allele was found to be an independent factor predicting OR; patients with the FCGR3A V/V allele were found to have a more than 7-fold higher probability of OR (P = .048, HR 7.686, 95% CI 1.028-61.748). In addition, the FCGR3A V/V allele was also identified as a predictive factor for rapid achievement of CR (P = .022, HR 1.654, 95% CI 1.076-2.542), together with early-stage disease (P = .036, HR 1.640, 95% CI 1.033-2.603) and low IPI score (P = .012, HR 2.764, 95% CI 1.255-6.088). A similar outcome was also identified on the analysis for the time to OR: the FCGR3A V/V allele (P = .005, HR 1.802, 95% CI 1.193-2.723) and early-stage disease (P = .050, HR 1.522, 95% CI 1.004-2.314).
However, in a Cox multivariate analysis of survival, neither the FCGR3A nor FCGR2A alleles were independently associated with survival after R-CHOP therapy for DLBCL. On the other hand, low IPI score was found to be a favorable prognostic factor for OS (P = .007, HR 0.199, 95% CI 0.061-0.646) and EFS (P < .001, HR 0.107, 95% CI 0.042-0.271) after R-CHOP therapy for patients with DLBCL.
Discussion
This study demonstrates that the FCGR3A SNP is predictive of response to R-CHOP chemotherapy in patients with DLBCL, but is not predictive of OS or EFS. Based on the current observation, the additive effect of rituximab through ADCC, which is mediated via FcR, may be one of the important action mechanisms of R-CHOP in patients with DLBCL in addition to the synergistic activity of rituximab with CHOP by overcoming bcl-2-mediated chemoresistance.
The cumulative incidence of achievement of CR and OR to frontline R-CHOP therapy according to FCGR3A allele.
The cumulative incidence of achievement of CR and OR to frontline R-CHOP therapy according to FCGR3A allele.
Although the precise mechanism of action of rituximab is not fully elucidated, it likely includes ADCC, complement-dependent cytotoxicity, and induction of apoptosis.5 The benefit of R-CHOP therapy has been demonstrated in a randomized trial of R-CHOP versus CHOP in elderly patients with DLBCL1 ; in that trial, the benefit of rituximab was primarily limited to patients with overexpression of bcl-2, suggesting that rituximab may act by overcoming bcl-2-mediated chemoresistance in part.3 This observation was supported by Chow et al,16 who demonstrated that combining rituximab with different cytotoxic drugs such as doxorubicin or mitoxantrone significantly decreased the dose of chemotherapeutic drugs necessary for the induction of apoptosis.
However, several investigators have suggested that the mechanism of action of rituximab is not solely confined to overcoming bcl-2-mediated chemoresistance. Ansell et al4 reported that a high response rate was observed in lymphoma trials when rituximab was combined with nonchemotherapeutic agents such as IL-12, which can enhance cellular cytotoxicity, and that the ORR was 64% (7 of 11 patients), with 5 CRs in CD20-positive DLBCL patients who were previously treated with anthracycline-containing chemotherapy. This study suggested that tissue macrophages may be critical for the treatment of DLBCL with rituximab treatment as well as for FL.4 In addition, Manches et al17 reported that rituximab can induce lymphoma cell phagocytosis by tissue-scavenging cells such as macrophages or NK cells, important mediators of ADCC.
The binding of rituximab to CD20-positive DLBCL cells allows the recruitment of effector cells of ADCC such as NK cells and macrophages, which express Fc receptor.6 FcγRIIIa is predominantly expressed on NK cells and macrophages, whereas FcγRIIa is expressed on neutrophils and macrophages. Previous investigations suggested that the FCGR3A/FCGR2A gene SNPs can influence the response to rituximab therapy in FL2,7 and in Waldenström macroglobulinemia.8 The FCGR3A/FCGR2A gene SNPs may influence treatment outcome in several ways. First, the FCGR3A genotype may affect rituximab pharmacokinetics, thus influencing the response rate of R-CHOP.11 In addition, NK cell-mediated ADCC may contribute to the action of rituximab on DLBCL cells. After binding to NK cells through FcγRIIIa, granzyme release may activate caspase, thus overcoming chemoresistance of lymphoma cells.16
The major polymorphism of FCGR3A is a point mutation that results in the expression in the extracellular domain at amino acid position 158 of either V (V158) or F (F158). The FCGR3A-V158 genotype has a higher affinity for IgG than does the F158 genotype.18 Recent investigations also suggested that the individuals expressing FCGR3A-158 VV and VF showed increased NK cell-surface expression of FcγRIIIa and binding of rituximab, and demonstrated higher levels of ADCC activity in response to rituximab in healthy individuals.19
Our results demonstrate that FCGR3A SNPs correlate with the response to R-CHOP in patients with DLBL (especially the V allele for better and faster response; Figures 1, 2). The same phenomenon was not observed in the CHOP group without any correlation of the FCGR3A SNPs to response to CHOP chemotherapy. These observations imply that ADCC may contribute in part to the mechanism of R-CHOP. However, no association of FCGR3A SNPs with survival was noted in the present study, for the following reasons: (1) the relatively small number of patients; (2) the relatively short period of follow-up may not be enough to see significant difference of survival; and (3) ADCC itself may not be a predominant mechanism of R-CHOP in DLBCL patients in terms of survival. Other mechanisms such as bcl-2-mediated chemoresistance are likely important in the mechanism of action of R-CHOP, and this would be expected to affect survival of patients with DLBCL. Accordingly, further study will be necessary to reach a clear conclusion on this issue, especially on OS or EFS.
Survival curve after frontline R-CHOP therapy according to FCGR3A allele.
The major FCGR2A polymorphism is a point mutation affecting amino acid position 131, coding for either arginine (R131) or histidine (H131). The FCGR2A-H131 genotype has higher affinity for human IgG3. However, we could not determine the role of the FCGR2A allele in predicting the response and prognosis in patients with DLBCL who received R-CHOP therapy.
In the current study, the frequency of the V/V allele was relatively higher than that of previous reports in the white population.2,7 However, the frequency distribution of the Chinese population was very similar to that of our study (48.9% of patients had the V/V genotype, compared with 47% in the current study).20 The different distribution of allelic frequencies may result from different ethnic backgrounds.
In conclusion, the present study suggests that the FCGR3A SNP is a predictive of response to R-CHOP, but may not be a prognostic factor for patients with DLBCL. Besides the synergistic role of rituximab when combined with CHOP in overcoming bcl-2-mediated chemoresistance, the additive effect of rituximab through ADCC that was mediated by FcR may be one of the important action mechanisms of R-CHOP in DLBCL.
Prepublished online as Blood First Edition Paper, April 11, 2006; DOI 10.1182/blood-2006-01-009480.
Supported by BioMedical Research Institute grant, Kyungpook National University Hospital (2005).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully give thanks to Dr John Kuruvilla and Dr Grace Wu for their critical review of the manuscript.