Abstract
In contrast to lentiviral infections of humans and macaques, simian immunodeficiency virus (SIV) infection of natural hosts is nonpathogenic despite high levels of viral replication. However, the mechanisms underlying this absence of disease are unknown. Here we report that natural hosts for SIV infection express remarkably low levels of CCR5 on CD4+ T cells isolated from blood, lymph nodes, and mucosal tissues. Given that this immunologic feature is found in 5 different species of natural SIV hosts (sooty mangabeys, African green monkeys, mandrills, sun-tailed monkeys, and chimpanzees) but is absent in 5 nonnatural/recent hosts (humans, rhesus, pigtail, cynomolgus macaques, and baboons), it may represent a key feature of the coevolution between the virus and its natural hosts that led to a nonpathogenic infection. Beneficial effects of low CCR5 expression on CD4+ T cells may include the reduction of target cells for viral replication and a decreased homing of activated CD4+ T cells to inflamed tissue.
Introduction
It is now clear that human immunodeficiency virus type 1 (HIV-1) evolved from a closely related virus of chimpanzees (Pan troglodytes) named simian immunodeficiency virus (SIVcpz).1-3 Similarly, HIV-2 originated from a related virus, SIVsmm, which naturally infects sooty mangabeys (SMs).4-6 In marked contrast to HIV infection, which almost invariably progresses to AIDS unless treated with antiretrovirals, natural SIVcpz infection of chimpanzees and SIVsmm infection in SMs rarely result in clinical symptoms.7-12 Similarly, other natural hosts for SIV, such as African green monkeys (AGMs), mandrills, and several other African nonhuman primate (NHP) species, generally live normal lifespans despite many years of infection with a highly replicating virus.13-23
Importantly, the inoculation of SIV, which naturally infects African monkeys, into NHPs that are not natural hosts for SIV, such as macaques and baboons, may result in chronic infection that eventually progresses to a disease closely resembling AIDS.16,24-28 Given that the earliest documented exposure of a human to HIV-1 infection was in 1959,29 it is thought that the introduction of primate lentiviruses into a new host species results in AIDS because of the inability of a naive immune system to cope with a virus recently transmitted from another species. Although it is agreed that the rarity of disease likely reflects a pacific coevolution between SIV and its natural hosts resulting from many thousands of years of endemic infection, the exact mechanisms underlying this absence of disease are still poorly understood. It should be noted, however, that the preservation of the immune system in natural SIV hosts does not simply result from immune control of viral replication, a task obviously not achieved in these animals given their high level of viral load (VL).7,8,12,14,19-23 In addition, the differences in virulence between the SIVs naturally infecting African NHP hosts and the pathogenic HIV/SIV strains are not related to differences in virus biology. As do most HIV/SIVmac/SIVsmm strains, SIVs from African species use CD4 as a receptor30 and CCR5 as a major coreceptor for entry,31-33 suggesting that their target cells are CD4+CCR5+ T cells. Instead, increasing experimental evidence suggests that lower levels of immune activation may protect natural hosts from SIV disease progression.8,12,13,18,20,23,34 Ultimately, however, it is unknown to what extent the difference in the outcome of HIV/SIV infection between natural and nonnatural/recent hosts reflects differences in the viruses or in the immunology of the hosts themselves. Intensive research is under way in nonprogressing natural host species to determine why these animals resist the development of AIDS.
Numerous laboratories have now confirmed that SIV and HIV both selectively infect and destroy a highly specialized type of T-helper cells known as memory CD4+ T cells.35-40 Memory CD4+ T cells are necessary for stimulating cell-mediated and humoral immune responses. Selective loss of and failure to regenerate adequate numbers of these cells result in the severe immunodeficiency that follows HIV infection in humans and SIV infection of rhesus macaques (RMs).35-40 In humans and RMs, a subset of memory CD4+ T cells known to express high levels of CCR5 and CCR5 has been used as a surrogate marker to identify “memory” CD4+ T cells.37 During HIV infection of humans and SIV infection of RMs, memory CD4+ T cells are targeted because of their high expression of CCR5 (the major coreceptor for HIV and SIV) and their high levels of activation because activated cells support higher levels of lytic viral replication than resting cells.41-43 In healthy, uninfected humans and RMs, large numbers of CD4+CCR5+ memory T cells reside in the intestinal tract and other mucosal tissues.44 Several studies have now suggested that the massive and, to a large extent, irreversible depletion of these memory CD4+CCR5+ T cells during the early stages of pathogenic HIV and SIV infection is a common feature of lentiviral infections of humans and nonhuman primates.35-40 However, no comprehensive study has yet been performed focusing on CD4+CCR5+ T cells in various lymphoid compartments of nonprogressing host species.
Here we report the results of a systematic comparative analysis of CD4+CCR5+ T cells in several tissues obtained from progressing (human, RM, pigtail macaque [PTM], and baboon) and nonprogressing (SM, AGM, mandrill, sun-tailed monkey, and chimpanzee) primate host species. We demonstrate that, in contrast to progressing primate hosts, natural, nonprogressing hosts for SIV infection have markedly reduced numbers of CD4+CCR5+ T cells in peripheral blood, bone marrow, and lymph nodes (LNs). Importantly, we were able to sample mucosal tissues in nonnatural (RM and baboon) and natural (SM and AGM) hosts. We observed that natural hosts for SIV infection, both infected and uninfected, consistently show a paucity of CD4+CCR5+ T cells in all sampled tissues, including those mucosal tissues that represent the major site of viral replication and CD4+ T-cell depletion during pathogenic HIV/SIV infection. The strikingly low levels of CD4+CCR5+ T cells in nonprogressing natural hosts suggests that coevolution of these animals with SIV over millennia has resulted in the selection of individuals that no longer depend on CD4+CCR5+ T cells to maintain critical immune functions. Therefore, evolutionary pressure from the virus might have changed fundamental aspects of the immune system in these species that resulted in the ability to resist the development of AIDS despite persistent, high-level viremia in these infected hosts.
Materials and methods
Primates and specimens
Blood, LNs, intestinal biopsy specimens, and broncho-alveolar lavage (BAL) specimens were examined from healthy adult NHPs that are natural hosts of SIV: AGMs (Chlorocebus sabaeus), originating from St Kitts Island (n = 21) and Africa (n = 4), SMs (Cercocebus atys) (n = 30), mandrills (Mandrillus sphinx) (n = 25), sun-tailed monkeys (Cercopithecus solatus) (n = 2), and chimpanzees (Pan troglodytes) (n = 3). Samples from 5 nonnatural/recent hosts in which SIV infection is pathogenic were also included: rhesus macaques (Macaca mulatta) (n = 24), pigtail macaques (Macaca nemestrina) (n = 37), cynomolgus monkey (Macaca fascicularis) (n = 1), and baboons (Papio papio) (n = 2). Eleven healthy HIV-uninfected human volunteers were also tested. All SIV-uninfected animals were confirmed to be free of SIV at the time of examination by reverse transcription–polymerase chain reaction (RT-PCR) and Western blot analysis for anti–SIV antibodies (SIVsm strips, Zeptometrix, Buffalo, NY). The animals were housed at the Tulane National Primate Research Center (TNPRC), the Yerkes National Primate Research Center (YNPRC) (both AAALAC International accredited facilities), or the Centre Internationale de Recherches Medicales in Franceville (CIRMF; Gabon). Housing and handling of animals were performed in accordance with the Guide for the Care and Use of Laboratory Animals (US Public Health Service) and the Animal Welfare Act. All protocols and procedures for the animal studies were reviewed and approved by the Tulane University and Emory University Institutional Animal Care and Use Committee (IACUC), and all protocols and procedures for the human studies were approved by the Emory University Institutional Review Board (IRB). Informed consent was obtained in accordance with the Declaration of Helsinki.
Tissue collection
Blood (5 mL EDTA anticoagulated) was collected from the femoral vein and processed within 1 hour of collection. For LN biopsy, the skin over the inguinal or axillary LN was prepared for aseptic surgery. A small skin incision was made over the node, and blunt dissection was used to isolate and remove the LN. The subcutis and skin were then closed with absorbable sutures. Either ketamine (10 mg/kg) or tiletamine/zolazepam (Telazol; 4 mg/kg) was used for anesthesia, with frequency of administration determined by the veterinarian performing the procedure so as to maximize animal safety and comfort. For BAL, a fiberoptic bronchoscope was manipulated into the trachea after local anesthetic (2% lidocaine) was applied to the larynx. The scope was directed into the right primary bronchus and wedged into a distal subsegmental bronchus (right caudal lung lobe) that approximates the diameter of the bronchoscope. Saline was collected by aspiration between each individual lavage before a new aliquot was instilled. After collection was complete, the animal was maintained on oxygen by face mask for 5 minutes before return to its cage. Endoscope-guided pinch biopsy samples from the proximal jejunum and the rectum were collected as previously described.44
Isolation of lymphocytes
Lymphocytes from the intestine and LNs were isolated and stained for flow cytometry as previously described.44 Briefly, lymphocytes were isolated from intestinal biopsy samples using EDTA followed by collagenase digestion and Percoll density-gradient centrifugation.44 Lymphocytes were isolated from the axillary LNs by gentle mincing and pressing of tissues through nylon mesh screens.
Flow cytometry
Mononuclear cells derived from intestinal biopsy samples, BAL sample, and LNs were stained for flow cytometric analysis using 4-color staining combinations with the monoclonal antibodies CD3-fluorescein isothiocyanate (FITC), CCR5-phycoerythrin (PE), CD8-peridinin chlorophyll A protein (PerCP), and CD4-allophycocyanin (APC; BD Biosciences, PharMingen, San Diego, CA). Cells were incubated with an excess amount of monoclonal antibodies at 4°C for 30 minutes, followed by a phosphate-buffered saline (PBS) wash (400g, 7 minutes) and fixation in 2% paraformaldehyde. Whole blood was stained according to a whole blood lysis technique previously described.21 CCR5 expression on monocytes was determined by using a combination of the following 4 antibodies: CD14-FITC, CCR5-PE, CD8-PerCP, and CD4-APC. Data were acquired with a FACSCalibur flow cytometer (BD Biosciences Immunocytometry Systems) and analyzed with CellQuest software (BD Bioscience Immunocytometry Systems) or FlowJo software (Treestar, Ashland, OR). CD4+ and CD8+ T-cell percentages were obtained first by gating on CD3+ T cells. CCR5 expression on T-cell subsets was determined by gating on lymphocytes and then on total CD4+ or CD8+ T cells. CCR5 expression on monocytes was determined by gating first on monocytes and then on CD14+ cells or by gating on monocytes alone. Appropriate negative controls (unstained cells and fluorochrome isotype controls) and positive controls (staining with only one antibody-fluorochrome at a time) were used for determining gates during data analyses.
Statistical analyses
Percentages of cells were compared between groups using one-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test for multiple groups or the Mann-Whitney U test for comparison between 2 groups. Data were considered significant when P was below .05.
Results
Low levels of CCR5 expression on circulating and LN-derived CD4+ T cells of natural SIV hosts
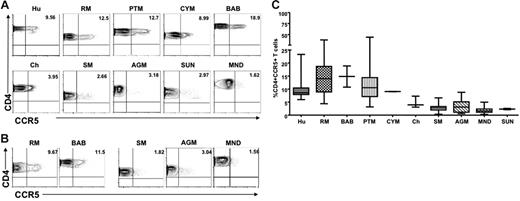
We first examined CCR5 expression levels on CD4+ T cells derived from the blood of healthy, uninfected individuals of 10 different primate species: 5 natural host species (AGMs, SMs, mandrills, sun-tailed monkeys, and chimpanzees) and 5 nonnatural/recent hosts in which HIV/SIV infection is pathogenic (humans, RMs, PTMs, cynomolgus macaques, and baboons). Figure 1A shows representative contour plots indicating the lower levels of CD4+CCR5+ cells in nonpathogenic hosts compared with pathogenic hosts in peripheral blood, and Figure 1B shows the levels of CD4+CCR5+ in LNs from 3 nonpathogenic (SM, AGM, and mandrill) and 2 pathogenic (RM and baboon) host species. A summary of the available data relative to CCR5 expression in CD4+ T cells is shown in Figure 1C. In all, these data indicate that circulating LN-derived CD4+ T cells from natural SIV hosts express lower levels of CCR5 than human and nonnatural SIV hosts.
Reduced CCR5 expression on peripheral blood– and LN-derived CD4+ T cells in natural hosts of SIV. (A) Representative contour plots showing CD4 and CCR5 staining of peripheral blood–derived T cells of healthy uninfected individuals belonging to 5 nonnatural/recent hosts of HIV/SIV—humans (Hu), rhesus macaques (RM), pigtail macaques (PTM), cynomolgus macaques (CYM), and baboons (Bab)—and 5 natural hosts of SIV—sooty mangabeys (SM), Caribbean African green monkeys (AGM), mandrills (MND), sun-tailed monkeys (SUN), and chimpanzees (Cy). (B) Representative contour plots showing CD4 and CCR5 staining of T cells isolated from the LNs of RMs, baboons, SM, AGM, and mandrills. (C) Summary of CCR5 expression on blood-derived CD4+ T cells of the primate species listed in panel A. Statistically significant differences in CCR5 expression on CD4+ T cells from natural and nonnatural hosts of SIV was determined by one-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test. For nonnatural human hosts of SIV versus natural AGM hosts of SIV, P < .05; for all other nonnatural hosts versus natural hosts, P < .001.
Reduced CCR5 expression on peripheral blood– and LN-derived CD4+ T cells in natural hosts of SIV. (A) Representative contour plots showing CD4 and CCR5 staining of peripheral blood–derived T cells of healthy uninfected individuals belonging to 5 nonnatural/recent hosts of HIV/SIV—humans (Hu), rhesus macaques (RM), pigtail macaques (PTM), cynomolgus macaques (CYM), and baboons (Bab)—and 5 natural hosts of SIV—sooty mangabeys (SM), Caribbean African green monkeys (AGM), mandrills (MND), sun-tailed monkeys (SUN), and chimpanzees (Cy). (B) Representative contour plots showing CD4 and CCR5 staining of T cells isolated from the LNs of RMs, baboons, SM, AGM, and mandrills. (C) Summary of CCR5 expression on blood-derived CD4+ T cells of the primate species listed in panel A. Statistically significant differences in CCR5 expression on CD4+ T cells from natural and nonnatural hosts of SIV was determined by one-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test. For nonnatural human hosts of SIV versus natural AGM hosts of SIV, P < .05; for all other nonnatural hosts versus natural hosts, P < .001.
CD8+ T cells from natural SIV hosts express levels of CCR5 comparable to those of nonnatural hosts
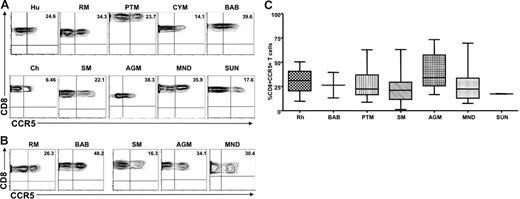
To ensure that the differences observed between natural and nonnatural hosts did not result from reduced recognition of the monoclonal antibodies used, CCR5 expression was also examined on CD8+ T cells derived from peripheral blood and LNs of the same set of primate species. We found that CD8+ T cells from the peripheral blood and LNs of natural SIV hosts expressed levels of CCR5 on CD8+ T cells similar to those observed in humans and nonnatural SIV host species; the only exception was that SMs showed a mild but significant decrease in the percentage of CD8+CCR5+ T cells. Figure 2 shows representative contour plots of CCR5 expression on peripheral blood–derived (Figure 2A) and LN-derived (Figure 2B) CD8+ T cells from the same set of primate species shown in Figure 1, and Figure 2C summarizes the available data. Taken together, these data indicate that the low levels of CCR5 expression on CD4+ T cells observed in natural SIV host species was not the result of lack of cross-recognition by a CCR5 antibody designed to detect human CCR5. In addition, these data indicate that the striking difference in the level of CCR5 expression observed between natural and nonnatural SIV hosts was specific to CD4+ T cells.
Similar levels of CCR5 are expressed on the surfaces of CD8+ T cells from natural and nonnatural hosts of SIV. (A) Representative contour plots showing CD8 and CCR5 staining of peripheral blood–derived T cells of healthy humans, RMs, PTMs, cynomolgus macaques, baboons, SMs, Caribbean AGMs, mandrills, sun-tailed monkeys, and chimpanzees. (B) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the LNs of RMs, baboons, SMs, AGMs, and mandrills. (C) Summary of CCR5 expression on peripheral blood–derived CD8+ T cells of the primate species listed in panel A. One-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test found no statistically significant differences (P > .05) in CCR5 expression on CD8+ T cells obtained from natural and nonnatural hosts of SIV. The only exception was SMs with lower levels of CD8+CCR5+ T cells than RMs (P < .05).
Similar levels of CCR5 are expressed on the surfaces of CD8+ T cells from natural and nonnatural hosts of SIV. (A) Representative contour plots showing CD8 and CCR5 staining of peripheral blood–derived T cells of healthy humans, RMs, PTMs, cynomolgus macaques, baboons, SMs, Caribbean AGMs, mandrills, sun-tailed monkeys, and chimpanzees. (B) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the LNs of RMs, baboons, SMs, AGMs, and mandrills. (C) Summary of CCR5 expression on peripheral blood–derived CD8+ T cells of the primate species listed in panel A. One-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test found no statistically significant differences (P > .05) in CCR5 expression on CD8+ T cells obtained from natural and nonnatural hosts of SIV. The only exception was SMs with lower levels of CD8+CCR5+ T cells than RMs (P < .05).
Low levels of CCR5 expression in MALT-associated CD4+ T cells from natural SIV hosts
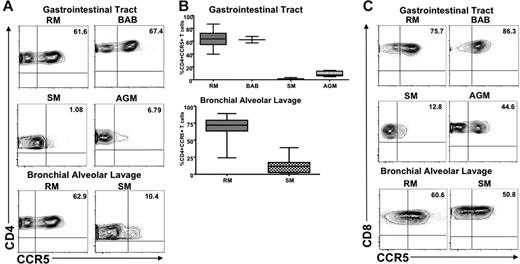
We next determined the level of CCR5 expression on mucosa-associated lymphoid tissue (MALT)–associated CD4+ T cells of 2 natural host species (SMs and AGMs) and 2 nonnatural host species (RMs and baboons). Small intestine samples were collected from 12 Caribbean AGMs, 24 RMs, and 2 baboons. Gut-associated lymphoid tissue was by rectal biopsy (RB) in 13 healthy, uninfected SMs. In addition, we sampled lung-associated lymphoid tissue (BAL) in 10 SMs and 15 RMs. Figure 3A shows representative dot plots of CCR5 expression on intestine-derived and BAL-derived CD4+ T cells. A summary of the available data on CCR5 expression on MALT CD4+ T cells is shown in Figure 3B. These data clearly indicate that natural hosts for SIV infection express dramatically lower levels of CCR5 on their MALT-associated memory CD4+ T cells compared with nonnatural hosts. In particular, the average levels of CCR5 expression on CD4+ T cells in the gastrointestinal tract were 9.13% in AGMs and 1.2% in SMs; conversely, more than 50% of the CD4+ T cells in RMs and baboons expressed high levels of CCR5. Only approximately 11% of T cells in the intestines in AGMs were CD4+ compared with 46% in macaques, 54% in baboons, and 36% in mangabeys (data not shown). As such, the low levels of CD4+ T cells in the intestine of uninfected AGMs resembled the levels reported in chronically SIV-infected macaques40 and HIV-infected humans.35,38 Similar to what has been observed in peripheral blood and LN-derived T cells, CCR5 expression on MALT-derived CD8+ T cells was significantly higher than that on CD4+ T cells in SMs and AGMs (Figure 3C), thus confirming that the differences in CCR5 on CD4+ T cells were not the result of impaired recognition or specificity of the antibody we were using to detect CCR5.
CCR5 expression on T cells derived from MALT in natural and nonnatural hosts. (A) Representative contour plots showing CD4 and CCR5 staining of T cells isolated from the gastrointestinal tract of healthy uninfected RMs, baboons, SMs, and AGMs (top) and the lung (through bronchial alveolar lavage) of healthy uninfected RMs and SMs (bottom). (B) Summary of CCR5 expression on CD4+ T cells isolated from the gastrointestinal tract of the species listed in panel A. Statistically significant differences in CCR5 expression on CD4+ T cells from natural and nonnatural hosts of SIV were observed with the use of one-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test and the Mann-Whitney U test. For nonnatural RM hosts of SIV versus natural SM hosts of SIV, P < .001; and versus natural SGM hosts of SIV, P < .01. For nonnatural BAB hosts versus natural SM and AGM hosts, P < .05. (C) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the gastrointestinal tract of healthy uninfected RMs, baboons, SMs, and AGMs (top) and the lung (through bronchial alveolar lavage) of healthy uninfected RMs and SMs (bottom).
CCR5 expression on T cells derived from MALT in natural and nonnatural hosts. (A) Representative contour plots showing CD4 and CCR5 staining of T cells isolated from the gastrointestinal tract of healthy uninfected RMs, baboons, SMs, and AGMs (top) and the lung (through bronchial alveolar lavage) of healthy uninfected RMs and SMs (bottom). (B) Summary of CCR5 expression on CD4+ T cells isolated from the gastrointestinal tract of the species listed in panel A. Statistically significant differences in CCR5 expression on CD4+ T cells from natural and nonnatural hosts of SIV were observed with the use of one-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test and the Mann-Whitney U test. For nonnatural RM hosts of SIV versus natural SM hosts of SIV, P < .001; and versus natural SGM hosts of SIV, P < .01. For nonnatural BAB hosts versus natural SM and AGM hosts, P < .05. (C) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the gastrointestinal tract of healthy uninfected RMs, baboons, SMs, and AGMs (top) and the lung (through bronchial alveolar lavage) of healthy uninfected RMs and SMs (bottom).
Natural hosts and nonnatural species have equivalent numbers of CCR5 molecules on their target cells
To measure the surface molecule density of CCR5 on SIV target cells, we compared the fluorescence intensities of CCR5 on CD4+ T cells and monocytes with a reference cell population in the different NHP species. We selected CD8+ T cells as the reference population because these cells had the brightest expression of CCR5. We next measured the ratio of the expression of CCR5 on CD4+ T cells or monocytes and the expression of the same molecule on CD8+ T cells and did not find any difference between pathogenic or nonpathogenic models; all ratios were close to 1.
SIV-infected natural hosts are also characterized by low levels of CCR5 expression on CD4+ T cells from blood, LNs, and MALT
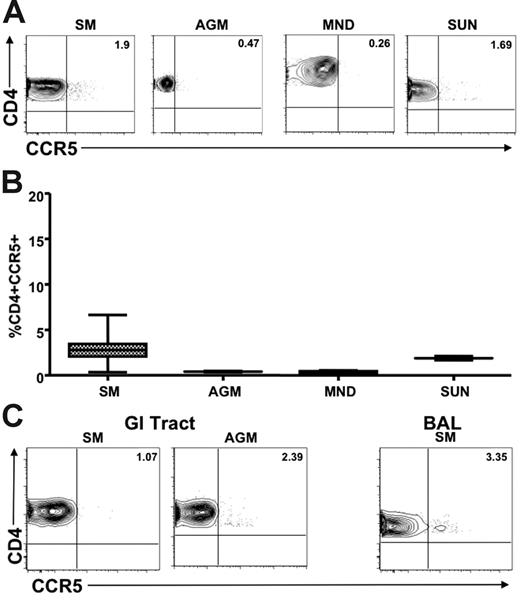
We next sought to determine whether the low levels of CCR5 expression on CD4+ T cells observed in healthy uninfected natural SIV hosts were also present in chronically SIV-infected animals. Figure 4A shows representative plots of CCR5 levels on CD4+ T cells from the peripheral blood and LNs of SIV-infected AGMs, SMs, mandrills, and sun-tailed monkeys. A summary of the available data on these animals is shown in Figure 4B. These findings indicate clearly that low levels of CD4+CCR5+ T cells are also a feature of SIV-infected natural hosts, thus ruling out the possibility that genetic factors have restricted the low CCR5 expression to uninfected animals. In addition, we measured CCR5 expression levels on MALT-associated CD4+ T cells in chronically SIV-infected animals of 2 natural host species (SMs and AGMs). We performed intestinal biopsy in chronically SIV-infected SMs (n = 18) and AGMs (n = 12). In addition, we performed BAL in 16 SIV-infected SMs. Figure 4C shows representative dot plots of CCR5 expression on RB- and BAL-derived CD4+ T cells. These data indicate clearly that natural hosts of SIV express dramatically lower levels of CCR5 on their MALT-associated CD4+ T cells regardless of their SIV infection status, thus confirming that the low CCR5 levels of uninfected animals do not represent a genetic restriction factor that protects from infection. Interestingly, in the 18 chronically SIV-infected SMs in which we tested CCR5 levels on CD4+ T cells in blood, LNs, intestinal biopsy specimens, and BAL specimens did not show any correlation between the level of viral replication on the percentage of CD4+CCR5+ T cells in any of the tested tissues (data not shown).
Limited CCR5 expression on CD4+ T cells in SIV-infected natural hosts. (A) Representative contour plots showing CD4 and CCR5 staining of peripheral blood–derived T cells of chronically SIV-infected SMs, AGMs, mandrills, and sun-tailed monkeys. (B) Summary of CCR5 expression on CD4+ T cells from the blood of the species listed in panel A. (C) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the gastrointestinal tract of chronically SIV-infected SMs and AGMs (left) and the lung (through bronchial alveolar lavage) of an SIV-infected SM (right).
Limited CCR5 expression on CD4+ T cells in SIV-infected natural hosts. (A) Representative contour plots showing CD4 and CCR5 staining of peripheral blood–derived T cells of chronically SIV-infected SMs, AGMs, mandrills, and sun-tailed monkeys. (B) Summary of CCR5 expression on CD4+ T cells from the blood of the species listed in panel A. (C) Representative contour plots showing CD8 and CCR5 staining of T cells isolated from the gastrointestinal tract of chronically SIV-infected SMs and AGMs (left) and the lung (through bronchial alveolar lavage) of an SIV-infected SM (right).
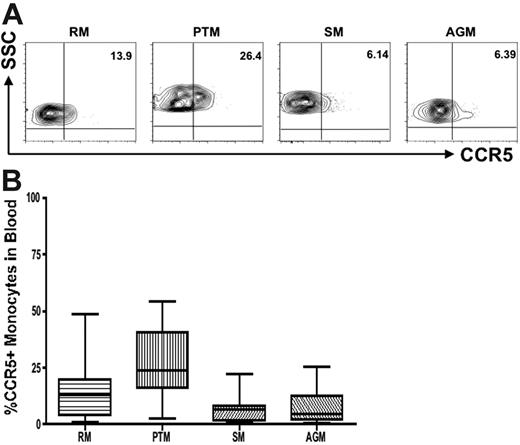
Monocytes also show lower CCR5 expression in natural hosts than in progressing species
We compared CCR5 expression on monocytes in natural and nonnatural hosts of SIV. Because of difficulties related to sampling (experiments performed in Central Africa or on endangered species), most of the CCR5 analyses were performed with a monocyte gate only. However, to confirm that this monocyte gating can be used as a surrogate for assessing the frequency and density of CCR5 on CD14+ cells, we also performed 4-color staining with a monocyte marker (CD14) in 9 uninfected AGMs and 5 uninfected RMs, and we further refined our analyses by gating first on monocytes and then on CD14. This experiment showed similar levels of CCR5 when using the 2 different gating strategies (data not shown). Figure 5A shows representative contour plots indicating CCR5 levels on monocytes in nonpathogenic hosts (AGMs and SMs) compared with pathogenic hosts (RMs and PTMs). In addition, a summary of the available data relative to CCR5 expression in monocytes is shown in Figure 5B. These data indicate that although CCR5 expression levels are lower on monocytes in natural hosts, the difference for susceptible hosts is not as dramatic as that observed for CD4+ T cells. In all, these data confirm our initial observation and support the idea that the reduction of CCR5 expression levels on target cells is a key feature of the coevolution between the virus and its natural host.
Reduced CCR5 expression on monocytes in natural hosts of SIV. (A) Representative contour plots showing CCR5 expression on monocytes in healthy uninfected individuals belonging to progressing (RMs and PTMs) and nonprogressing (SMs and AGMs) NHPs. (B) Summary of CCR5 expression on monocytes of the primate species listed in panel A. One-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test and the Mann-Whitney U test. For nonnatural RM hosts of SIV versus natural SM and AGM hosts, P > .05; for nonnatural PTM hosts of SIV versus natural SM and AGM hosts, P < .001.
Reduced CCR5 expression on monocytes in natural hosts of SIV. (A) Representative contour plots showing CCR5 expression on monocytes in healthy uninfected individuals belonging to progressing (RMs and PTMs) and nonprogressing (SMs and AGMs) NHPs. (B) Summary of CCR5 expression on monocytes of the primate species listed in panel A. One-way nonparametric ANOVA (Kruskal-Wallis) followed by the Dunn multiple comparison test and the Mann-Whitney U test. For nonnatural RM hosts of SIV versus natural SM and AGM hosts, P > .05; for nonnatural PTM hosts of SIV versus natural SM and AGM hosts, P < .001.
Discussion
Mechanisms underlying the strikingly different outcomes of primate lentiviral infection in natural hosts (in which the infection is generally nonpathogenic) compared with nonnatural/recent hosts (in which the infection is almost always pathogenic) represent one of the key enigmas in contemporary AIDS research. Here we describe and characterize a major, and previously undisclosed, immunologic feature of natural SIV hosts—the remarkably lower level of CCR5 expression on CD4+ T cells compared with nonnatural hosts. Moreover, CCR5 expression on monocytes of natural hosts is also lower than in progressive hosts, though not as dramatic as in CD4+ T cells. These observations are highly significant because most of the SIVs from African monkeys have been shown to use CCR5 as coreceptors for viral entry into the target cells.45 We observed this low CCR5 expression in SIV-infected and uninfected animals and in CD4+ T cells isolated from various tissues, particularly MALT (intestinal and lung mucosa), in which a large proportion of CD4+ T cells expressed CCR5 in nonnatural/recent hosts. Because the low levels of CD4+CCR5+ T cells were found in 5 different and relatively unrelated natural host species (SMs, AGMs, mandrills, sun-tailed monkeys, and chimpanzees), these observations identified an impressive example of convergent evolution of the immune system that likely reflects coadaptation between these species and their endemic primate lentiviruses. The possibility that this phenomenon is an evolutionary feature related to SIV infection is further supported by the observation that baboons—African monkey species that are not natural hosts of SIV but are susceptible to AIDS when inoculated with HIV-224,46 —have high levels of CCR5 on their CD4+ T cells. Interestingly, chimpanzees, which acquired SIV more recently than African monkey hosts,9 expressed CCR5 levels on CD4+ T cells slightly higher than those observed in the other natural SIV hosts.
The possibility that these low CD4+CCR5+ T-cell levels are artifacts resulting from poor recognition of CCR5 by antibodies designed to identify the human molecule was clearly ruled out by the expression of CCR5 on CD8+ T cells of natural hosts. The fact that CCR5 expression is low on CD4+ but not on CD8+ T cells also identifies a clear difference with the so-called CCR5-δ32 phenotype in humans or the CCR5-δ24 phenotype previously reported in red-capped mangabeys (RCMs),47 whereby CCR5 is not expressed in any cell of the body. From a virologic perspective, the main difference between the CCR5-δ32 phenotype in humans and the low CD4+CCR5+ phenotype of natural hosts is that the former appears to be refractory to viral transmission and viral replication. In RCMs, the CCR5-δ24 phenotype resulted in coreceptor switch by the SIVrcm toward the exclusive use of CCR2 as a coreceptor.47 This last example, together with our observation, suggests that the CCR5 molecule is expressed on the CD4+ T cells of natural hosts of SIVs at an optimal level to allow virus persistence without inducing major deleterious effects in the host species.
The low CCR5 expression on SIV target cells does not protect from transmission or viral replication. On the contrary, viral replication in natural host species is as high as that observed during pathogenic infection.35-40 This finding is consistent with the theoretical possibility that CD4+ T cells that appear to be CCR5− by flow cytometry may in fact express low levels of this molecule and still be infected by the virus.37 From an immunologic point of view, the low CD4+CCR5+ phenotype of natural SIV hosts indicates that these animals have evolved to be no longer dependent on large numbers of MALT-associated CD4+CCR5+ T cells, whereas their CD8+ T cells still express (and likely use) this molecule. Although our current understanding of the function of CCR5 is limited, conceivably the absence of this receptor on CD4+ T cells could have profound differences in the way natural, nonprogressing SIV hosts respond to an infection that specifically uses this coreceptor for viral attachment and entry. Importantly, our data indicate that NHP species considered nonprogressing hosts of SIV inherently have significantly lower levels of the very CD4+CCR5+ T cells these viruses use for optimal infection and replication during pathogenic HIV/SIV infections.
Although this finding suggests that VLs should be low in natural hosts for SIV infection, all the available experimental data indicate that this is not the case, thus posing the problem of defining the source of viral replication in natural hosts and the reasons this down-modulation of CCR5 on CD4+ T cells is such an important evolutionary feature of natural SIV hosts beyond restricting the level of viral replication. In terms of sources of virus replication in natural SIV hosts, one possibility could be that the high viremia level of these animals originates from a pool of long-lived infected cells, most likely macrophages. This is an intriguing possibility. In fact, macrophage infection has been shown in tissues isolated from naturally SIV-infected SMs (I.P., unpublished observations, 2004). However, studies of the turnover of virus and infected cells performed in SIV-infected SMs by measuring the kinetics of changes in viral replication after antiviral therapy indicate that, in these animals, the bulk of viral replication is sustained by infected cells with lifespans of approximately 1.1 days, similar to what has been reported in pathogenic HIV/SIV infections48-51 (S.G., J. Engram, R. Dunham, R. Ribeiro, I.P., B. Lawson, D. Sodora, S. Staprans, A. Perelson, and G.S., unpublished observations, 2005; I.P., C.A., R. Gautam, A.C. Carter, M. Barnes, M. Pattison, C. Stoulig, A.S. Perelson, and R.M. Ribeiro, unpublished observations, 2006). This apparent contradiction could be resolved if one postulates that the main difference in the type of viral turnover between natural and nonnatural hosts is not the type or the lifespan of infected cells but, rather, the amount of virus (ie, burst size) produced on a per cell basis. In this model, the high VL observed in natural SIV hosts would be the result of fewer target cells (hence, the low levels of CD4+CCR5+ T cells) that produce more virus per infected cell despite a similarly short half-life. This combination of greater viral fitness and reduced availability of target cells would represent an elegant type of coevolutionary adaptation between virus and host, whereby the virus maintains high levels of replication while the host achieves the important goal of limiting the damage that the virus poses to immune system function by reducing the number of cells killed at any given time.
A consistent feature of SIV infection of natural hosts is the presence of markedly lower levels of immune activation than those observed in nonnatural/recent hosts in which the infection is pathogenic (for reviews, see Grossman et al,52 McCune,53 Silvestri and Feinberg,54 and Hazenberg et al55 ). In pathogenic HIV infection, the level of immune activation is considered as important as, and perhaps even more important than, the level of viral replication in predicting the progression of the infection to full-blown AIDS.56 As such, we and others have proposed that the lack of chronic, generalized immune activation is a key mechanisms that helps natural SIV hosts from developing progressive CD4+ T-cell depletion and AIDS.8,10,12,14,18,20,21,23 In this context, it is conceivable that an additional beneficial effect of low levels of CCR5 expression on MALT-associated CD4+ T cells of natural SIV hosts is represented, in these animals, by a reduced net influx of activated CD4+ T cells to the mucosal tissues. Although low CCR5 levels on CD4+ T cells may not affect the homing of activated CD4+ T cells to mucosal tissues in general, they may be instrumental in reducing the specific homing of activated CD4+ T cells to areas of the mucosal immune system that are already involved in a process of immune activation and inflammation. Because the level of inflammation in MALT may be part of a potentially dangerous cycle of recruitment of activated CD4+ T cells (likely CCR5 dependent), increased availability of target CD4+ T cells, and thus higher local VLs, our data are consistent with the possibility that low levels of CCR5 expression on CD4+ T cells protects from continuous recruitment of activated CD4+ T cells in “inflamed” mucosal sites that are active sites of virus replication.
Although the etiology of the divergent outcome of SIV infection between natural and nonnatural hosts remains a mystery, the presented findings identify a striking and fundamental difference between the immunologic make-up of natural and nonnatural hosts—the presence in natural SIV hosts (infected and uninfected) of very low numbers of CD4+CCR5+ T cells known to be the main cellular targets for HIV and SIV replication during pathogenic infections of humans and RMs, respectively. This observation represents, to our knowledge, a prominent and potentially significant difference between the immune system of natural and nonnatural SIV hosts. The fact that this feature is strikingly conserved across a number of relatively unrelated primate species indicates that the low levels of CCR5 expression on CD4+ T cells of natural hosts are examples of convergent evolution that followed the encounter between SIV and its natural hosts. Unfortunately, these data also suggest that evolution could not devise or select an immune response that would effectively suppress viral transmission or replication but, rather, shaped the immune system of natural SIV hosts to be less dependent on the presence of CD4+CCR5+ T cells. Although the implications of these findings in terms of AIDS vaccine development are obviously daunting, there is no doubt that a better understanding of how these natural SIV hosts cope with their chronic lentiviral infections may eventually prove helpful in combating HIV infection and AIDS in humans.
In summary, these data suggest a possible beneficial dual adaptation resulting from the low levels of CCR5 on CD4+ T cells. The host might have adapted by restricting the pool of target cells for viral replication or by contributing to low immune activation through reduced homing of activated CD4+ T cells to the mucosal tissue. The high levels of viral replication in natural hosts of SIV suggest that the virus might also have adapted by increased fitness or by infecting predominantly other cell populations not as critical to the immune system as CD4+CCR5+ T cells, thus avoiding profound immunodeficiency. This unique interaction allows the host to survive the viral infection and simultaneously offers the virus the ability to multiply at high levels.
Authorship
Contribution: I.P., C.A., P.A.M., A.A.L., V.M.H., and R.S.V. collected and analyzed the data from African green monkeys, baboons, pig-tailed macaques, rhesus macaques, and cynomolgus macaques. S.G., B.S., A.K., I.P., C.A., and G.S. collected and analyzed data from sooty mangabeys. B.S., P.R., and G.S. collected data on mandrills, chimpanzees, and sun-tailed monkeys. I.P., C.A., and G.S. designed the research. I.P., C.A., P.A.M., A.A.L., R.S.V., and G.S. analyzed the data. I.P., C.A., A.A.L., and G.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ivona Pandrea, Department of Comparative Pathology, Tulane National Primate Research Center, 18703 3 Rivers Rd, Covington, LA 70433; e-mail: ipandrea@tulane.edu.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants R01 AI64066 (I.P.), R01 AI49080 (R.S.V.), R01 AI65325 (C.A.), R01 AI49809 (A.K.), R01 AI52755 and AI66998 (G.S.), RR00164 (Tulane National Primate Research Center [TNPRC]), and RR00165 (Yerkes National Primate Research Center [YNPRC]).
We thank Drs Daniel Douek and Louis Picker for helpful discussion; Mary Jane Dodd, Janel Leblanc, Melissa Patisson, and Kelsi Rasmussen for technical support; Dr Ann Chahroudi for critical reading of the manuscript; and the veterinary staff of the TNPRC and the YNPRC for assistance with animal work.