Abstract
C/EBP epsilon is a transcription factor involved in myeloid cell differentiation. Along with C/EBP-α, -β, -γ, -δ, and -ζ, C/EBP-ϵ belongs to the family of CCAAT/enhancer binding proteins that are implicated in control of growth and differentiation of several cell lineages in inflammation and stress response. We have previously shown that C/EBP-ϵ preferentially binds DNA as a heterodimer with other C/EBP family members such as C/EBP-δ, CHOP (C/EBP-ζ), and the b-zip family protein ATF4. In this study, we define the consensus binding sites for C/EBP-ϵ dimers and C/EBP-ϵ–ATF4 heterodimers. We show that the activated NFkappaB pathway promotes interaction of the C/EBP-ϵ subunit with its cognate DNA binding site via interaction with RelA. RelA-C/EBP interaction is enhanced by phosphorylation of threonine at amino acid 75 and results in increased DNA binding compared with the wild-type nonphosphorylated C/EBP both in vitro and in vivo. We suggest that interaction of the activated NFkappaB pathway and C/EBP-ϵ may be important in selective activation of a subset of C/EBP-ϵ–responsive genes.
Introduction
Specific regulation of gene expression is achieved through interaction of multiple transcriptional activators and repressors with their cognate DNA sites. This activity is subject to several levels of control, including posttranslational modification by phosphorylation, acetylation, selective degradation, and interaction with coactivators and corepressors
Previous studies of CCAAT/enhancer binding protein family of transcriptional activators had identified domains responsible for dimerization, specific DNA recognition, transcriptional activation, and intramolecular repression.1–6 Several members of the C/EBP protein family (C/EBP-α, C/EBP-β, C/EBP-δ, C/EBP-ϵ) contain a transactivation domain and a basic “leucine zipper” domain that serve for DNA recognition and dimer formation. Others like C/EBP-γ and C/EBP-ζ (CHOP, GADD153) are involved in transcriptional regulation through dimerization with other basic leucine zipper proteins from the C/EBP, ATF/CREB, and Fos/Jun families.
Recent studies directed at identification of C/EBP-responsive genes resulted in identification of a diverse and degenerate collection of putative C/EBP binding sites. At the same time, in vitro experiments with site selection typically produce very common short consensus sequences. Identification of binding sites for each individual member of the C/EBP family is complicated by common coexpression of several of these proteins in the same cell and their ability to form heterodimeric complexes with each other and with structurally related ATF/CREB proteins.7–12
Activity of C/EBP proteins is regulated by phosphorylation mediated through several major kinase pathways that affect DNA binding, subcellular localization in the nucleus, and interaction with cell-cycle proteins. Our recent studies of C/EBP-ϵ phosphorylation13,14 identified several sites, including 1 for p38 MAPK within the transactivation domain of C/EBP-ϵ. As a result of phosphorylation at threonine 75 (T75), C/EBP-ϵ DNA binding and specific transcriptional activity was significantly enhanced.
In this study, we identified consensus binding sequences for C/EBP-ϵ and for heterodimers of C/EBP-ϵ with its most common dimerization partner ATF4.15,16 Furthermore, we find that C/EBP-ϵ–DNA interaction is highly up-regulated by interaction with the activated NFkappaB pathway protein p65RelA. This interaction depends on the T75 phosphorylation status of C/EBP-ϵ and can be enhanced by introduction of phosphomimetic Thr to Asp mutation. By using selective removal of p65RelA- and siRNA-mediated p65 knock-down, we demonstrate that the activated NFkappaB pathway is required for high-level expression of the C/EBP-ϵ–responsive promoter. This novel interaction may play an important role in regulation of C/EBP-ϵ–dependent genes in myeloid cells.
Materials and methods
CMV–C/EBP-ϵ, ATF4, and siRNA expression constructs
C/EBP-ϵ, C/EBP-ϵT75A, and C/EBP-ϵT75D expression and pMim-luciferase reporter plasmids were described previously.6,13,14 ATF4 (CREB2) expression plasmid was a generous gift of Dr Jeff Leiden. RelA knock-down and control cell lines were generated by lentiviral transduction of MiaPaca2 cells with siRNA targeting human RelA under the control of H1 RNA promoter-based cassette placed into the long-terminal repeat (LTR) of self-inactivating lentivirus, as described previously.17 Briefly, for lentiviral siRNA knock-out of p65RelA we used pLSLPw vector. Phosphorylated oligonucleotides targeting human RelA mRNA (hRelA-1 si-direct, 5′ to 3′ GATCCGGAGCACAGATACCACCAAGCTTCCTGTCACTTGGTGGTATCTGTGCTCTTTTTG; hRelA-1 si-reverse, 5′ to 3′ AATTCAAAAAGAGCACAGATACCACCAAGTGACAGGAAGCTTGGTGGTATCTGTGCTCCG) or mouse RelA mRNA (mRelA-1 si-direct, 5′ to 3′ GATCCGGAAGCACAGATACCACCAAGCTTCCTGTCACTTGGTGGTATCTGTGCTTCTTTTTG; mRelA-1 si-reverse, 5′ to3′ AATTCAAAAAGAAGCACAGATACCACCAAGTGACAGGAAGCTTGGTGGTATCTGTGCTTCCG) were made double-stranded by annealing and were then cloned into BamHI-EcoRI sites of pLSLPw, placing anti-RelA siRNA under control of H1RNA promoter within the 3′LTR region of prolentiviral construct, producing pLSLPsiRelA. This vector also contains the puromycin resistance marker. For production of VSV-G–pseudotyped lentiviral particles, 293T cells were transiently cotransfected with plasmid DNA of lentiviral vector, pVSV-G expression plasmid for pseudotyping, and pCMV-Delta R8-1-2, providing trans-factors necessary for lentiviral replication. Virus-containing supernatants were collected between 48 and 72 hours after transfection, and effective viral titer was determined by infecting HEK293 monolayer cultures with diluted viral stock, selection with 1 μg/mL of puromycin for 7 days, and counting of puromycin-resistant foci derived from lentiviral transduction. For production of stable MiaPaca2 clones carrying anti-p65RelA siRNA, cells were plated at approximately 50% confluence and transduced in the presence of 4 μg/mL polybrene (Sigma, St Louis, MO) with lentiviral stocks at low multiplicity of infection (MOI) of 1 to 2 viral infectious units (as determined by titering). Starting at 48 hours after transduction, cells were subjected to selection with 1 μg/mL of puromycin and individual puromycin-resistant clones were identified. The extent of p65RelA protein knock-down was analyzed by Western blotting with specific anti-p65 antibodies, and significant clonal variation in knock-down efficiency was found. For further analysis for the purposes of this study, we have used puromycin-resistant clone with no detectable p65RelA. Puromycin-resistant clone that showed no decrease of p65RelA protein was used as control.
For transient p65RelA knock-down experiments, high MOI of 10 to 20 was used, insuring near-100% efficiency of transduction. A parallel mock experiment using identical conditions but with a GFP-tagged lentiviral construct was conducted to monitor different steps of viral production, transduction, and selection.
Transient transfections and reporter assays
Transient transfection assays were performed in COS1, Jurkat, and MiaPaca cell lines using lipofectamin-2000 (Invitrogen, Carlsbad, CA). For luciferase or β-galactosidase reporter assays, cells were transfected with 3 μg of either pMim-luciferase or C/EBP-LacZ reporter and 0.3 μg of the C/EBP expression vectors, wild type or mutants, as previously described.14,15 The internal control for transfection efficiency was 1 μg pCMV–β-galactosidase or 0.1 μg pCMVrenilla-luciferase. At 12 hours after transfection, the Jurkat cells were activated with TPA (12-O-tetradecanoyl phorbol 13-acetate; 2 ng/mL) and the calcium ionophore A23187 (250 ng/mL) for 24 hours. After this time, the transfections were harvested and the luciferase and β-galactosidase assays were performed according to the manufacturer's instructions (Promega Biotech, Madison, WI). Chloramphenicol acetyl transferase (CAT) assays were performed using a composite ATF4/CEBP-ϵ binding site dimer cloned into the pCATB reporter plasmid as described previously.13,18
Mouse 32Dcl3 cells were grown in RPMI1640 medium supplemented with 10% fetal calf serum and 10% of WEHI3B cell–conditioned medium as a source of IL-3. To induce neutrophil differentiation, cells were plated in medium without IL-3, supplemented with 100 ng/mL G-CSF. Transduction with C/EBP-ϵ–expressing pBABEpuro vectors was done as previously described.14 Following transduction, 32Dcl3-derived cells were selected with 2 μg/mL of puromycin for 5 days, after which they were transduced with lentiviral vectors carrying RelA-specific siRNA or control vectors at MOI of 5 to 10. IL-3 containing WEHI3B-conditioned media was removed 12 hours after transduction and total RNA was harvested 3 days after transduction and isolated using TRIzol method; chromatin immunoprecipitation (ChIP) assay samples were also harvested at day 3 after lentiviral transduction. Morphologic maturation of 32Dcl3-derived cells was determined by examination of Wright-Giemsa–stained cytospins. Viable cells were counted on the basis of trypan blue exclusion and the number of terminally differentiated cells was determined and expressed as a percentage of total live cells (% neutrophils).
MBP–C/EBP-ϵ and GST–C/EBP-ϵ fusion proteins
Full-length C/EBP-ϵ, C/EBP-ϵT75A, and C/EBP-ϵT75D were subcloned into pGEX-5X-1, and the resulting constructs were transformed into BL-21 cells. The glutathione-S–transferase (GST)–C/EBP fusion proteins were isolated according to the manufacturer's instructions (Amersham-Pharmacia, Piscataway, NJ) with modifications. Following overnight growth, the culture was divided 1:50 and then grown for a further 2 hours. The GST-C/EBP fusion protein was induced by 0.5 mM IPTG (isopropyl-β-D galactopyranoside) for 3 hours. The GST fusion proteins were isolated by sonication in hypertonic conditions and stored bound to the glutathione-sepharose beads in 50% glycerol. Expression of the GST fusion proteins was monitored by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and either Coomassie staining or Western immunoblotting with a specific C/EBP-ϵ antibody (Santa Cruz Biotechnology, Santa Cruz, CA). For MBP–C/EBP-ϵ protein purification, C/EBP-ϵ cDNA was inserted into pMALC1 expression vector, and bacterial culture growth, induction, and purification on amylose resin (New England Biolabs, Beverly, MA) were done essentially as described by the manufacturer.
Nuclear lysate preparation for EMSA
COS1 cells were transfected with 10 μg of the C/EBP-ϵ expression plasmids, wild type and mutated, per 10-cm dish, using Lipofectamine 2000 (Invitrogen). The empty vector pcDNA3.1 was used as the negative control for these experiments. Nuclear proteins were prepared 24 to 36 hours after transfection as previously described.19 Briefly, 5 × 106 COS1 cells were washed 3 times with ice-cold phosphate-buffered saline (PBS). After the last wash, adherent cells were scraped off the dish with a rubber policeman and resuspended in 500 μL extraction buffer B (20 mM HEPES, pH 7.9; 20% glycerol; 10 mM NaCl; 0.2 mM EDTA; 1.5 mM MgCl2; 0.1% Triton X; 1 mM dithiothreitol [DTT]; 1 mM phenylmethyl sulfonyl fluoride [PMSF]; 40 μL/mL Complete [Boehringer, Indianapolis, IN]). After 15 minutes of incubation on ice, the nuclei were pelleted at 250g for 10 minutes. Nuclei were resuspended in extraction buffer B, and NaCl was added dropwise with mixing to a final concentration of 300 mM NaCl. Nuclei were rocked for 60 minutes at 4°C. Samples were microcentrifuged at 10 000g and supernatants were frozen at −80°C. A similar procedure was used to prepare nuclear lysates from transfected Jurkat cells. To activate Jurkat cells, TPA (12-O-tetradecanoyl phorbol 13-acetate; 2 ng/mL) and the calcium ionophore A23187 (250 ng/mL) were added for 24 hours. Preclearing of p65RelA from nuclear lysates was achieved by immunoabsorption of p65 with specific purified antibodies and 10 μg per 10μL nuclear lysate, with normal rabbit IgG serving as control (sc-372; Santa Cruz Biotechnology), and protein A/G agarose beads (Sigma). The nuclear extracts were initially separated by SDS-PAGE and analyzed by Western immunoblot to confirm expression of C/EBP, ATF4, or RelA proteins. DNA binding by the C/EBP proteins was determined by electrophoretic mobility shift assay (EMSA) using double-stranded oligonucleotides containing the C/EBP consensus site and adjacent sequences of the mim promoter.19 A standard reaction for the EMSA contained 1 ng 32P-labeled oligonucleotide, 10 μg nuclear extract, 2 μg poly dIdC (poly-deoxyinosinic-deoxycytidylic acid), and 5 μg bovine serum albumin in a 20-μL volume. Competing cold oligonucleotides at 100-fold molar excess or antibodies (1 μg) were added where indicated. Electrophoresis was performed on a 5% polyacrylamide gel at 30 mA for 3 to 5 hours. The gel was dried and the results visualized by autoradiography. Binding sites included C/EBP consensus binding site (5′-TGCAGATTGCGCAATCTGCA-3′), C/EBP-mutated site (5′-TGCAGAGACTAGTCTCTGCA-3′), and ATF/CEBP heterodimer binding site (ATEBP; 5′-TGCAGATGATGCAATCTGCA-3′).
Random oligonucleotide binding-site selection
The following synthetic oligonucleotides were used for identification of high-affinity binding sites in CASTing experiments: N-16, 5′-CGCTCGAGGGATCCGAATTCNNNNNNNNNNNNNNNNTCTAGAAAGCTTGTC; A-16, 5′-CGCTCGAGGGATCCGAATTC-3′; and B-16, 5′-GCGTCGACAAGCTTTCTAGA-3′.
Double-stranded molecules were generated by annealing oligonucleotides containing 16 random nucleotides (N-16) with primers A-16 and B-16 and then extending by Taq polymerase (Promega Biotech). Enrichment for binding sites was performed by a modification of the filter-binding method.20–22 The binding reaction (35 to 100 μL) contained 20 to 50 μL of COS or KCL22 cell nuclear lysate (adjusted to 10 mg/mL) or 2 μg of purified fusion proteins, 0.3 μg of double-stranded N-16 oligonucleotide, and 2.7 μg of poly (dI-dC) in nuclear lysate preparation buffer. Salt concentration was adjusted to 150 mM. Each binding reaction was incubated for 30 minutes at room temperature. For C/EBP and ATF4 derived from either COS1 or KCL22 cells, specific rabbit polyclonal anti–C/EBP-ϵ or anti-ATF4 antibodies (1 μg) (Santa Cruz Biotechnology) were added to the binding reaction along with the appropriate amount of protein A/G sepharose beads. Protein A/G sepharose beads were preadjusted with nuclear lysate preparation buffer. Agarose beads with immobilized C/EBP-ϵ or ATF4 and bound DNA were washed 3 times with 3 mL of 1 × binding buffer. Bound nucleotides were recovered by proteinase K digestion and phenol extraction. Extracted DNA was amplified by polymerase chain reaction (PCR) using primers A-16 and B-16, and 0.3 μg of amplified dsDNA was subjected to the next round of site-selection procedure. After 6 or 7 rounds of site selection, the amplified products were cloned into pTZ19R (Pharmacia, Uppsala, Sweden) for sequencing.
In vitro pull down, coimmunoprecipitation assays, and antibodies used in this study.
Pull-down assays were done using the fusion GST and MBP proteins as previously described.17 Nuclear lysates were mixed with indicated amounts of GST fusion proteins loaded on glutathione-sepharose beads in 0.5 mL of binding buffer (20 mM Tris-Cl, pH 7.5; 150 mM NaCl; 0.5% NP40; 0.1 mM EDTA; 1 mM dithiothreitol [DTT]; and complete protease inhibitor cocktail; Roche Molecular Biochemicals, Indianapolis, IN). Proteins were allowed to interact, with continuous mixing, for 1 hour at 4°C. Binding reactions were washed 3 times with 1.4 mL of binding buffer, denatured in a sample buffer, and analyzed by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis. Gels were subjected to Western blotting, immunodetection, and BioMax MR film (Kodak, Rochester, NY) at −80°C. Antibodies used in this study included C/EBP-ϵ–specific rabbit polyclonal antiserum raised against GST–C/EBP-ϵ protein.13 Other antibodies were purchased from Santa Cruz Biotechnology, including anti–C/EBP-ϵ (C-22), anti-ATF4 (CREB2, C-20), anti-p65RelA (C-20, sc-372), anti-p52 (K-27), anti-p50 (E-10), and anti–C/EBP-γ (H-50).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were done as described,23 with several modifications. Briefly, 2 × 107 cells were cross-linked by addition of formaldehyde into the medium at a final concentration of 1% and incubated for 30 minutes at 37°C. Prior to cross-linking, an aliquot of the cells was removed for analysis of input chromatin DNA. Cells were then washed with ice-cold PBS and resuspended in 200 μL of ChIP lysis buffer with protease inhibitors (Roche, Indianapolis, IN), incubated on ice for 30 minutes, and sonicated to produce DNA fragments 500 to 800 nucleotides (NTs) in size. Sonicated lysates were then diluted to 3 mL with ChIP dilution buffer (0.01% SDS; 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris-HCl, pH 8.0; and 167 mM NaCl), followed by preclearing with 100 μL protein A/G agarose beads for 60 minutes at 4°C with rotation. The precleared lysates were then immunoprecipitated using polyclonal antibodies for either C/EBP-ϵ, p65RelA (sc-372; Santa Cruz Biotechnology; 10 μg), or normal rabbit IgG controls (Santa Cruz Biotechnology) at 4°C overnight with rotation. Immune complexes were collected with 80 μL protein A/G agarose and washed once with 1.5 mL each of the following buffers: low-salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 150 mM NaCl), high-salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 1500 mM NaCl), LiCl wash buffer (250 mM LiCl; 1% nonidet P-40 [NP-40]; 1% sodium deoxycholate; 1 mM EDTA; and 10 mM Tris-HCl, pH 8.0), and 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Immune complexes were next eluted using freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). Cross-links were reversed by heating at 65°C in the presence of NaCl followed by proteinase K treatment. The DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation and resuspended in 20 μL distilled water. ChIP DNA (5 μL) was next used as a template for PCR using the appropriate oligonucleotides: human LF (Dir, 5′-TGGCGGGGAGTGGGAGGGAA; Rev, 5′-AAGCTTGTCGACCGACTTGGCAAACGAAG); HNP (Dir, 5′-GTCAACTGTGTTAGGAGCCAT; Rev, 5′-CGTGCACAAGTGGACTTC); IkBα (Dir, 5′-GACGACCCCAATTCAAATCG; Rev, 5′-TCAGGCTCGGGGAATTTCC).
Results
Selection of C/EBP-ϵ binding sites
Previous studies indicated that C/EBP-ϵ can readily dimerize with coexpressed factors such as C/EBP-δ, CHOP, ATF4, E2F, and Rb.15,19,24 Therefore, we attempted to identify DNA sites that are preferentially bound by C/EBP-ϵ homodimers and C/EBP-ϵ–ATF4 heterodimers.
To select C/EBP-ϵ binding sites, we have used a modification of the cyclic amplification of selected target sites (CASTing) method.22 We initially used bacterially produced maltose-binding protein–C/EBP-ϵ fusion protein as a “bait” for site selection. This approach insures that we only study homodimers formed by C/EBP-ϵ. As previously shown for other C/EBP family members, the C/EBP-binding site consists of palindromic sequences containing half-sites 5′-ATTGC-3′.1,23 Three-hundred nanograms of randomized oligonucleotides contain approximately 4 × 1012 molecules, sufficient to ensure that all possible combinations of the 16-nucleotide randomized region (416 = 4.3 × 109) would be represented in each binding reaction. A total of 6 cycles of random nucleotide selections were performed in each experiment prior to cloning of the selected site library into plasmid vector and sequencing. The PCR-amplified DNA recovered after each cycle of selection was subjected to EMSA analysis. Sequences of cloned oligonucleotides were optimally aligned by inspection, a task that was facilitated by the presence of the characteristic 5′-N T T N-3′ core within most sequences. However, to our surprise, we were unable to determine a highly specific consensus binding site using C/EBP-ϵ fusion protein expressed in bacteria. A total of 131 individual sites were identified in 2 independent sets of CASTing experiments. About 50% of selected sites were nonpalindromic, sometimes containing direct repeats. Palindromic sites had a highly degenerated central region, where spacing between 5′-nTTn-3′ half-sites varied between 0 and 5. A minority of selected sites (about 15%) contained 5′-A/GTTnnnnAAT/C palindromes. These results suggested that homodimeric C/EBP-ϵ either has a low degree of specificity in DNA binding or lacks some necessary modification that occurs in vivo.
Next, we used nuclear lysates derived from the C/EBP-transfected COS1 cell line and myeloblast KCL22 cell line as a source of endogenously produced C/EBP-ϵ. The KCL22 cell line does not express detectable amounts of ATF4, C/EBP-α, C/EBP-β, or C/EBP-δ as determined by Western blotting and EMSA-supershift analysis with specific antibodies.19,20 Two independent CASTing experiments using nuclear lysates derived from either transfected COS1 or KCL22 cells produced a highly homogenous population of palindromic DNA sites shown in Figure 1A.
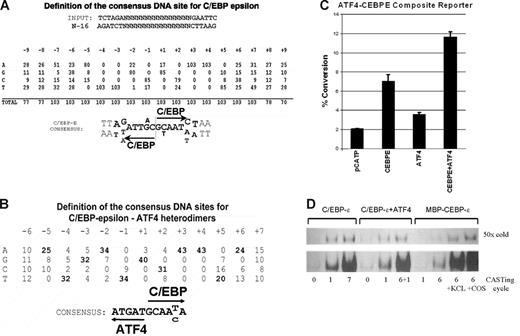
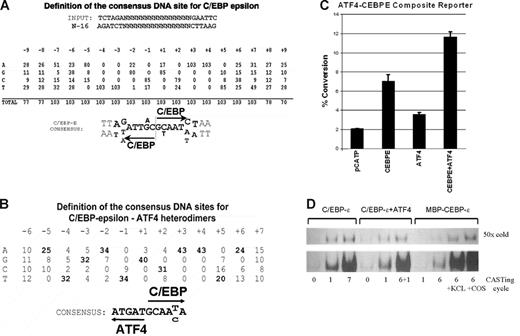
Cyclic amplification of selected sequences yields consensus C/EBP-ϵ and C/EBP-ϵ–ATF4 binding sites. (A) Definition of the consensus C/EBP-ϵ binding DNA sequences. Analysis of 103 individual clones derived from 7 cycles of site selection by anti–C/EBP-ϵ antibodies. Invariable AA and TT sequences within each C/EBP-binding half-site were used to align the selected sequences. Half-sites are indicated by arrows. (B) Definition of the consensus C/EBP-ϵ/ATF4 heterodimer binding DNA sequences. Analysis of 43 individual clones derived from site selection experiments using C/EBP-ϵ/ATF4 heterodimers produced in COS1 cells. Selected sequences were aligned using an invariable AA sequence within the C/EBP-binding half-site. (C) Activation of promoter containing C/EBP-ϵ–ATF4 heterodimer binding site by coexpressed ATF4 and C/EBP-ϵ in Jurkat cells. Jurkat cells were transfected with C/EBP-ϵ and ATF4 expression constructs and pCATP-ATCEBP reporter in chloramphenicol acetyl transferase (CAT) reporter. The cells were harvested 48 hours after transfection and CAT and β-galactosidase assays were performed. These results are shown as percentage of chloramphenicol conversion to acetylated form, adjusted for transfection efficiency as monitored by cotransfected β-galactosidase reporter assay. Experiments were done in triplicate; and the mean and SD are shown by black bars and brackets, respectively. (D) DNA binding of C/EBP-ϵ to DNA is enhanced in the presence of nuclear lysates. Samples of dsDNA prior to and after several cycles of selection were analyzed by EMSA. The number of CASTing cycles for each radiolabeled probe is indicated. C/EBP-ϵ and C/EBP-ϵ–ATF4 were derived from nuclear lysates of transfected COS1 cells, whereas MBP–C/EBP-ϵ fusion protein was produced and purified from E coli. Specificity of binding was confirmed by addition of 50-fold molar excess of unlabeled (“cold”) C/EBP-ϵ binding oligonucleotide. In the 2 right lanes, nuclear lysates derived from KCL22 and COS1 cells were added to EMSA reaction containing bacterially produced MBP–C/EBP-ϵ.
Cyclic amplification of selected sequences yields consensus C/EBP-ϵ and C/EBP-ϵ–ATF4 binding sites. (A) Definition of the consensus C/EBP-ϵ binding DNA sequences. Analysis of 103 individual clones derived from 7 cycles of site selection by anti–C/EBP-ϵ antibodies. Invariable AA and TT sequences within each C/EBP-binding half-site were used to align the selected sequences. Half-sites are indicated by arrows. (B) Definition of the consensus C/EBP-ϵ/ATF4 heterodimer binding DNA sequences. Analysis of 43 individual clones derived from site selection experiments using C/EBP-ϵ/ATF4 heterodimers produced in COS1 cells. Selected sequences were aligned using an invariable AA sequence within the C/EBP-binding half-site. (C) Activation of promoter containing C/EBP-ϵ–ATF4 heterodimer binding site by coexpressed ATF4 and C/EBP-ϵ in Jurkat cells. Jurkat cells were transfected with C/EBP-ϵ and ATF4 expression constructs and pCATP-ATCEBP reporter in chloramphenicol acetyl transferase (CAT) reporter. The cells were harvested 48 hours after transfection and CAT and β-galactosidase assays were performed. These results are shown as percentage of chloramphenicol conversion to acetylated form, adjusted for transfection efficiency as monitored by cotransfected β-galactosidase reporter assay. Experiments were done in triplicate; and the mean and SD are shown by black bars and brackets, respectively. (D) DNA binding of C/EBP-ϵ to DNA is enhanced in the presence of nuclear lysates. Samples of dsDNA prior to and after several cycles of selection were analyzed by EMSA. The number of CASTing cycles for each radiolabeled probe is indicated. C/EBP-ϵ and C/EBP-ϵ–ATF4 were derived from nuclear lysates of transfected COS1 cells, whereas MBP–C/EBP-ϵ fusion protein was produced and purified from E coli. Specificity of binding was confirmed by addition of 50-fold molar excess of unlabeled (“cold”) C/EBP-ϵ binding oligonucleotide. In the 2 right lanes, nuclear lysates derived from KCL22 and COS1 cells were added to EMSA reaction containing bacterially produced MBP–C/EBP-ϵ.
Definition of DNA sites bound by heterodimeric C/EBP-ϵ–ATF4
We have previously identified several dimerization partners of C/EBP-ϵ, among them ATF4 (CREB2), a member of the b-zipfamily of transcription factors.10,15,16 To define the consensus DNA binding site for C/EBP-ϵ–ATF4 heterodimers, we used CASTing with nuclear lysates derived from COS1 cells cotransfected with ATF4 and C/EBP-ϵ expression plasmids. Target sites were identified by immunoselection of C/EBP-ϵ–bound sites for 5 cycles of CASTing and then selection with anti-ATF4 antibodies at cycle 6, anti–C/EBP-ϵ antibodies at cycle 7, and, again, anti-ATF4 antibodies at cycle 8, thus enriching for DNA bound to heterodimeric ATF4–C/EBP-ϵ complexes. Figure 1B shows a consensus DNA site derived from this procedure. To access the effects of ATF4–C/EBP-ϵ dimer formation on transcriptional activity of a promoter containing the consensus heterodimer binding site, 2 copies of the ATEBP (see “Nuclear lysate preparation for EMSA” in “Materials and methods”) site were cloned into the pCATP minimal promoter–containing reporter. Cotransfection of this reporter together with the C/EBP-ϵ and ATF4 expression plasmids produced an approximately 5-fold activation over background promoter activity (Figure 1C). Transfection of expression plasmids for either ATF4 or C/EBP-ϵ typically results in 1.5- and 3-fold activation, suggesting that this composite site is preferentially bound by heterodimers of ATF4 and C/EBP-ϵ. Of note, COS1 cells do express a small amount of endogenous ATF4, which may contribute to higher levels of transactivation of this reporter when C/EBP-ϵ is expressed in these cells.
RelA increases specificity of C/EBP-ϵ DNA binding
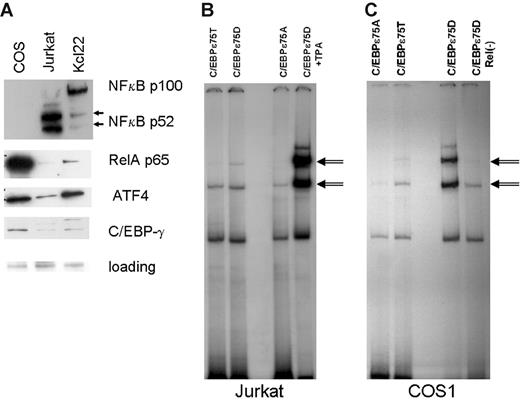
Using EMSA, we observed that addition of either COS1- or KCL22-derived nuclear lysate to MBP–C/EBP-ϵ expressed from Escherichia coli resulted in a dramatic increase in DNA binding to C/EBP consensus sites (Figure 1D). This increase could be explained by the presence of another C/EBP-ϵ–interacting protein in the nuclear lysate derived from these cells. We have previously shown that C/EBP-ϵ can readily form heterodimers with C/EBP-δ, C/EBP-ζ (CHOP), and ATF415,16 and can interact with RB and E2F proteins18,25 as well as a few others. However, anti–C/EBP-δ or anti-CHOP antibodies did not produce specific supershifts in our EMSA experiments (data not shown), and no changes were observed in mobility of the C/EBP-ϵ–DNA complex that would be expected if either E2F or RB proteins were involved in this interaction. Previous studies reported that members of the Rel family of transcription factors can facilitate DNA binding of C/EBP-α and C/EBP-β proteins in vitro.26–35 Also, our prior experiments indicated that phosphorylation at the Thr75 residue of C/EBP-ϵ results in a more than 10-fold increase in affinity of C/EBP-ϵ to consensus DNA binding sites.14 Both events, Rel interaction with either C/EBP-α or C/EBP-β as well as C/EBP-ϵ phosphorylation by several kinases, result in increased affinity of C/EBP-ϵ to DNA without inducing a novel shifted DNA-C/EBP complex.
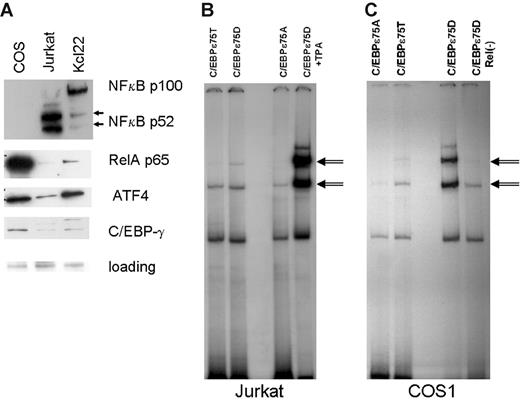
Western-blot analysis of COS1 and KCL22 nuclear lysate indicated the presence of significant amounts of p65RelA protein, especially in the COS1 lysate (Figure 2A). COS1 contains very little p52 NFkappaB protein and no p50 NFkappaB, suggesting that p65RelA is found mostly in the homodimeric state in these cells. To directly address the role of RelA in increased DNA binding of C/EBP-ϵ, we induced nuclear expression of p65RelA in Jurkat cells transfected with C/EBP-ϵ expression vectors (Figure 3B). Gel retardation (EMSA) experiments were done using the consensus C/EBP-ϵ palindromic binding sequence (Figure 2B). Both the wild-type T75 and constitutively active D75 mutant of C/EBP-ϵ formed specific complexes with DNA, whereas the A75 mutant showed decreased DNA binding consistent with our previous data.14 In TPA-activated Jurkat cells, DNA binding of C/EBP-ϵ was significantly enhanced (specific bands are indicated by arrows, Figure 2B).
Activated nuclear NFkappaB (RelA) facilitates C/EBP-ϵ DNA binding to consensus sites. (A) Western-blot analysis of NFkappaB subunits and ATF4 localized in the nucleus of COS1, Jurkat, and KCL22 cells. Antibodies against proteins listed in the right panel were used to probe Western blots derived from nuclear lysates (20 μg per lane) prepared from COS1, Jurkat, and KCL22 cells. Equal gel loading controls were Ponceau stained after transfer. (B) Gel shift analysis was performed on nuclear lysates derived from the Jurkat cells transfected with either the wild-type C/EBP-ϵ (C/EBPϵ75T) or mutant C/EBPϵ (75D or 75A) expression constructs. In the right lane, the nuclear lysate was derived from Jurkat cells transfected with C/EBPϵ75D and activated with TPA. C/EBP-ϵ–specific bands are indicated by double arrows. (C) Gel shift analysis was performed as shown in panel B, except that RelA-containing COS1 cells were used for transfection instead of Jurkat cells. In the right lane, nuclear lysate derived from C/EBP-ϵ75D–transfected cells was cleared of endogenous RelA by immunoprecipitation with a specific antibody.
Activated nuclear NFkappaB (RelA) facilitates C/EBP-ϵ DNA binding to consensus sites. (A) Western-blot analysis of NFkappaB subunits and ATF4 localized in the nucleus of COS1, Jurkat, and KCL22 cells. Antibodies against proteins listed in the right panel were used to probe Western blots derived from nuclear lysates (20 μg per lane) prepared from COS1, Jurkat, and KCL22 cells. Equal gel loading controls were Ponceau stained after transfer. (B) Gel shift analysis was performed on nuclear lysates derived from the Jurkat cells transfected with either the wild-type C/EBP-ϵ (C/EBPϵ75T) or mutant C/EBPϵ (75D or 75A) expression constructs. In the right lane, the nuclear lysate was derived from Jurkat cells transfected with C/EBPϵ75D and activated with TPA. C/EBP-ϵ–specific bands are indicated by double arrows. (C) Gel shift analysis was performed as shown in panel B, except that RelA-containing COS1 cells were used for transfection instead of Jurkat cells. In the right lane, nuclear lysate derived from C/EBP-ϵ75D–transfected cells was cleared of endogenous RelA by immunoprecipitation with a specific antibody.
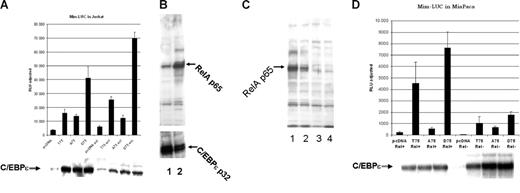
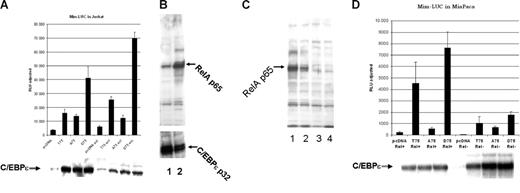
Mutation of Thr75 within the p38 MAP kinase site of C/EBPϵ affects C/EBP-ϵ–specific transactivation of a myeloid-specific promoter in the presence of activated NFkappaB. (A) The C/EBP-ϵ expression constructs for the wild-type (T75) or mutant (A75 and D75) proteins were transfected into Jurkat cells along with Mim promoter in a luciferase reporter vector. Half of the transfected cells were activated by addition of TPA and calcium ionophore. Cells were harvested 48 hours after transfection and luciferase (LUC) activity was analyzed using dual luciferase assay kit (Promega). Cotransfected renilla-luciferase expression plasmid was used as internal control for transfection efficiency. Results are presented in relative luciferase units (RLUs) as a mean of 3 independent experiments (▪), adjusted for transfection efficiency, with standard deviation (brackets). (B) Western-blot analysis of nuclear accumulation of p65RelA prior to (lane1) and after Jurkat cell activation with TPA and calcium ionophore (lane 2). (C) Level of C/EBP-ϵ transcriptional activation is dependent on the presence of p65RelA. Western-blot analysis of nuclear expression of p65RelA in parental MiaPaca2 cells (lane1) and after lentiviral vector–driven siRNA knock down of p65RelA expression in 3 different clones. Clones with maximal suppression of RelA expression, analyzed in lanes 3 and 4, were used for reporter assay experiments. (D) The C/EBP-ϵ expression constructs for either the wild-type (T75) or mutant (A75 and D75) proteins were transfected into MiaPaca cells that express high levels of nuclear RelA (Rel+) or the MiaPaca cells in which RelA expression was suppressed by specific siRNA (Rel-). C/EBP-dependent myeloid Mim luciferase reporter was cotransfected with either expression plasmids or control vector (pcDNA3.1). Each experimental point was done in triplicate and error bars represent standard deviation (SD).
Mutation of Thr75 within the p38 MAP kinase site of C/EBPϵ affects C/EBP-ϵ–specific transactivation of a myeloid-specific promoter in the presence of activated NFkappaB. (A) The C/EBP-ϵ expression constructs for the wild-type (T75) or mutant (A75 and D75) proteins were transfected into Jurkat cells along with Mim promoter in a luciferase reporter vector. Half of the transfected cells were activated by addition of TPA and calcium ionophore. Cells were harvested 48 hours after transfection and luciferase (LUC) activity was analyzed using dual luciferase assay kit (Promega). Cotransfected renilla-luciferase expression plasmid was used as internal control for transfection efficiency. Results are presented in relative luciferase units (RLUs) as a mean of 3 independent experiments (▪), adjusted for transfection efficiency, with standard deviation (brackets). (B) Western-blot analysis of nuclear accumulation of p65RelA prior to (lane1) and after Jurkat cell activation with TPA and calcium ionophore (lane 2). (C) Level of C/EBP-ϵ transcriptional activation is dependent on the presence of p65RelA. Western-blot analysis of nuclear expression of p65RelA in parental MiaPaca2 cells (lane1) and after lentiviral vector–driven siRNA knock down of p65RelA expression in 3 different clones. Clones with maximal suppression of RelA expression, analyzed in lanes 3 and 4, were used for reporter assay experiments. (D) The C/EBP-ϵ expression constructs for either the wild-type (T75) or mutant (A75 and D75) proteins were transfected into MiaPaca cells that express high levels of nuclear RelA (Rel+) or the MiaPaca cells in which RelA expression was suppressed by specific siRNA (Rel-). C/EBP-dependent myeloid Mim luciferase reporter was cotransfected with either expression plasmids or control vector (pcDNA3.1). Each experimental point was done in triplicate and error bars represent standard deviation (SD).
Next, we cleared the COS1 nuclear lysate of p65 by immunoprecipitation with specific anti-RelA antibodies (sc-372; Santa Cruz Biotechnology). Precleared lysate was unable to facilitate DNA binding by bacterially produced C/EBP-ϵ (data not shown); and in C/EBP-ϵ–expressing transfected COS1 cells, C/EBP-ϵ–specific DNA binding was markedly decreased (Figure 2C, compare 2 lanes on the right side).
Threonine 75 phosphorylation of C/EBP-ϵ is important for interaction with activated RelA
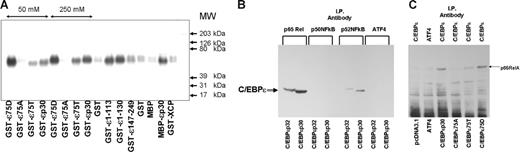
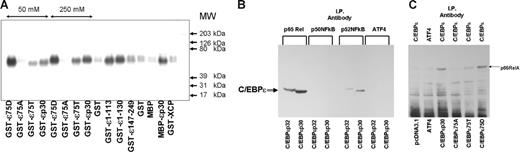
We have used the in vitro pull-down assay to define the interaction between C/EBP-ϵ and RelA. GST–C/EBP-ϵ fusion proteins used in this study contained p30 and p32 isoforms of C/EBP-ϵ. Also, we analyzed the effects of either Ala or Asp residue substitutions in the known C/EBP-ϵ phosphorylation site at Thr75 on RelA–C/EBP-ϵ interaction. All GST-C/EBP fusion proteins were immobilized on GSH-sepharose beads and incubated with RelA-containing nuclear lysate. Following extensive washing and elution, bound proteins were subjected to denaturing SDS–polyacrylamide gel electrophoresis, Western blotting, and analysis with antibodies against p65, p50, and p52 NFkappaB subunits. As shown in Figure 4A, p65RelA readily bound to wild-type and phosphomimetic 75D mutant C/EBP-ϵ fusion proteins, whereas little or no binding was seen in control MBP and GST protein–containing lanes.
C/EBP-ϵ interacts with p65RelA. (A) Specific binding of GST–C/EBP-ϵ fusion proteins to p65RelA is facilitated by the presence of phosphomimetic Asp75 (D) residue within the p38 MAPkinase target site. Equal quantity (40 μg) of immobilized bacterially produced GST–C/EBP-ϵ or MBP–C/EBP-ϵ protein and its mutant or truncated derivatives and isoforms were incubated with nuclear lysates containing p65RelA. Beads were washed extensively in conditions of either normal salt (150 mM), 50 mM, or 250 mM salt (where indicated by line with arrows). Bacterially produced GST and the GST fusion protein containing the full-length resistin-like hXCP1 protein (GST-XCP) were used as negative controls. Eluted proteins were analyzed by Western blotting using specific anti-p65RelA antibodies (Santa Cruz Biotechnology). (B) C/EBP-ϵ–NFkappaB interaction in nuclear lysates of COS1 cells transfected with expression constructs for p30 and p32 isoforms of C/EBP-ϵ. Immunoprecipitation (IP) was performed with antibodies specific to proteins listed above the lanes. Coimmunoprecipitated C/EBP-ϵ was identified by Western blotting with rabbit polyclonal anti–C/EBP-ϵ antiserum. (C) Endogenous p65RelA is coimmunoprecipitated with C/EBP-ϵ, but not ATF4, in COS1 cells transfected with expression constructs listed below each lane or control pcDNA3.1 expression vector. Following immunoprecipitation with either C/EBP-ϵ– or ATF4-specific antibody, p65RelA was identified by Western blotting.
C/EBP-ϵ interacts with p65RelA. (A) Specific binding of GST–C/EBP-ϵ fusion proteins to p65RelA is facilitated by the presence of phosphomimetic Asp75 (D) residue within the p38 MAPkinase target site. Equal quantity (40 μg) of immobilized bacterially produced GST–C/EBP-ϵ or MBP–C/EBP-ϵ protein and its mutant or truncated derivatives and isoforms were incubated with nuclear lysates containing p65RelA. Beads were washed extensively in conditions of either normal salt (150 mM), 50 mM, or 250 mM salt (where indicated by line with arrows). Bacterially produced GST and the GST fusion protein containing the full-length resistin-like hXCP1 protein (GST-XCP) were used as negative controls. Eluted proteins were analyzed by Western blotting using specific anti-p65RelA antibodies (Santa Cruz Biotechnology). (B) C/EBP-ϵ–NFkappaB interaction in nuclear lysates of COS1 cells transfected with expression constructs for p30 and p32 isoforms of C/EBP-ϵ. Immunoprecipitation (IP) was performed with antibodies specific to proteins listed above the lanes. Coimmunoprecipitated C/EBP-ϵ was identified by Western blotting with rabbit polyclonal anti–C/EBP-ϵ antiserum. (C) Endogenous p65RelA is coimmunoprecipitated with C/EBP-ϵ, but not ATF4, in COS1 cells transfected with expression constructs listed below each lane or control pcDNA3.1 expression vector. Following immunoprecipitation with either C/EBP-ϵ– or ATF4-specific antibody, p65RelA was identified by Western blotting.
Similar results were obtained in coimmunoprecipitation experiments as shown in Figure 4B. COS1 cells expressing endogenous nuclear RelA were transfected with 10 μg per 10-cm dish of expression plasmids containing p32 and p30 wild-type C/EBP-ϵ isoforms. Nuclear lysates were harvested 36 hours after transfection and analyzed by Western blotting to insure equal expression of C/EBP isoforms. Following immunoprecipitation with anti-RelA and control antibodies, SDS gel electrophoresis, and Western blotting, C/EBP-ϵ–specific bands were visualized by chemiluminescence with anti–C/EBP-ϵ antibodies and protein A/G–HRP conjugate (Figure 4B). These results indicate that both of the major naturally occurring isoforms of C/EBP-ϵ, p32 and p30, can efficiently interact with p65RelA. Besides p65RelA, another member of the NFkappaB family, p52, can also interact with C/EBP-ϵ. Further experiments are required to determine whether this is a direct interaction.
The reverse coimmunoprecipitation experiment is shown in Figure 4C. Nuclear lysates isolated from COS1 transfected with either C/EBP-ϵ or ATF4 expression plasmids were subjected to immunoprecipitation with either C/EBP-ϵ– or ATF4-specific polyclonal antibodies. Following SDS-PAGE and Western blotting, coimmunoprecipitated p65RelA was identified with specific secondary antibodies and HRP-conjugated protein A/G. Very little p65RelA was coimmunoprecipitated with either 75A mutant C/EBP-ϵ or ATF4. In contrast, wild-type and 75D mutant C/EBP-ϵ produced p65RelA-specific bands, indicated by an arrow, in Figue 4C.
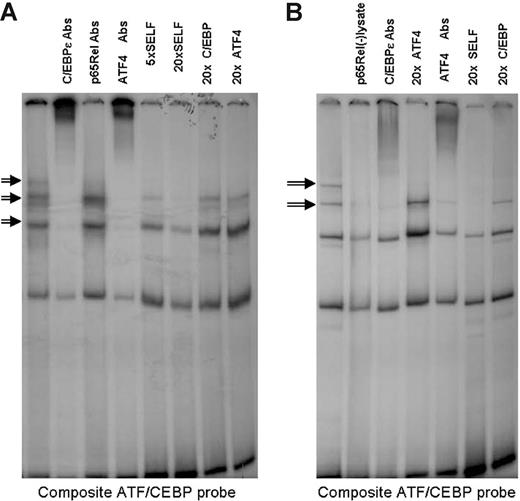
Similar experiments were performed to analyze the effects of the p65RelA–C/EBP-ϵ interaction on DNA binding of the C/EBP-ϵ/ATF4 heterodimeric complex. EMSA analysis of C/EBP-ϵ–ATF4 composite binding site is presented in Figure 3. Specific bands containing heterodimers of C/EBP-ϵ and ATF4 are suppressed by addition of increasing amounts of unlabeled “Self” oligonucleotide, lack of competitive suppression after addition of unlabeled oligonucleotides containing palindromic binding sites for C/EBP-ϵ or ATF4, as well as specific “supershifts” produced in the presence of anti–C/EBP-ϵ or anti-ATF4 antibodies. As shown in Figure 5A, in nuclear lysates derived from transfected Jurkat cells, the ATF4–C/EBP-ϵ complex bound to a composite ATF–C/EBP-ϵ binding site can be supershifted with either anti–C/EBP-ϵ or anti-ATF4 antibodies but not anti-RelA antibodies. Specificity of this heteromeric complex is demonstrated by its ability to be suppressed by the addition of 5- and 20-fold excess of unlabeled self-oligonucleotide but not 20-fold excess of oligonucleotides containing either C/EBP or ATF4 consensus sites.
Specific binding of C/EBP-ϵ–ATF4 heterodimers to a high-affinity binding site is facilitated by the presence of nuclear RelA. Gel shift analysis was performed using a composite ATF/CEBP binding probe and nuclear lysates from cells transfected with the wild-type C/EBP-ϵ and ATF4 expression vectors. Specific heterodimer binding (indicated by arrows) was confirmed by competition with 5- and 20-fold molar excess of unlabeled self-oligonucleotide and lack of competition with 20-fold molar excess of consensus C/EBP-ϵ or ATF4 homodimer binding oligonucleotides. Specific complexes contain both ATF4 and C/EBP-ϵ, but not p65RelA, as indicated by supershifts produced by addition of specific antibodies (Abs). (A) Nuclear lysates were derived from transfected and TPA- and Ca++ ionophore–activated Jurkat cells. (B) Nuclear lysates were derived from transfected COS1 cells. Elimination of endogenous p65RelA from nuclear lysates by preclearing with 10 μg of α-p65RelA per 10 μL of nuclear lysate (second lane from the left) results in disruption of specific binding of ATF4–C/EBP-ϵ heterodimers.
Specific binding of C/EBP-ϵ–ATF4 heterodimers to a high-affinity binding site is facilitated by the presence of nuclear RelA. Gel shift analysis was performed using a composite ATF/CEBP binding probe and nuclear lysates from cells transfected with the wild-type C/EBP-ϵ and ATF4 expression vectors. Specific heterodimer binding (indicated by arrows) was confirmed by competition with 5- and 20-fold molar excess of unlabeled self-oligonucleotide and lack of competition with 20-fold molar excess of consensus C/EBP-ϵ or ATF4 homodimer binding oligonucleotides. Specific complexes contain both ATF4 and C/EBP-ϵ, but not p65RelA, as indicated by supershifts produced by addition of specific antibodies (Abs). (A) Nuclear lysates were derived from transfected and TPA- and Ca++ ionophore–activated Jurkat cells. (B) Nuclear lysates were derived from transfected COS1 cells. Elimination of endogenous p65RelA from nuclear lysates by preclearing with 10 μg of α-p65RelA per 10 μL of nuclear lysate (second lane from the left) results in disruption of specific binding of ATF4–C/EBP-ϵ heterodimers.
In Figure 5B, the same composite ATF4–C/EBP-ϵ binding site was used in EMSA with nuclear lysates prepared from transfected COS1 cells. Specific binding (indicated by arrows in Figure 5B) was abolished in nuclear lysates precleared with anti-RelA antibodies. It should be noted that these precleared lysates still contain large quantities of C/EBP-ϵ and ATF4, as indicated by Western blotting (data not shown). Taken together, these data indicate that C/EBP-ϵ interaction with RelA is required for efficient binding of C/EBP-ϵ to both palindromic C/EBP consensus sites and asymmetric ATF4–C/EBP-ϵ consensus elements.
Modulation of C/EBP-ϵ–dependent promoter activity by activated NFkappaB
To study in vivo cross-talk between NFkappaB and C/EBP-ϵ transcription factors, we used a transcriptional reporter construct based on the C/EBP-ϵ–dependent Mim promoter.13,14 Jurkat cells (5 × 106 cells per experiment) were cotransfected with MIM-luciferase reporter construct and either wild-type 75T C/EBP-ϵ, 75A C/EBP-ϵ mutant, or phosphomimetic 75D C/EBP-ϵ expression plasmid. An empty pcDNA3.1 expression vector was used in control transfection, and renilla luciferase reporter plasmid served as internal control for transfection efficiency and cell viability (Figure 3A). Twelve hours after transfection, one half of each set of transfected cells was treated with TPA and a calcium ionophore, resulting in significant induction of nuclear NFkappaB (Figure 3B lane 2). At 36 hours after transfection, luciferase reporter assays were performed on both the induced and uninduced cultures using the dual luciferase assay system (Promega Biotech). As shown in Figure 3A, induction of NFkappaB in the Jurkat cell line was accompanied by significant up-regulation of the C/EBP-ϵ–dependent reporter. This was particularly evident in the case of the wild-type T75 C/EBP-ϵ and the D75 phosphomimetic mutant. This activation was not due to increased expression of C/EBP-ϵ, as indicated by Western blotting and detection with anti–C/EBP-ϵ (Figure 3A-B bottom panels). Activity of A75 C/EBP-ϵ mutant (lacking the ability to be phosphorylated at this site) was less than that of the wild-type and D75 C/EBP-ϵ. The A75 C/EBP-ϵ was not significantly affected by NFkappaB activation, further suggesting that T75 phosphorylation is required for efficient interaction of C/EBP-ϵ and NFkappaB.
Since NFkappaB activation involves a plethora of pathways and may result in effects that are nonspecific with regard to C/EBP-ϵ–dependent transcriptional activation, we studied transactivating properties of C/EBP-ϵ in MiaPaca cells (do not express C/EBP-ϵ, -α, -β, -δ; data not shown). These cells were subjected to siRNA-mediated specific knock down of p65 RelA protein. The siRNA was delivered by transduction with a self-inactivating lentiviral vector that also carries the puromycin-resistance gene. Each integration event results in H1 RNA promoter-driven expression of siRNA from 2 sites within the viral LTRs. Following transduction, several puromycin-resistant MiaPaca2 clones were analyzed with specific anti-p65RelA antibodies to make sure that p65 knock down was efficient (Figure 3C).
MIM-LUC luciferase reporter construct was then cotransfected into RelA− MiaPaca cells (clone no. 4; Figure 3C, lanes 3 and 4, respectively) as well as into the parental cell line having abundant nuclear expression of p65RelA (Figure 3C lane 1). As seen in Figure 3D, transcriptional activity of the MIM promoter is C/EBP-ϵ dependent and can be significantly up-regulated in the presence of the wild-type T75 and phosphomimetic D75 C/EBP-ϵ. This up-regulation appears to be dependent on the presence of p65RelA and it is greatly diminished in RelA knock-down cells. As predicted, activity of A75 mutant of C/EBP-ϵ, which is defective in RelA interaction, is not down-regulated in p65RelA-deficient cells.
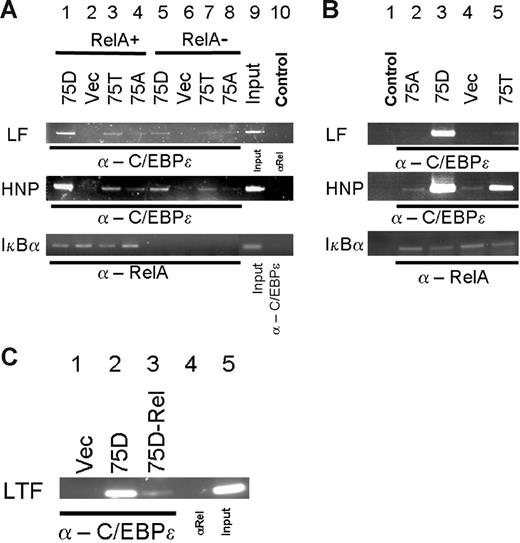
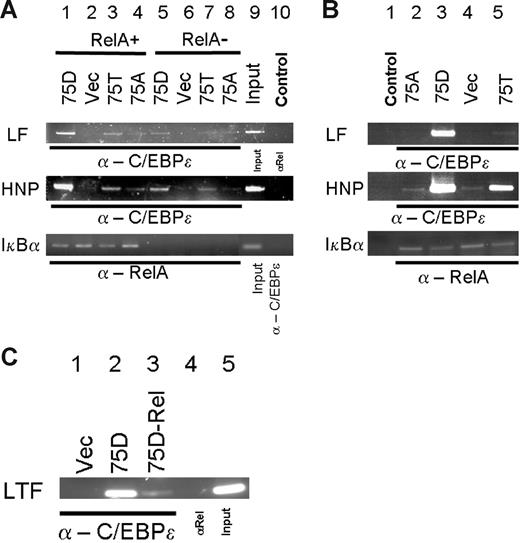
To analyze the effect of C/EBP-ϵ–p65RelA in vivo, in Figure 6 we used chromatin immunoprecipitation assay (ChIP). To achieve specific knock down of p65RelA protein, MiaPaca2 cells were transiently transduced with lentivirus carrying a RelA-specific siRNA expression cassette, whereas the control cells were transduced with lentivirus carrying a mutant siRNA expression cassette. No p65RelA protein was detected in nuclear lysates at day 5 after transduction, as assessed by ChIP and PCR analysis of the p65RelA binding site within the IkBα promoter (Figure 6A lanes 5-8, and lanes 1-4 in bottom panel) or by immunoblotting (data not shown). At day 5 after transduction, expression constructs for the wild-type (75T) or mutant 75D and 75A C/EBP-ϵ proteins or an empty vector were introduced into siRNA-transduced cells. We then used ChIP with α–C/EBP-ϵ or α-p65RelA and PCR with promoter-specific primers to detect interaction of C/EBP-ϵ and RelA with 2 of the known C/EBP-ϵ–responsive promoters, lactoferrin (LF) and defensin (HNP), as well as RelA-responsive IkBα promoter. As seen in Figure 6A, RelA knock down results in much decreased binding of C/EBP-ϵ to LF and HNP promoter sites (compare lanes 1 and 5, 3 and 7). Control experiments with α-p65RelA antibodies (Figure 6A lane 10) suggest that RelA is not able to interact directly or indirectly with these promoter sites. Likewise, C/EBP-ϵ did not bind to IkBα promoter site (Figure 6A lane 10 bottom panel).
ChIP analysis of effects of p65RelA on C/EBP-ϵ binding to lactoferrin (LF) and defensin (HNP) promoters. (A) Chromatin immunoprecipitations were performed from MiaPaca2 cells treated with control lentiviral vector (lanes 1-4, 9-10) or MiaPaca2 cells with siRNA-induced knock down of p65RelA (lanes 5-8). Five days after transduction, cells were transfected with expression plasmids directing expression of 75D, 75A, or 75T variants of C/EBP-ϵ or an empty vector control. At 36 hours after transfection, chromatin immunoprecipitations were performed, using α–/EBP-ϵ or α-p65RelA. The precipitated chromatin was analyzed using primers specific for C/EBP sites in the LF promoter, HNP promoter, or IkBα promoter. Input chromatin (1:20 dilution) is presented in lane 9. In lane 10, input chromatin was derived from immunoprecipitation with a control antibody (α-p65RelA in case of LF and HNP detection, α–C/EBP-ϵ in case of IkBα detection). (B) In vivo effects of C/EBP-ϵ mutations at codon 75 on DNA binding within the LF and HNP promoters. Chromatin immunoprecipitations were performed on RelA expressing HEK293 cells, transfected with expression plasmids for the wild-type C/EBP-ϵ (75T, lane 5), phosphomimetic 75D mutant (lane 3), or 75A mutant of C/EBP-ϵ. Empty vector–transfected control cells were used for ChIP in lane 4. In negative control experiments presented in lane 1, input chromatin was derived from immunoprecipitation of C/EBP-ϵ–transfected HEK293 cells with a control antibody (α-p65RelA in case of LF and HNP detection, α–C/EBP-ϵ in case of IkBα detection). (C) Chromatin immunoprecipitations (ChIPs) and detection of lactoferrin (LF) promoter were performed in mouse 32Dcl3 cells that were transduced with retroviral vector expressing C/EBP-ϵ phosphomimetic D75 mutant (lanes 2-5) or an empty vector control (lane 1). Five days after transduction, cells were treated with control lentiviral vector (lanes 2, 4, 5) or with RelA-specific siRNA vector. ChIPs were performed 3 days after siRNA transduction, with α–C/EBP-ϵ (lanes 1-3) or control α-p65RelA (lane 4).
ChIP analysis of effects of p65RelA on C/EBP-ϵ binding to lactoferrin (LF) and defensin (HNP) promoters. (A) Chromatin immunoprecipitations were performed from MiaPaca2 cells treated with control lentiviral vector (lanes 1-4, 9-10) or MiaPaca2 cells with siRNA-induced knock down of p65RelA (lanes 5-8). Five days after transduction, cells were transfected with expression plasmids directing expression of 75D, 75A, or 75T variants of C/EBP-ϵ or an empty vector control. At 36 hours after transfection, chromatin immunoprecipitations were performed, using α–/EBP-ϵ or α-p65RelA. The precipitated chromatin was analyzed using primers specific for C/EBP sites in the LF promoter, HNP promoter, or IkBα promoter. Input chromatin (1:20 dilution) is presented in lane 9. In lane 10, input chromatin was derived from immunoprecipitation with a control antibody (α-p65RelA in case of LF and HNP detection, α–C/EBP-ϵ in case of IkBα detection). (B) In vivo effects of C/EBP-ϵ mutations at codon 75 on DNA binding within the LF and HNP promoters. Chromatin immunoprecipitations were performed on RelA expressing HEK293 cells, transfected with expression plasmids for the wild-type C/EBP-ϵ (75T, lane 5), phosphomimetic 75D mutant (lane 3), or 75A mutant of C/EBP-ϵ. Empty vector–transfected control cells were used for ChIP in lane 4. In negative control experiments presented in lane 1, input chromatin was derived from immunoprecipitation of C/EBP-ϵ–transfected HEK293 cells with a control antibody (α-p65RelA in case of LF and HNP detection, α–C/EBP-ϵ in case of IkBα detection). (C) Chromatin immunoprecipitations (ChIPs) and detection of lactoferrin (LF) promoter were performed in mouse 32Dcl3 cells that were transduced with retroviral vector expressing C/EBP-ϵ phosphomimetic D75 mutant (lanes 2-5) or an empty vector control (lane 1). Five days after transduction, cells were treated with control lentiviral vector (lanes 2, 4, 5) or with RelA-specific siRNA vector. ChIPs were performed 3 days after siRNA transduction, with α–C/EBP-ϵ (lanes 1-3) or control α-p65RelA (lane 4).
Next, we directly compared the effect of mutations affecting phosphorylation site at codon 75 of C/EBP-ϵ and NFkappaB interaction on DNA binding to LF and HNP promoter sites by ChIP assay in HEK293 cells transiently transfected with C/EBP-ϵ expression vectors (Figure 6B). In agreement with our in vitro EMSA data and results of the reporter assays, “phosphomimetic” 75D mutation that has increased capacity to bind p65RelA also showed increased binding to LF and HNP promoter sites (Figure6B compare lanes 3 and 5). Inactive 75A mutant C/EBP-ϵ showed very little interaction with either the LF or HNP promoters.
In order to study the effect of the C/EBP-ϵ–RelA interaction on DNA binding in myeloid cells, we have used 32Dcl3 mouse cells that can be induced to differentiate into neutrophil-like cells expressing secondary granule genes (Figure 6C). We have transduced these cells with the retrovirus carrying the D75 phosphomimetic mutant C/EBP-ϵ or an empty vector control (Figure 6C lanes 2-5 vs lane 1), as described.14 C/EBP-ϵ–expressing 32Dcl3 cells were then transiently transduced with lentiviral siRNA expression construct specific for mouse RelA (Figure 6C lane 3) or a control scrambled siRNA vector (Figure 6C all other lanes). The results of chromatin immunoprecipitation of mouse lactoferrin gene promoter indicate that strong binding of C/EBP-ϵ to this promoter (Figure 6C lane 2) is greatly reduced when the p65RelA is down-regulated (Figure 6C lane 3). Taken together, ChIP assay data support our observations of transfected Mim-reporter assays and in vitro DNA binding experiments.
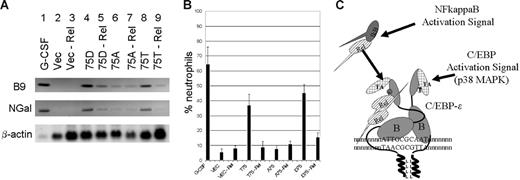
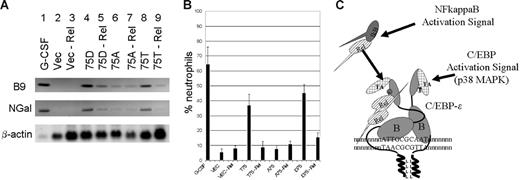
RelA–C/EBP-ϵ interaction alters expression of secondary granule genes and neutrophil differentiation
It has previously been shown that overexpression of C/EBP-ϵ and especially of D75 mutant of C/EBP-ϵ in mouse myeloid 32Dcl3 cells is capable of inducing neutrophil differentiation and expression of the secondary granule genes, even in the absence of G-CSF induction.14,23,36 To examine the role of RelA–C/EBP-ϵ interaction in granulopoiesis, 32Dcl3 cells were transduced with the wild-type (T75) or mutant (D75 and A75) C/EBP-ϵ expression constructs or the control vector (Figure 7A). Following the selection with puromycin, cells were transiently transduced with the lentiviral vectors carrying RelA-specific siRNA cassette (Figure 7A lanes 3, 5, 7, and 9) or the control siRNA (Figure 7A lanes 2, 4, 6, and 8). Following the siRNA introduction, cells were cultured without IL-3 for 3 days and total RNA was extracted and analyzed using Northern blotting and probes specific for 2 secondary granule genes, B9 and NGal. As a control, RNA derived from the wild-type 32Dcl3 cells differentiated for 5 days in the presence of 100 ng/mL of G-CSF was analyzed in lane 1 (Figure 7A). Comparison of the hybridizing bands in lanes 4 and 5, as well as 8 and 9 (Figure 7A), suggests that RelA knock down results in significant reduction of C/EBP-ϵ–induced secondary granule gene expression. The low level of expression of these genes induced by the A75 mutant (shown to be defective in RelA interaction) appears to be unaffected by RelA down-regulation in 32Dcl3 cells (Figure 7A lanes 6-7). In a parallel experiment (Figure 7B), we have analyzed the morphology of 32Dcl3 cells expressing C/EBP-ϵ in the presence or absence of RelA. C/EBP-ϵ–expressing and control vector-transfected cells were treated with lentivirus expressing RelA-specific or scrambled siRNA and allowed to differentiate in the absence of IL-3 for 5 days. Graphs shown in Figure 7B represent average percentage of band-neutrophil–like cells. Down-regulation of RelA in 32Dcl3 cells is also accompanied with increased apoptosis; apoptotic cells were excluded from counting (data not shown). As seen in Figure 7B, down-regulation of RelA in 32Dcl3 cells expressing the wild-type (T75) or phosphomimetic (D75) C/EBP-ϵ results in significant decrease in the number of differentiated neutrophil-like cells. Furthermore, these data are in agreement with the ChIP results and parallel the results of Northern-blot analysis of the secondary granule genes shown in Figure 7B. Taken together, these results indicate that RelA–C/EBP-ϵ interaction is important for effective DNA binding to promoters, activation of secondary granule genes, and C/EBP-ϵ–induced differentiation in myeloid cells.
Interaction of C/EBP-ϵ with p65RelA is important for neutrophil differentiation. (A) Northern-blot analysis of secondary granule B9 and NGal gene expression in murine 32Dcl3–derived cells. Twenty micrograms of total RNA derived from 32Dcl3 cells transduced with retroviral vectors expressing the wild-type (T75), mutant (D75 and A75) C/EBP-ϵ, or an empty vector control was analyzed with probes for the secondary granule B9 and NGal genes. Cells were transiently transduced with lentivirus targeting RelA (lanes 3, 5, 7, 9) or nonspecific siRNA (lanes 2, 4, 6, 8). Lane 1 contained 20μg total RNA derived from the wild-type 32Dcl3 cells that were terminally differentiated for 6 days in the presence of 100 ng/mL G-CSF. (B) Neutrophilic differentiation of 32Dcl3 cells transduced with C/EBP-ϵ expression vectors in the presence or absence of p65RelA. Maturation of 32Dcl3 cells transduced with the wild-type C/EBP-ϵ (T75), D75 and A75 mutants, or control vector was observed in the presence or absence or p65RelA. Following infection with the lentivirus expressing RelA-specific siRNA, cells were cultured for 5 days in the absence of IL-3. Graphs represent the percentage of band-neutrophil–like differentiated cells. Only living cells were counted. Graphs show the average and SD of 3 transduction experiments. (C) Schematic representation of RelA activation pathway leading to increased affinity of C/EBP-ϵ to its cognate DNA binding sites. The basic region of C/EBP-ϵ subunits involved in DNA recognition is indicated as B. TA denotes transcriptional activation domain, whereas the leucine zipper dimerization domain is indicated as LLLL. Phosphorylation of the Thr75 site on C/EBP-ϵ results in increased interaction with activated nuclear NFkappaB, which in turn may produce changes in the conformation of C/EBP-ϵ subunits within the dimer to improve DNA recognition.
Interaction of C/EBP-ϵ with p65RelA is important for neutrophil differentiation. (A) Northern-blot analysis of secondary granule B9 and NGal gene expression in murine 32Dcl3–derived cells. Twenty micrograms of total RNA derived from 32Dcl3 cells transduced with retroviral vectors expressing the wild-type (T75), mutant (D75 and A75) C/EBP-ϵ, or an empty vector control was analyzed with probes for the secondary granule B9 and NGal genes. Cells were transiently transduced with lentivirus targeting RelA (lanes 3, 5, 7, 9) or nonspecific siRNA (lanes 2, 4, 6, 8). Lane 1 contained 20μg total RNA derived from the wild-type 32Dcl3 cells that were terminally differentiated for 6 days in the presence of 100 ng/mL G-CSF. (B) Neutrophilic differentiation of 32Dcl3 cells transduced with C/EBP-ϵ expression vectors in the presence or absence of p65RelA. Maturation of 32Dcl3 cells transduced with the wild-type C/EBP-ϵ (T75), D75 and A75 mutants, or control vector was observed in the presence or absence or p65RelA. Following infection with the lentivirus expressing RelA-specific siRNA, cells were cultured for 5 days in the absence of IL-3. Graphs represent the percentage of band-neutrophil–like differentiated cells. Only living cells were counted. Graphs show the average and SD of 3 transduction experiments. (C) Schematic representation of RelA activation pathway leading to increased affinity of C/EBP-ϵ to its cognate DNA binding sites. The basic region of C/EBP-ϵ subunits involved in DNA recognition is indicated as B. TA denotes transcriptional activation domain, whereas the leucine zipper dimerization domain is indicated as LLLL. Phosphorylation of the Thr75 site on C/EBP-ϵ results in increased interaction with activated nuclear NFkappaB, which in turn may produce changes in the conformation of C/EBP-ϵ subunits within the dimer to improve DNA recognition.
Discussion
Transcriptional regulators, such as members of the C/EBP family of proteins, activate their cognate target genes in concert with several distinct interacting partners. As it was previously shown by us and by others, C/EBP-ϵ can readily form heterodimers with the transcriptional activator C/EBP-δ (also called NF-IL6β for its ability to interact with C/EBP-β [NF-IL6] and C/EBP-ζ [CHOP]). In turn, C/EBP-ζ has been implicated in stress-induced modulation of transcription via a unique set of C/EBP-ζ–ATF3 heterodimer binding sites.37,38 These heterodimers are, in turn, subject to modulation by specific protein phosphorylation (C/EBP-δ) that inactivates effective binding to DNA or to SUMOylation (C/EBP-ϵ) that targets protein for degradation.
The regulatory network of interactions between basic leucine zipper–containing proteins of the C/EBP family also includes the related ATF/CREB group of proteins. Members of the ATF/CREB protein family, in turn, can interact with the Fos/Jun family of transcriptional regulators containing basic leucine zipper DNA binding and dimerization domains. Using yeast 2-hybrid libraries and pull-down assays, we have previously identified ATF4/CREB as a predominant C/EBP-ϵ heterodimerization partner (Gombart et al16 ; A. F. Gombart, unpublished, 2005). C/EBP-ϵ protein tends to form heterodimers, which are more stable than homodimers.1,9 Therefore, we hypothesized that this interaction might change the DNA binding specificity of the resulting ATF4–C/EBP-ϵ heterodimer. Using several cycles of site selection from a random collection of sequences, we identified consensus binding sites for C/EBP-ϵ homodimers and ATF4–C/EBP-ϵ heterodimers (Figure 1). Of note, our attempt to use bacterially expressed, purified C/EBP-ϵ protein for site selection produced only short sites or palindromic sites with a degenerate central region.
By using C/EBP-ϵ and ATF4–C/EBP-ϵ complexes produced in eukaryotic cells, we were able to define the high-affinity binding sites for C/EBP-ϵ homodimers and heterodimers. The C/EBP-ϵ homodimer binding site appears to be a palindrome with central symmetry, where each C/EBP-ϵ is bound to a half-site GCAAT. This is in agreement with prior studies that identified C/EBP-α and C/EBP-β binding sites.1,20 Interestingly, our analysis of available data on human and murine genomes indicates that this sequence is found only very rarely in the genome and nowhere close to C/EBP-ϵ–regulated genes. At the same time, genes that are known to be regulated by C/EBP-ϵ, such as lactoferrin (LF), defensins (HNPs), and other granule proteins, tend to have suboptimal binding sites (such as ATTGG/GCAAT and TGAGG/GCAAT in case of LF) or composite binding sites rather than high-affinity palindromic sequences. We hypothesize that weaker binding can easily be modulated via interactions with coregulators, thus offering another level of control in gene regulation. Sites bound by ATF4–C/EBP-ϵ heterodimers consist of a symmetrically arranged half-site for the C/EBP-ϵ subunit (GCAAT) and the ATF4 subunit (ATGAT) (Figure 1B). This is reminiscent of previously identified CHOP-ATF3 heterodimer binding sites CATGAT/GCAAT.20,33,34,39
While performing DNA site-selection experiments, we have found that weak DNA binding ability of C/EBP-ϵ protein produced in E coli can be greatly enhanced by addition of nuclear lysates prepared from COS1 and KCL22 cells. Several potential C/EBP-ϵ–interacting proteins could be involved in modulation of affinity of C/EBP-ϵ to DNA. Western-blot analysis indicated that COS1 and Kcl22 do not express proteins of the C/EBP family, with an exception of minute amounts of C/EBP-γ and ATF4. Another group of C/EBP-interacting proteins is composed of proteins that couple transcription machinery to cell-cycle control. We have previously shown that C/EBP-ϵ can form functional complexes with the retinoblastoma (Rb) protein that is abundantly expressed in COS1 cells and the transcriptional activator E2F.18 However, our prior experiments did not detect increased C/EBP-ϵ–DNA interaction associated with either Rb or E2F complexes.
Yet another level of regulation of transcription by C/EBP proteins has been reported to be due to interaction with activated NFkappaB proteins, in particular p65RelA. RelA was shown to greatly increase the affinity of C/EBP-α, C/EBP-β, and C/EBP-δ to DNA without producing additional supershifting bands on EMSA gels. Several lines of evidence point to the importance of RelA-C/EBP interaction in regulation of promoter activity.27,29–35 Our first hypothesis was that addition of p65RelA derived from either COS1 or Kcl22 nuclear lysates might stabilize C/EBP-ϵ–DNA interaction by directly binding to C/EBP-ϵ. Indeed, Western-blot analysis confirmed that p65RelA is expressed in the nucleus of both cell lines. Our previous studies of C/EBP-ϵ phosphorylation by p38 MAPK have identified a specific site of phosphorylation, threonine 75, resulting in a 10- to 15-fold increase of C/EBP-ϵ affinity to DNA.14 This effect could also be achieved by mutating the T75 residue to D75, thus introducing a constitutive phosphomimetic negative charge at this site. Our second hypothesis was that phosphorylation-dependent changes in affinity of C/EBP-ϵ to DNA could be due to interaction of phospho-T75 or D75-containing domain with a member of the NFkappaB family, RelA, resulting in stabilization of the C/EBP-ϵ–DNA interaction.
To address these 2 hypotheses, we employed an in vitro pull-down assay using bacterially produced wild-type and mutant C/EBP-ϵ proteins fused to either GST or MBP tags together with COS1 nuclear lysate. Following interaction, C/EBP-ϵ–bound p65RelA was visualized using specific antibodies and chemiluminescence assay. As shown in Figure 4A, wild-type T75 and mutant D75 C/EBP-ϵ proteins bound p65RelA, whereas A75 C/EBP-ϵ mutant and a number of control proteins did not. Also, the N-terminal segment of C/EBP, rather than the C-terminal region, was involved in p65RelA binding. This interaction was further confirmed by reciprocal immunoprecipitation of C/EBP-ϵ and NFkappaB in nuclear lysates (Figure 4B-C).
Thr75 phosphorylation of C/EBP-ϵ frequently occurs under normal conditions in cells that express C/EBP-ϵ, but data suggest that it may also be enhanced under stress conditions. It probably plays an important role in neutrophilic differentiation. In C/EBP-ϵ knock-out mice and individuals with specific granule deficiency who carry homozygous mutations of C/EBP-ϵ, granulocytes have a markedly decreased ability to produce superoxide and migrate, and they do not produce several important bactericidal and inflammatory components of secondary granules.40–43 Inhibition of p38 MAP kinase that is involved in T75 phosphorylation of C/EBP-ϵ in human neutrophils also results in reduction of chemotaxis, adhesion, and superoxide production.44–49 NFkappaB activation that is predominantly regulated by phosphorylation, acetylation, and associated effects on protein stability and subcellular localization is a classic pathway of rapid induction of proinflammatory genes.50,51
Therefore, we studied the consequences of the C/EBP-ϵ–p65RelA interaction on induction of myeloid-specific C/EBP-ϵ–regulated Mim promoter. First, we transfected the Mim-promoter–driven luciferase reporter plasmid into the Jurkat cell line where the NFkappaB pathway can be induced by treatment with TPA and calcium ionophore. As shown in Figures 3A and 6B, both the wild-type T75 and phosphomimetic D75 C/EBP-ϵ proteins synergize with activated NFkappaB to stimulate the Mim reporter through C/EBP-ϵ sites. Importance of C/EBP-ϵ T75 phosphorylation for interaction with the NFkappaB pathway was demonstrated by lack of synergy with A75 mutant that cannot be phosphorylated. To demonstrate further the role of p65RelA in C/EBP-ϵ regulation of Mim promoter, we established clones of the MiaPaca cell line in which abundant endogenous p65RelA was knocked-down by RelA-specific siRNA (Figure 3C-D). These cells lack constitutive expression of C/EBP proteins. Cotransfection of C/EBP-ϵ expression plasmids with the Mim-LUC reporter into the parental (RelA+) cell line and its RelA− derivative resulted in a dramatic decrease in C/EBP-ϵ inducibility of Mim promoter in case of the wild-type T75 and mutant D75 proteins. As predicted, activity of the A75 C/EBP-ϵ mutant that cannot properly interact with RelA was not affected by specific p65RelA knock-down.
These data were further expanded to include endogenous C/EBP-ϵ–regulated genes, LF, and HNPs. Interestingly, both genes lack “good” palindromic binding sites for C/EBPs and we were able to demonstrate significant dependency of C/EBP-ϵ binding to these sites on the intact threonine 75 phosphorylation site (Figure 6). Not only did the 75D mutant of C/EBP-ϵ bind these sites better than did the wild-type 75T protein (Figure 6B), but binding to sites in LF and HNP promoters was also significantly affected by the presence of nuclear p65RelA, as shown by ChIP assay using siRNA knock down of p65RelA (Figure 6A).Very similar results were also obtained in mouse 32Dcl3 cells overexpressing activated D75 variant of C/EBP-ϵ (Figure 6C)
C/EBP-ϵ is normally expressed in several types of cells, including short-lived and rapidly activated granulocytes, and in other cell lines, such as differentiating CD34+ cells in a temporally restricted fashion. In both cases, activation of specific gene subsets probably depends on specificity of the C/EBP-ϵ–containing complexes which is, in turn, determined by reversible and regulated heterodimer formation of proteins such as ATF4, CHOP, and C/EBPδ. We now report that affinity of C/EBP-ϵ DNA binding is also subject to regulation by cross-talk with the NFkappaB pathway that is regulated by proinflammatory stimuli. Our results presented in Figure 7A-B suggest that cross-regulation of RelA and C/EBP-ϵ plays a role in induction of the secondary granule genes that are C/EBP-ϵ dependent and in the process of neutrophil differentiation itself. Activity of RelA-p65 protein is subject to complex regulation by phosphorylation and reversible acetylation. It remains to be determined whether C/EBP-ϵ–RelA interaction depends on these modifications. It is also unclear whether association of these transcription factors takes place in the nucleus or in the cytoplasm. It is also possible that C/EBP-ϵ, which can associate with acetyltransferases such as CBP-p300, may itself be involved in regulation of activity of RelA-dependent promoters via enhanced acetylation of RelA. We hypothesize that the interaction of phospho-T75 C/EBP-ϵ protein with nuclear p65 Rel results in conformation changes of C/EBP-ϵ dimers and increased interaction with its cognate DNA sequences (Figure 7). A recent study reported that IkappaB kinase epsilon, which is also involved in the activation of the NFkappaB pathway, is essential for effective binding of C/EBP-δ to DNA and activation of target genes.52 We hypothesize that coordinated phosphorylation of C/EBP-ϵ by the kinase signaling pathway and interaction with active nuclear p65RelA is important for C/EBP-ϵ–regulated transcription.
Authorship
Contribution: A.M.C. designed research, performed research, and wrote the paper. A.S. performed research. E.A.W. contributed vital new reagents and analytical tools. H.P.K. cowrote the paper and coanalyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexey M. Chumakov, Division of Hematology/Oncology, Cedars-Sinai Research Institute, 5005 Davis Research Building, 8700 Beverly Blvd, Los Angeles, CA 90048; e-mail: chumakov@gmail.com.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants (5 RO1 CA26038-21 and 5 RO1 CA26038-25). H.P.K. is a member of the Jonsson Comprehensive Cancer Center and the Molecular Biology Institute at the University of California at Los Angeles (UCLA).