Abstract
Acute graft-versus-host disease (aGvHD) contributes significantly to morbidity and mortality after allogeneic hematopoietic stem cell transplantation (allo-HSCT). Diagnosis of GvHD is mainly based on clinical features and tissue biopsies. A noninvasive, unbiased laboratory test for GvHD diagnosis does not exist. Here we describe the application of capillary electrophoresis coupled online with mass spectrometry (CE-MS) to 13 samples from 10 patients with aGvHD of grade II or more and 50 control samples from 23 patients without GvHD. About 170 GvHD-specific polypeptides were detected and a tentatively aGvHD-specific model consisting of 31 polypeptides was chosen, allowing correct classification of 13 of 13 (sensitivity 100.0% [95% confidence interval {CI} 75.1 to 100.0]) aGvHD samples and 49 of 50 (specificity 98.0% [95% CI 89.3 to 99.7]) control samples of the training set. The subsequent blinded evaluation of 599 samples enabled diagnosis of aGvHD greater than grade II, even prior to clinical diagnosis, with a sensitivity of 83.1% (95% CI 73.1 to 87.9) and a specificity of 75.6% (95% CI 71.6 to 79.4). Thus, high-resolution proteome analysis represents an unbiased laboratory-based screening method, enabling diagnosis, and possibly enabling preemptive therapy.
Introduction
Allogeneic hematopoietic peripheral blood stem cell or bone marrow transplantation (allo-HSCT) is applied with success to the treatment of hematopoietic malignancies, hematopoietic failure, and autoimmune diseases as well as to genetic disorders. To date, the application of allo-HSCT is limited due to severe life-threatening complications such as severe acute graft-versus-host disease (aGvHD).1–3 Although reduction of conditioning regimen–related toxicity led to a wider application of HSCT, especially to the inclusion of older patients, there was no beneficial influence on the incidence of aGvHD.4,5 Depending on the type of transplantation, the immunosuppressive treatment, and underlying diseases, between 35% and 70% of patients develop GvHD after allo-HSCT, with more than 35% of these patients requiring immunosuppressive therapy.6 To date, diagnosis of aGvHD is mainly based on clinical features, such as skin rash, gastrointestinal complications, and elevation of liver enzymes, and is verified with tissue biopsies and histopathologic examination. Differential diagnosis of acute GvHD includes discrimination of other common complications after HSCT, such as bacterial or viral infections and/or medication-induced toxicities. Early diagnosis of aGvHD, preferably based on unbiased laboratory screening tools, may increase the safety of allo-HSCT and thus further broaden its application to even larger patient populations and to a broader donor pool. In the past, many efforts were made to use single-protein biomarkers, which were specific for infection or inflammation after allo-HSCT or specific for aGvHD.7–10 Although some of these reports seem to hold promise, in many cases there is a high probability that 1 single marker will probably be increased in more than 1 incidence, thus making differential diagnosis of similar diseases difficult. It is reasonable that the simultaneous monitoring of more than 1 protein or peptide within a sample holds greater promise for the differential diagnosis of diseases, including aGvHD. Recently, the application of proteomic tools allowing screening for differentially expressed or excreted proteins in body fluids is becoming more important.11,12 Proteomic screening of body fluids with capillary electrophoresis coupled online to mass spectrometry (CE-MS) allows the simultaneous analysis of more than 1000 different proteins and peptides in 1 sample within a short examination period.13,14 These proteins and peptides are identified via their specific mass per charge (m/z) ratio, the migration time in the capillary electrophoresis, as well as the signal intensity, which gives a measure for the relative abundance of the peptides.15 The method allows compilation of data generated from different samples as well as patients and thus allows proteomic patterns specific for different pathologic conditions to be established. The generation of specific patterns that may also be modulated during disease progression or response to therapy seems much more suitable to the requirements of today's diagnosis and follow-up.16 Support vector machine (SVM)–based model prediction17 allows the best possible separation of disease groups and controls
We have recently published a pilot study demonstrating the feasibility of the early diagnosis of acute GvHD based on CE-MS applied to the screening of urine samples collected and analyzed at the time of GvHD development.18 Meanwhile we have improved the sample preparation method to meet the need of better comparability of data sets, especially in the case of pathologic proteinuria.19 Here we report the establishment of an aGvHD-specific proteomic model based on a robust sample-preparation protocol and its application to the screening of samples collected prospectively from 141 patients after allo-HSCT and analyzed in a blinded fashion to evaluate the feasibility of differential diagnosis of aGvHD from other complications occurring after allo-HSCT. The aim of the present study was the improvement and thorough validation of this approach in a blinded multicenter setting.
Patients, materials, and methods
Patients
Midstream urine samples from 141 patients after allo-HSCT were prospectively collected after informed consent was obtained in accordance with the Declaration of Helsinki. The study protocol for diagnosis of aGvHD by application of CE-MS screening was approved by the ethics committee of the Hannover Medical School (Hannover, Germany). The majority of the patients (n = 132) received a transplant for hematologic malignancies (acute myeloid leukemia [AML], n = 58; acute lymphoid leukemia [ALL], n = 23; secondary AML [sAML], n = 15; chronic myelogenous leukemia [CML], n = 7; chronic lymphocytic leukemia [CLL], n = 5; multiple myeloma [MM], n = 9; non-Hodgkin lymphoma [NHL], n = 7; Hodgkin disease [HD], n = 4; myelodysplastic syndromes [MDSs], n = 4), whereas 9 received a transplant because of hematopoietic failure syndromes (severe and very severe aplastic anemia [sAA], n = 5; osteomyelofibrosis [OMF], n = 3; paroxysmal nocturnal hemoglobinuria [PNH], n = 1). Forty-five patients were treated according to dose-reduced conditioning regimens (28 of these according to the FLAMSA [FLAMSA: Dose reduced conditioning regimen: Fludarabin, AMSACRIN (= FLAMSA) followed by low dose total body irradiation (4Gy), cyclophosphamid (60 mg/kg), and antithymocyte globulin (20 mg)] protocol20 ). GvHD prophylaxis consisted of antibodies (antithymocyte globulin [ATG] or thymoglobulin, n = 90; alemtuzumab, n = 3) together with cyclosporin A (CsA) and methotrexate (MTX) or mycophenolic acid (MMF); 48 patients did not receive antibodies as GvHD prophylaxis in this study population. Seventy-nine patients received transplants from matched unrelated donors (MUDs) and 56 received stem cells from matched related donors (MRDs, SIB), 3 from haploidentical donors, 2 from mismatched donors, and 1 from a syngeneic donor. Seven hundred twenty-six urine samples from 141 consecutive allo-HSCT patients from 5 centers, (Hannover Medical School; Ludwig-Maximilians-University, Munich; University Clinic Hamburg Eppendorf, Hamburg; University of Regensburg, Germany; University of Michigan) were collected prospectively from day −14 up to day +365. These samples were divided into a training set including 63 samples from 33 individuals and a blinded set of 663 samples from 141 individuals. Different urine samples from the same patient taken at different time points after HSCT might constitute either the training or the blinded set.
Training set (n = 63).
Urine samples from 33 consecutive allo-HSCT patients were collected from day +5 to +231. Thirteen samples from 10 individuals (mean age, 40 years; range, 18-66 years) with aGvHD grade II or more at the time of collection (day +7 to day +57) and 50 samples from 23 patients (mean age, 48 years; range, 18-69 years) showing no evidence of disease (Table 1) constituted the training set. Characteristics of all patients in the study are given in Table S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Blinded analysis of sensitivity and specificity of aGvHD diagnosis (n = 663).
Six hundred sixty-three urine samples from 141 allo-HSCT patients were collected prospectively from day −14 up to +365. All samples were analyzed in a blinded fashion. More detailed characteristics of all patients in the study are shown in Table S1.
Quality control (n = 71).
Urine samples from healthy volunteers and patients with chronic renal diseases were used to eliminate nonspecific biomarkers. These included healthy controls (n = 20) and patients with diabetic nephropathy ([DN] n = 6), vasculitis (n = 11), focal-segmental glomerulosclerosis ([FSGS] n = 6), IgA nephropathy ([IgAN] n = 11), minimal change disease ([MCD] n = 8), membranous nephropathy ([MGN] n = 5), and systemic lupus erythematosus ([SLEs], n = 4) collected at different centers.
Urine sample collection
Ten milliliters of midstream urine was obtained from each participant as described previously,15,18 and samples were immediately frozen and stored at −20°C until analysis. A 0.7-mL aliquot of urine was thawed shortly before use and diluted with 0.7 mL of 2 M urea, 10 mM NH4OH containing 0.02% SDS. Proteins of higher molecular weight, such as albumin or immunoglobulin G, were removed by filtration through Amicon ultracentrifugation filter devices (20 kDa molecular weight cutoff [MWCO]; Millipore, Bedford, MA) at 3000g until 1.1 mL of filtrate was obtained. The filtrate was then applied onto a PD-10 desalting column (Amersham Bioscience, Uppsala, Sweden) equilibrated in 0.01% NH4OH in high-performance liquid chromatography (HPLC)–grade H2O (Roth, Germany) to decrease matrix effects by removing urea, electrolytes, and salts and to enrich polypeptides present. Finally, all samples were lyophilized, stored at 4°C, and suspended in HPLC-grade H2O shortly before CE-MS analysis, as described.16,21
CE-MS analysis
CE-MS analysis was performed as described16,21,22 using a P/ACE MDQ (Beckman Coulter, Fullerton, CA) system online coupled to an electrospray ionization–time-of-flight (ESI-TOF) MS (Mariner Biospectrometry Workstation; Applied Biosystems, Farmington, CT) using sheath flow coupling (30% methanol, 0.5% formic acid in H2O). The potential of the ESI sprayer (Agilent Technologies, Palo Alto, CA) was set between 3 and 4 kV. Data acquisition and MS acquisition methods were automatically controlled by the CE via contact-close relays. Spectra were accumulated every 3 seconds, over a range of 400 to 3000 m/z.
Data processing and cluster analysis
Mass spectral ion peaks representing identical molecules at different charge states were deconvoluted into single masses using MosaiquesVisu 2.1.0 software (Mosaiques-Diagnostics GmbH, Hannover, Germany.21 In addition, the migration time and ion signal intensity (amplitude) were normalized using internal polypeptide standards.19 The resulting peak list characterizes each polypeptide by its molecular mass (Da), normalized migration time (min), and normalized signal intensity. All detected polypeptides were deposited, matched, and annotated in a Microsoft SQL database, allowing further analysis and comparison of multiple samples (patient groups). Polypeptides within different samples were considered identical if the mass deviation was less than 200 ppm and the migration time deviation was less than 2 minutes. CE-MS data of all individual samples can be accessed in Table S2.
Statistical methods
Estimates of sensitivity and specificity were calculated based on tabulating the number of correctly classified samples.16 Confidence intervals (95% CI) were based on exact binomial calculations and were carried out in MedCalc version 8.1.1.0 (MedCalc Software, Mariakerke, Belgium). The receiver operating characteristic (ROC) plot was obtained by plotting all sensitivity values (true positive fraction) on the y-axis against their equivalent (1 minus specificity) values (false-positive fraction) for all available thresholds on the x-axis (MedCalc Software). The area under the ROC curve (AUC) was evaluated, as it provides a single measure of overall accuracy that is not dependent upon a particular threshold.23
Multivariate analysis was performed using data sets of the test set and SAS software package ( http://www.spss.com). Only data sets of urine samples collected after day 2 after HSCT were included. Since the number of control samples was about 3 times larger than the number of cases, control samples were randomly excluded until equal group sizes were achieved (95 in each group). Classification result was used as the dependent variable and was analyzed for the factors age; sex; conditioning regimen with or without total body irradiation (TBI), ATG, or other antibodies in the GvHD prophylaxis; as well as standard versus reduced-intensity conditioning.
Definition of biomarkers and sample classification using MosaiquesCluster
For biomarker panel definition, only polypeptides that were found in more than 40% of the urine samples in at least 1 of the different groups of the training set (aGvHD or controls) were included. This selection should help to reduce artifacts due to polypeptides present only in a small number of samples, which would only marginally improve specificity while unproportionally increasing the number of polypeptides in the panel. In the first step, a list of predefined polypeptides was obtained considering all available data sets of the sample groups compared (eg, aGvHD vs controls). Only polypeptides that showed a difference in frequency of greater than 0.4 or a difference in amplitude of greater than 2 between the compared groups were predefined. The predefined set of polypeptides was further validated by randomly excluding 30% of available samples. This kind of bootstrapping procedure was repeated up to 10 times to use only markers that are of high statistical significance. Discriminatory polypeptides were included to a SVM–derived prediction model using MosaiquesCluster software.24
Sequencing of polypeptides
Candidate biomarkers were subsequently sequenced using liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis as recently described in detail (Q-TOF).25 Briefly, a complete CE run was spotted onto the Matrix-assisted laser desorption ionization (MALDI) target (1 spot every 15 s) with the matrix solution (5 mg/mL sinapinic acid in 50% acetonitrile and 0.1% TFA) added as sheath liquid at 4 μL/min.
Results
Sample collection and sample preparation for CE-MS
Urine samples from 141 patients undergoing allo-HSCT for hematologic malignancies (n = 132) or hematologic failure syndromes (n = 9) were collected prospectively and blinded at 5 different transplant centers. The clinical details concerning primary disease, type of transplantation, conditioning regimen, stem cell source, onset of GvHD, and grading of aGvHD are summarized in “Patients” and shown in Table S1. Early in this and other studies it became evident that larger molecules present in urine, such as albumin, transferrin, and others, significantly influence the reproducibility of the data generated with CE-MS.19 These molecules also influence the diagnosis of aGvHD according to the proteomics pattern published earlier.21 Early after HSCT, large molecules are excreted in varying amounts into urine, possibly due to the often reversible impairment of renal function, induced by chemotherapy and the conditioning regimen. This led to a loss of reproducibility in the intrapatient CE-MS measurements. Therefore, a modification of the sample preparation protocol excluding larger, more abundant molecules from urine became necessary. The modified preparation protocol aimed toward removal of all molecules larger than 20 kDa to further enhance the reproducibility of the generated CE-MS data and the improved sample preparation protocol was applied to the preparation of all samples analyzed in this study.
In addition to the training set of aGvHD patients, control urine samples from healthy volunteers and from patients with chronic renal disease were included as additional control groups for the evaluation (Table 1). Diagnosis of acute GvHD was based on histopathologic examination of tissue biopsies and grading of aGvHD was based on the grading system of the 1994 consensus conference on acute GvHD grading.26
Generation of aGvHD-specific proteomic patterns and models after allo-HSCT
The aGvHD-specific biomarker definition was performed by comparing the data sets obtained with the optimized preparation protocol of 13 urine samples from patients with GvHD to those of 50 urine samples from patients after allo-HSCT without any evidence for GvHD (Table 1). In the first step, a pattern consisting of the polypeptides defined in our pilot study18 was rebuilt and, after model optimization, was able to distinguish the 63 samples of the training set with sensitivity and specificity of 77% and 100%, respectively. This result demonstrates the reproducibility of the marker definition process performed in the pilot study. However, most likely due to the optimized preparation protocol, the obtained sensitivity of 77% in the training set was worthy of improvement. We sought to enhance the obtained sensitivity by a more precise definition of HSCT patients with aGvHD grade II: the inclusion of patients with renal failure and/or chronic renal disease in the control set.
Initially, a list of polypeptides differentially excreted in the case and control groups was obtained, including all 63 available data sets of the aGvHD and HSCT control samples. In this first step, only polypeptides present with a difference in frequency of greater than 0.4 or a difference in signal intensity (= amplitude) of more than 2-fold between the 2 groups were selected. This yielded an initial set of 170 polypeptides, differentially excreted by patients with or without GvHD. The polypeptides described in our pilot study were also found in the 170 preselected peptides. Further bootstrapping of these 170 markers by randomly excluding 30% of available samples of both groups and repeated application of the selection criteria resulted in a list of 50 preselected polypeptides of higher consistency. In an effort to exclude peptides relevant for renal insufficiency, 71 samples of patients with chronic renal disease were included into the training set analyses. Comparison of the data sets of the 13 aGvHD urine samples with 71 samples from patients with chronic renal diseases and healthy controls (Table 1) led to further elimination of 19 polypeptides not exclusively specific for aGvHD. Thus, 31 potentially relevant peptides (Table S3) could be combined to an aGvHD-specific proteomic pattern (Figure 1). Since the generated data require special statistical features for evaluation and SVMs have shown good performance in the evaluation of multidimensional data,17,27 an SVM-based prediction model of the 31 polypeptides (Table S3) was established by using the MosaiquesCluster software.24 The result of SVM classification is a dimensionless number representing the Euclidian distance of data points to the separating hyperplane. In our case, the proteomic data sets of patients are represented in the n dimensions of the aGvHD biomarker space.
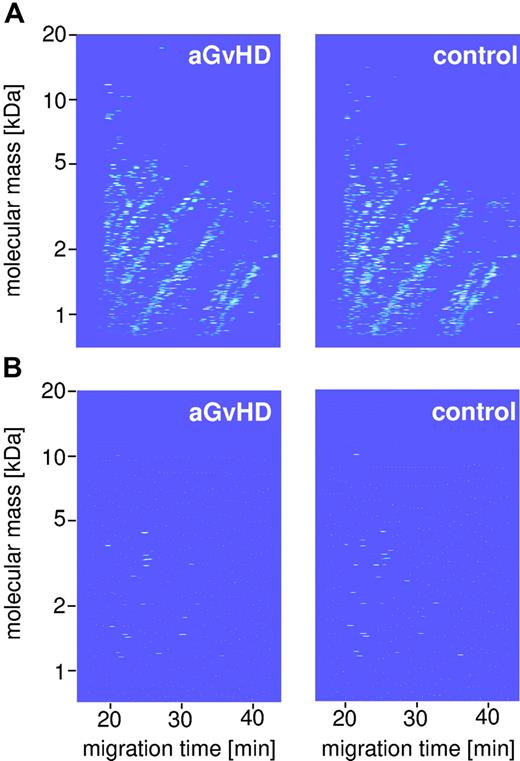
Polypeptide pattern distinguishing HSCT patients with aGvHD from HSCT controls. This figure shows the compiled data sets of 13 GvHD samples (A, top left panel) and 50 HSCT control (top right panel) of the training set (Table 1). Normalized molecular weight is plotted against normalized migration time. The mean signal intensity is color coded. (B) The bottom panel depicts the 31 indicative polypeptides defining the specific pattern for GvHD (bottom left panel) and controls (bottom right panel).
Polypeptide pattern distinguishing HSCT patients with aGvHD from HSCT controls. This figure shows the compiled data sets of 13 GvHD samples (A, top left panel) and 50 HSCT control (top right panel) of the training set (Table 1). Normalized molecular weight is plotted against normalized migration time. The mean signal intensity is color coded. (B) The bottom panel depicts the 31 indicative polypeptides defining the specific pattern for GvHD (bottom left panel) and controls (bottom right panel).
The model obtained after application of SVM to the data distinguished samples at the time of aGvHD from control samples upon complete cross-validation with a sensitivity of 100.0% (95% CI 75.1 to 100.0) and a specificity of 98.0% (95% CI 89.3 to 99.7; Table 2; Figure 2A).
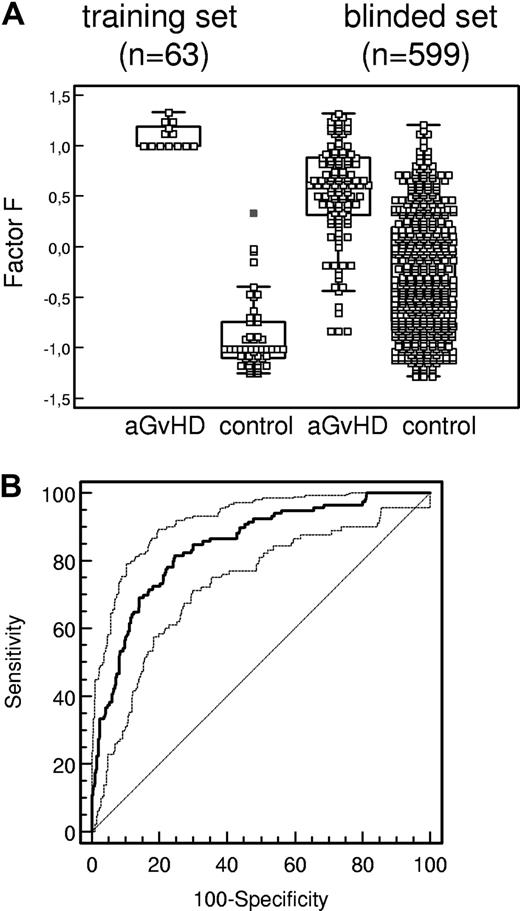
Sensitivity and specificity of aGvHD pattern. (A) Box-and-whisker plots of classification factor F obtained for classification of each individual HSCT sample (GvHD indicates graft-versus-host disease diagnosed according to Przepiorka et al26 ; control, HSCT sample showing no evidence of GvHD) in the training set and the blinded set (Table 1). The boxes depict the quartiles Q1 and Q3 of each distribution, and the statistical medians are shown as horizontal lines in the boxes. The whiskers indicate 3/2 times the interquartile range of Q1 and Q3. (B) ROC curve (bold line, AUC = 0.85) of the proteomics pattern diagnosis. Using the GvHD-specific polypeptide panel from Table 3, classification factor F is used as variable in ROC analysis in the 599 samples of the blinded set (Table 1). Ninety-five percent confidence intervals (95% CI) are indicated by thin lines.
Sensitivity and specificity of aGvHD pattern. (A) Box-and-whisker plots of classification factor F obtained for classification of each individual HSCT sample (GvHD indicates graft-versus-host disease diagnosed according to Przepiorka et al26 ; control, HSCT sample showing no evidence of GvHD) in the training set and the blinded set (Table 1). The boxes depict the quartiles Q1 and Q3 of each distribution, and the statistical medians are shown as horizontal lines in the boxes. The whiskers indicate 3/2 times the interquartile range of Q1 and Q3. (B) ROC curve (bold line, AUC = 0.85) of the proteomics pattern diagnosis. Using the GvHD-specific polypeptide panel from Table 3, classification factor F is used as variable in ROC analysis in the 599 samples of the blinded set (Table 1). Ninety-five percent confidence intervals (95% CI) are indicated by thin lines.
Validation of the aGvHD-specific pattern and model
Subsequently, the established pattern was validated in a blinded assessment of 663 prospectively collected urine samples from 141 patients undergoing allo-HSCT at 5 different transplant centers (Table 1). Of the 663 samples, 252 scored positive and 411 scored negative for the aGvHD-specific pattern.
After unblinding of the samples, a significant clustering of false-positive classification was observed around day 0 (Table S2, spreadsheet 2), a time point not covered by the training set. This observation may reflect the cytokine release during and after conditioning and/or an initial allogeneic response at the time of transplantation of the allogeneic cells. Since diagnosis of GvHD before day +2 is very unlikely, 64 samples of the 663 samples of the blinded study collected around day 0 were excluded retrospectively from data analysis (Table S2, spreadsheet 2). Of the remaining 599 samples, 99 of 119 aGvHD samples were scored correctly (sensitivity of 83.1% [95% CI 73.1 to 87.9]) by the aGvHD-specific pattern and 363 of 480 control samples were classified correctly (specificity of 75.6% [95% CI 71.6 to 79.4]; Table 2; Figure 2A-B). Analyzing the clinical history of incorrectly positive-classified samples revealed bacterial infections in combination with fever higher than 38°C as the major reason for false-positive results. In contrast, no evidence was found for an interference of viral infections with the aGvHD-specific pattern; importantly, reactivation of cytomegalovirus and other herpes viruses do not seem to interfere at significant levels with the aGvHD-specific pattern (Table 3; supplementary material). Forty-three samples were obtained at times of viral reactivation: 23 were taken at cytomegalovirus (CMV) reactivations, whereas 20 were collected during episodes of Epstein-Barr virus (EBV) reactivation. Six samples scored positive concerning classification factor F in the CMV-reactivation group, of those 4 were false-positive since there was no aGvHD ongoing (< 20%). Of samples collected from patients with reactivation of EBV, 5 were positive for the classification factor and only 1 sample was positive 20 days prior to the clinical diagnosis of aGvHD. Patient 3049 had aGvHD grade IV of the intestine, which persisted almost 9 months (Table 3; supplementary material).
In this study, 73 patients developed aGvHD and 39 had aGvHD grade II or more. The aGvHD-specific protein pattern preceded the development of clinical symptoms (median, 7 days; range, 1 to 13 days, prior to clinical symptoms).
To investigate if proteomic classification is influenced by age, sex, conditioning regimen and/or reduced conditioning, or ATG (or other antibodies) in the conditioning regimens, a multivariate analysis was performed (Table 4). The positivity of classification factor F was significant for the development of aGvHD (P < .001), whereas within this group of patients no other factors showed any influence on aGvHD.
Correlation of classification factor F to the clinical features of patients
Correlation of clinical data with SVM-based classification and a subsequent analysis of the classification time courses after HSCT were performed. Selected individual patients are shown to depict the performance of the GvHD pattern in a more detailed fashion. Figure 3 shows the correlation of the aGvHD-specific scores to the clinical follow-up of patients who show typical classification scores for varying clinical features.
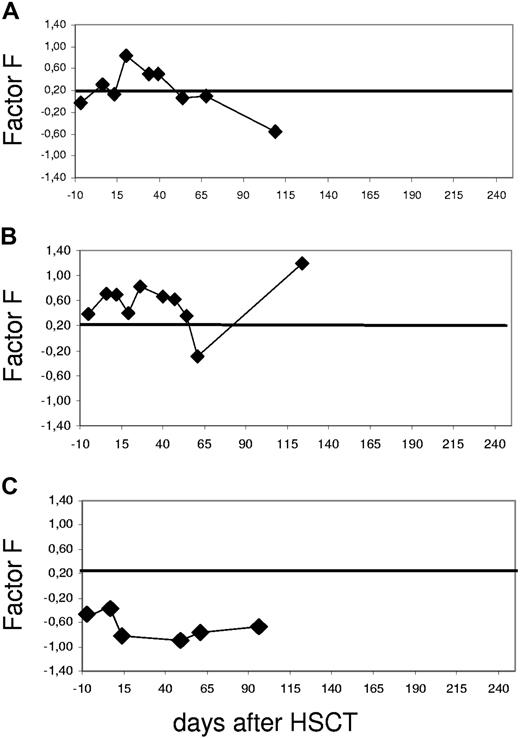
Examples of time courses of the proteomics diagnosis based on the aGvHD pattern. The SVM-based classification factor F (FGvHD) is plotted against the days before and after HSCT starting prior to conditioning treatment (day −10). The threshold was chosen at classification factor FGvHD = 0.2, as indicated by a line; values above indicate GvHD. The resulting curve is shown for individual patients. (A) Patient 2791 was diagnosed with GvHD grade II on day +27. The classification factor score was 0.8 on day +20 after HSCT, 7 days prior to clinical diagnosis, and returned to inconspicuous values at day +41, reflecting response to treatment. (B) Patient 2725 was diagnosed with GvHD grade III of the skin on day +25 after HSCT. Classification factor F rose to 0.5, thus indicating aGvHD. The initial response to steroid treatment led to the normalization of the factor value on day +55, with no change due to a reactivation of CMV at about day +60 after HSCT. The next rise of the classification factor was concerted with cGvHD (BOOP). (C) Patient 3264 did not develop aGvHD, thus representing an example of a negative control. The values remain around baseline well below the aGvHD cutoff number of 0.2 throughout the observation period.
Examples of time courses of the proteomics diagnosis based on the aGvHD pattern. The SVM-based classification factor F (FGvHD) is plotted against the days before and after HSCT starting prior to conditioning treatment (day −10). The threshold was chosen at classification factor FGvHD = 0.2, as indicated by a line; values above indicate GvHD. The resulting curve is shown for individual patients. (A) Patient 2791 was diagnosed with GvHD grade II on day +27. The classification factor score was 0.8 on day +20 after HSCT, 7 days prior to clinical diagnosis, and returned to inconspicuous values at day +41, reflecting response to treatment. (B) Patient 2725 was diagnosed with GvHD grade III of the skin on day +25 after HSCT. Classification factor F rose to 0.5, thus indicating aGvHD. The initial response to steroid treatment led to the normalization of the factor value on day +55, with no change due to a reactivation of CMV at about day +60 after HSCT. The next rise of the classification factor was concerted with cGvHD (BOOP). (C) Patient 3264 did not develop aGvHD, thus representing an example of a negative control. The values remain around baseline well below the aGvHD cutoff number of 0.2 throughout the observation period.
Patient 2791 (Figure 3A) is an example of a patient who developed aGvHD and responded to therapy. The patient was diagnosed with GvHD grade II of the skin on day +27 after HSCT from a MUD. The aGvHD SVM score (classification factor) rose to a value of 0.8 on day +20 after HSCT, 7 days prior to the clinical diagnosis. Upon response to therapy, classification factor F returned to inconspicuous values at day +41. At this time, all symptoms of GvHD had subsided. This illustrates that this approach allows not only early diagnosis of aGVHD but also monitoring of response to therapy of the disease.
The second patient, 2725 (Figure 3B), was chosen as an example for initial response to therapy and recurrence of GvHD at day +75 after HSCT. The patient was diagnosed with aGvHD grade III of the skin on day +25 after HSCT. At that time, the SVM-based classification factor F was 0.5, indicating aGvHD. The initial response to treatment led to normalization of the proteome pattern on day +55. Reactivation of CMV was diagnosed at about day +60 after HSCT. This reactivation did not influence the aGvHD pattern, as the classification factor remained negative, suggesting only minor interference of proteomic aGvHD recognition with CMV reactivation. On day +85 after HSCT, this patient developed severe complications involving the lung, suggesting chronic GvHD. The diagnosis was bronchiolitis obliterans organizing pneumonia (BOOP). This event went hand in hand with an increase of classification factor F. In this case, the inflammatory background underlying the cGvHD development was most likely similar to that of aGvHD recognized by the proteomic pattern. Of 21 patients developing cGvHD after HSCT in this cohort, only samples of 3 additional patients scored positive with the aGvHD-specific pattern. In all of these cases, the patients had GvHD of the intestine: 1 had progressive aGvHD since day +25 after HSCT, and the other 2 patients developed intestinal GvHD after day +100.
Patient 3264 (Figure 3C) did not develop aGvHD after HSCT and represents an example of a negative control. Throughout the observation period the classification factor remained inconspicuous.
Sequencing of biomarkers of interest
In order to identify the biomarkers that constitute the aGvHD-specific pattern, tandem mass spectrometry (MS/MS) was performed. The MS/MS analyses were done with a MALDI-TOF–TOF-MS (Ultraflex, Bruker Daltonik, Bremen, Germany) as described previously.18,25 Briefly, the complete CE-MS run was spotted onto target plates and the target was subsequently examined in MS mode for the polypeptides of interest, based on the data from the CE-MS analyses. Polypeptides of interest were sequenced in MS/MS mode, without the use of collision-induced dissociation (CID) gas. To date, 3 of the 31 peptides from the GvHD pattern gave interpretable sequence data (Table 5; Figure 4). All obtained sequences are fragments from collagen α-1 (I) and α-1 (III). Chain (I) was found to be down-regulated and chain (III) up-regulated in the aGvHD cases investigated.
The high-resolution MS/MS spectrum (bottom panel) of the polypeptide indicative for GvHD (Table 3) with the molecular weight of 4.306 kDa. This polypeptide was up-regulated in GvHD samples compared with control samples (Table 1). SwissProt database matching indicated that this is a fragment of collagen α-1 (III) chain (319-366; Homo sapiens) with a calculated mass of 4.306 kDa and a sequence as indicated (top panel). The masses of b-ion fragments (red) and y-ion fragments (blue) are correlated with the obtained sequence using fragment numbers.28,29
The high-resolution MS/MS spectrum (bottom panel) of the polypeptide indicative for GvHD (Table 3) with the molecular weight of 4.306 kDa. This polypeptide was up-regulated in GvHD samples compared with control samples (Table 1). SwissProt database matching indicated that this is a fragment of collagen α-1 (III) chain (319-366; Homo sapiens) with a calculated mass of 4.306 kDa and a sequence as indicated (top panel). The masses of b-ion fragments (red) and y-ion fragments (blue) are correlated with the obtained sequence using fragment numbers.28,29
Discussion
Our data clearly indicate the value of the CE-MS–based urinary proteome analysis for the early detection of aGvHD after HSCT in a laboratory-based, unbiased fashion. While proteomic screening of body fluids is becoming increasingly important in clinical research,30,31 application to the diagnosis of complications after stem cell transplantation in a prospective study does not currently exist. Here we describe the blinded evaluation of an aGvHD-specific proteome pattern on 599 samples collected prospectively and blinded in 5 different centers. Compared with our pilot study,18 the pattern of discriminatory peptides was changed due to alteration in the sample preparation, removing all proteins with a molecular weight greater than 20 kDa. This modification of the sample preparation was necessary to achieve high reproducibility of the data, a prerequisite when aiming for comparison of different samples, especially when obtained at different institutions. Ideally, crude, unprocessed samples should be analyzed, in order to minimize artificial losses or biases arising from sample preparation. Since all body fluids contain a large amount of different ions, lipids, carbohydrates, and other confounding material, these samples cannot be analyzed in the native form by mass spectrometry but have to be processed prior to screening. In our study, we found that the abundance of large molecules led to the diminished detection of smaller molecules, which may be even more relevant for diagnostic purposes. Fragmentation of the large molecules during storage and sample preparation leads to additional problems. Enlarging the patient population studied will also influence the discriminatory value of single peptides within the aGvHD-specific proteomic pattern, which contains a smaller amount of discriminatory peptides. This is not due to a limited degree of reliability of the screening method but rather displays the complexity of complications as well as their depiction in the excretion of peptides. The CE-MS screening and the subsequent evaluation of the data with the appropriate software solutions was applied successfully to the analyses of the blinded samples, resulting in a sensitivity of 83.1% (95% CI 73.1 to 87.9) and a specificity of 75.6% (95% CI 71.6 to 79.4). The high sensitivity and specificity of the proteome-based approach are very promising, especially since the blinded evaluation set was approximately 10 times larger then the training set. This also underlines the stability of the classification results. Sequencing of the marker molecules is possible but not particularly necessary for the diagnosis of diseases. The identification of the defined biomarkers presents some unique challenges, since we aim for sequencing of native undigested peptides. The sequence analysis has to be performed from a complex mixture and potential biomarkers are frequently prototypically processed and/or posttranslationally modified. Potential biomarkers detected by CE-MS are likely to be small fragments of larger proteins. Thus, to identify a 2- to 10-kDa (modified) portion of a protein with a possible molecular weight greater than 60 kDa requires extensive top-down peptide sequence analysis. Such an approach is more demanding than, eg, MudPIT approaches on tryptic digests (bottom-up approaches), where the parent ion mass of the tryptic peptide already serves as 1 good parameter for identification. The bottom-up approaches are able to provide theoretical parent protein masses in body fluids.23 Unfortunately, modifications that are generally observed (eg, oxidation, proteolytic processing, or glycosylation) prevent the direct correlation of such data to defined biomarkers.
The sequence information obtained was from collagen fragments from collagen α-1 (I) and α-1 (III). Chain (I) was down-regulated in aGvHD samples, whereas chain (III) was up-regulated in the aGvHD cases investigated. In concordance with these findings, Pihusch et al32 recently reported elevated serum procollagen (III) peptide (PNPIII) levels in hepatic GvHD. Increased bilirubin levels during HSCT, caused by toxic drugs, may be 1 reason for the elevation of PNPIII levels, as PNPIII is processed in the liver.33 In addition, a tubular damage by immunosuppressive drugs such as cyclosporine, also results in an increase of PNPIII, which is normally secreted via the kidneys.34 On the other hand, sepsis and microangiopathic hemolytic anemia, which affect renal and hepatic clearance function, can also lead to high serum levels of PNPIII.32
After unblinding of the study, it became evident that the false-positive classification rate was high in samples collected around day 0. Presumably, the inflammatory environment generated by the conditioning treatment is depicted. This environment may be similar to that of GvHD.35 Multivariate analysis applied on the data revealed that only classification factor F played a significant role for the diagnosis of GvHD (P < .001), whereas neither the conditioning treatment nor the application of antibodies was significant. While CMV or Epstein-Barr virus infections or reactivation did not interfere with the proteomic pattern classification (Table 3; Figure 3B), infections with fever above 38°C induced by bacterial or fungal infections required special attention. The ability to monitor response to therapy is shown by plotting classification factor F against the days after HSCT and correlating it with the clinical data of the patients. Response to therapy can be seen (Figure 3A) as a drop of classification factor F value to below 0.2, which was chosen as the cutoff for diagnosis of GvHD in this setting. This is currently further evaluated for the early recognition of patients with steroid-resistant aGvHD, who have a particularly poor prognosis.36 The early diagnosis of aGvHD-predicting changes within body fluids of patients will probably allow preemptive treatment of patients at risk for developing aGvHD. Initiating preemptive therapy according to the proteomics data even before severe clinical symptoms become evident may help to reduce the severity and probably the incidence of severe aGvHD.
To date, our study is the first to correlate proteomic data with the clinical diagnosis of aGvHD in a blinded, prospectively collected, multicenter approach. There are reports in the literature employing proteomic techniques to the screening of plasma, using an intact-protein–based quantitative analysis combined with protein tagging and immunodepletion of abundant proteins to quantitatively profile the plasma proteome in patients with acute GvHD after HSCT.30 In this paper, a large number of differentially expressed candidate peptides and proteins were described that could be indicative for aGvHD development. However, the data were generated on pooled samples of patients with or without GvHD, respectively, and sequencing was done after tryptic digestion of the peptides of interest. In addition, the relevance of the molecules for diagnosis of aGvHD by evaluating prospectively collected samples remains to be shown. Another paper describes the application of surface enhanced laser disorption ionization (SELDI) to serum collected from patients with and without GvHD after allo-HSCT.31 Although the data appear encouraging, both the technology used and the stability of the body fluid chosen are currently heavily debated.37–40 Unfortunately, a prospective or blinded evaluation of patient samples, which would have enabled validation of the data, is missing in this report.
The study presented here is the first to describe the application of proteomics toward the diagnosis of aGvHD on prospectively collected and blinded patient samples. The noninvasive character of the method together with a short data-evaluation time allows the time-near analysis of multiple patient samples and fast therapeutic intervention according to the obtained results. Such a fundamental change in aGvHD diagnosis might be of inestimable value for clinicians in daily routine allo-HSCT therapy.
A future revision of the aGvHD-specific pattern using the broad database of the classified blinded samples may help to further refine the current pattern, resulting in further improvement of sensitivity and specificity of the proteomic-based diagnosis of aGvHD. Our results demonstrate that aGvHD can be predicted with high sensitivity and specificity using urinary proteome analysis, even in a multicenter-screening approach, thus excluding center-specific biases of the method.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Annika Krons and Meike Hillmann for excellent technical assistance, Elke Dammann for help with the documentation of complications after bone marrow or stem cell transplantation, Julia Kontsendorn and Mohamed Dakna for help with the multivariate analysis, and Drs Michael Morgan and Harald Mischak for critically reading the manuscript.
This work was sponsored in part by a grant provided by the “Deutsche Jose Carreras Leukämie Stiftung” DJCLS R05/08 (E.M.W. and B.H.).
Authorship
Contribution: E.M.W designed the research, analyzed data, and wrote the paper. E.S. performed research and analyzed data. B.H. designed the research and analyzed data. J.L.F. collected samples, performed research, and analyzed data. E.H. performed research, collected samples, and analyzed data. M.S. collected samples and analyzed data. H.-J.K. collected samples, performed research, and analyzed data. A.Z. collected samples, performed research, and analyzed data. P.Z. performed research and analyzed data. M.K. performed research and analyzed data. A.G. performed study design and analyses of data.
Conflict-of-interest disclosure: Two of the authors (E.S. and P.Z.) are employed by Mosaiques-Diagnostics GmbH whose (potential) product was studied in the present work. E.M.W. is married to the co-owner and founder of Mosaiques Diagnostics.
E.M.W. and E.S. contributed equally to this study.
Correspondence: Eva M. Weissinger, Hannover Medical School, Department of Hematology, Hemostasis and Oncology, Carl-Neuberg-Str 1, 30625 Hannover, Germany; e-mail: mischak-weissinger.eva@mh-hannover.de.