Abstract
Production of tumor necrosis factor-α (TNFα) by the neutrophil (PMN) is a pivotal event in innate immunity, but the signals regulating TNFα induction in this primary cell are poorly understood. Herein, we use protein transduction to identify novel, opposing anti– and pro–cytokine-inducing roles for RhoA in the resting and lipopolysaccharide (LPS)–stimulated human PMN, respectively. In the resting cell, RhoA suppresses Cdc42 activation, IκBα degradation, nuclear factor-κB (NF-κB) activation, and induction of TNFα and NF-κB–dependent chemokines. Suppression of TNFα induction by RhoA is Rho kinase α (ROCKα) independent, but Cdc42 dependent, because TNFα induction by C3 transferase is attenuated by inhibition of Cdc42, and constitutively active Cdc42 suffices to activate NF-κB and induce TNFα. By contrast, we also place RhoA downstream of p38 mitogen-activated protein kinase and Cdc42 in a novel LPS-activated pathway in which p38, Cdc42, and ROCKα all promote TNFα protein expression. The p65 subunit of NF-κB coprecipitates with RhoA in a manner sensitive to the RhoA activation state. Our findings suggest a new, 2-faced role for RhoA as a checkpoint in innate immunity.
Introduction
The prototypical early-response cytokine TNFα has multiple regulatory effects on both inflammation and host defense. Depending on the context, “positive” effects of TNFα, such as enhanced bacterial killing and polymorphonuclear leukocyte (PMN) recruitment to sites of infection, may be overshadowed by negative consequences for the organism, such as aggravated organ injury.1,2 Likely, because of these mixed effects of TNFα, induction of TNFα by LPS and other stimuli is precisely regulated at multiple steps, including transcription, mRNA stability, translation, and protein stability.3-5 Further complicating this issue, TNFα produced by different cell types has been described to have distinct and nonredundant functions in vivo.6 Moreover, multiple examples exist of cell-type–dependent differences in the pathways regulating TNFα expression.5,7,8 Hence, findings derived from cell lines and even primary cells may not be applied universally, and, moreover, systemic pharmacotherapies aiming to modulate TNFα production are likely to be confounded by important differences among target tissues.
The human PMN is a pivotal acute-response effector cell in inflammation and host defense and an important source of TNFα in response to stimuli such as LPS. Regulation of PMN quiescence and activation is central to health. However, little is known about regulation of the resting PMN, including whether PMN quiescence itself is an active or passive state. Recent studies suggest that PMNs, perhaps because of their distinct role as short-lived sentinels in innate immunity, have unique posttranscriptional regulatory mechanisms providing for precise spatiotemporal control of proinflammatory gene product expression,9,10 and uniquely poised features for NF-κB activation.11 Nevertheless, because the human PMN is essentially nontransfectable, analysis of signaling pathways in the PMN have been grossly limited to the short list of molecular targets for which cell-permeant pharmacologic inhibitors are available.8 Although reports in cell lines have suggested a role for Rho GTPases in cytokine production12 and NF-κB activation,13-15 the inability, with available inhibitors such as the pan-Rho GTPase inhibitor Clostridium difficile toxin B16 to discriminate further among these G-proteins in primary cells, has left many basic questions unanswered.
The Rho GTPases, of which the best-described members include RhoA, Cdc42, and Rac1/2, are molecular switches that have classically been associated with chemotaxis, superoxide anion (O
Materials and methods
Reagents and antibodies
Endotoxin-free reagents and plastics were used throughout. Aprotinin, leupeptin, AEBSF, NaF, Na3VO4, DMSO, and protein A–Sepharose were from Sigma (St Louis, MO), and SB203580 was from Calbiochem (San Diego, CA). RhoA and Cdc42 assay kits and glutathione-S-transferase (GST) were from Upstate Cell Signaling Solutions (Lake Placid, NY). C3 transferase, Rhotekin-RBD-GST, L61Cdc42, GST-wt RhoA, GST-L63RhoA, and GST-Sepharose were from Cytoskeleton (Denver, CO). pET23–wild-type Wiskott-Aldrich syndrome protein (WASP)–CRIB (amino acids 201-321) and mutant non–Cdc42-binding WASP-CRIB (F271C, H246D, H249D) constructs were kind gifts from Klaus Hahn (University of North Carolina) and were expressed as C-terminal His6-tagged fusion proteins in Escherichia coli, purified, and quantified by Bradford assay.25 E coli 0111:B4 LPS was from List Biological Laboratories (Campbell, CA). Antibodies include rabbit anti-RhoA (Upstate, Lake Placid, NY), -ROCKα, -Cdc42, -IκBα, and -p38 (Santa Cruz Biotechnology, Santa Cruz, CA), and -PO4-p38 (Thr-180/Tyr-182; Cell Signaling, Beverly, MA). Rhodamine-phalloidin was from Molecular Probes (Eugene, OR). BioPORTER reagent was from Gene Therapy Systems (San Diego, CA). Human TNFα enzyme-linked immunoabsorbent assay (ELISA) was from ELISAtech (Aurora, CO). The NE-PER kit was from Pierce (Rockford, IL), and the TransAM NF-κB p65 Transcription Factor Assay Kit was from Active Motif (Carlsbad, CA).
PMN isolation and treatments
PMNs were isolated from whole blood of healthy donors by discontinuous plasma Percoll centrifugation, as reported.8 For LPS exposure experiments (100 ng/mL), the cells were resuspended in RPMI 1640 culture medium (BioWhittaker, Walkersville, MD) supplemented with 10 mM HEPES (pH 7.6) and 1% human heat-inactivated platelet-poor plasma. Approval was obtained from the National Jewish Medical and Research Center Institutional Review Board for the studies. Informed consent was provided according to the Declaration of Helsinki.
RhoA and Cdc42 activation assays
In vitro kinase assays
A modification of a published protocol for assaying ROCK kinase was used.28 Briefly, precleared lysates were immunoprecipitated with 1.5 μg rabbit anti-ROCKα antibody and 15 μL protein A–Sepharose (2 hours, 4°C). The beads were washed and incubated with kinase reaction buffer, as described.28 Reaction products were resolved on a 14% SDS-PAGE (polyacrylamide gel electrophoresis) gel, transferred to nitrocellulose, and imaged using a Storage Phosphor Screen and Storm 860 scanner (Molecular Dynamics, Sunnyvale, CA). P38 kinase activity was assayed as previously reported.8
Protein transduction
Recombinant proteins were transduced into PMNs using a modification of a published protocol.24 Briefly, 2 μL BioPORTER in methanol was divided into aliquots into 1.5-mL reaction tubes and dried overnight. Varying quantities (4-20 μg) of recombinant proteins plus PBS, pH 7.4, up to a final volume of 40 μL were used to rehydrate the BioPORTER per the manufacturer's protocol, whereupon 5 × 106 PMNs in Krebs Ringer Phosphate Dextrose buffer were added to a final volume of 1 mL. PMNs were then incubated (2-4 hours, 25°C), pelleted (500g, 3 minutes), and resuspended. PMNs were treated with C3 transferase (20 μg/mL, 4 hours, 37°C, 107 cells/mL), a modification of a previous report.29
PCR and ELISA
Nuclear/cytoplasmic fractionation
The NE-PER kit (Pierce) was used, per the manufacturer's protocol.
Rhodamine-phalloidin staining and microscopy
Rhodamine-phalloidin staining and microscopy were performed as previously reported.30 Briefly, PMNs were fixed with an equal volume of 4% paraformaldehyde, 3% sucrose/PBS (30 min, 37°C), settled on coverslips, permeabilized with 0.02% Tween 20/PBS (5 min, room temperature), washed once, stained with 5 units of rhodamine-phalloidin (Molecular Probes, Eugene, OR), and then mounted with Gel/Mount (Biomeda, Foster City, CA). Photographs were taken using a Zeiss Axiovert 200M microscope (Thornwood, NY) equipped with a 60×/1.40 numerical aperture oil objective and a Cooke Sensicam 2.0 camera (Cooke, Romulus, MI). Images were acquired and processed using SlideBook software version 4.0.1.16 (Intelligent Imaging Innovations, Denver, CO).
NF-κB activation assays
PMNs (20 × 106) were resuspended in 400 μL sonication buffer (20 mM imidazole, pH 7.4; 0.25 M sucrose; 5 mM EGTA; 2.5 mM MgCl2) supplemented with protease and phosphatase inhibitors, sonicated on ice (Fisher Scientific [Hampton, NH] Sonic Dismembrator Model 100, setting 2, 20 × 2-second pulses), salt-extracted (400 mM NaCl, 20 minutes, 4°C), and centrifuged (18 380g, 15 minutes, 4°C). Sonicate (15 μg) was assayed by TransAM p65 kit (Active Motif) per the manufacturer's instructions. Cytoplasmic fractions were normalized by protein assay25 and immunoblotted with rabbit anti-IκBα antibody.
Rho GTPase pulldown
PMNs (20 × 106) were lysed in 500 μL (50 mM Tris pH 7.5, 10% glycerol, 1% Ipegal in 0.9% NaCl), clarified by centrifugation, precleared with GST-Sepharose (15 μg, 1 hour, 4°C), and then incubated for 2 hours with glutathione-Sepharose beads and 15 μg either GST or GST fusion protein (wt RhoA, L63RhoA). Beads were then washed 3 times, eluted in 1 × Laemmli buffer, and boiled.
Statistical analysis
Data are reported as mean ± SE. In analyses involving multiple comparisons (Figures 1A-B; 2A,C,E; 3D,G), ANOVA was used (2-way ANOVA with Bonferroni post test analysis, or stratified 1-way ANOVA with Tukey post test analysis). In other cases, analysis was performed using a 2-tailed Student t test (Prism; GraphPad Software, San Diego, CA). P less than .05 was considered statistically significant.
Results
LPS activates RhoA and ROCKα by a p38 MAPK-mediated pathway
Our group has previously reported that p38 is the only MAPK activated by LPS in the suspended human PMN and that p38 regulates LPS-induced NF-κB activation and TNFα expression.8 In this context, we sought to define regulators downstream of p38 on the pathway to TNFα induction in the human PMN. Rho GTPases have classically been associated with multiple G-protein–coupled receptor-associated functions in leukocytes, including actin polymerization, O
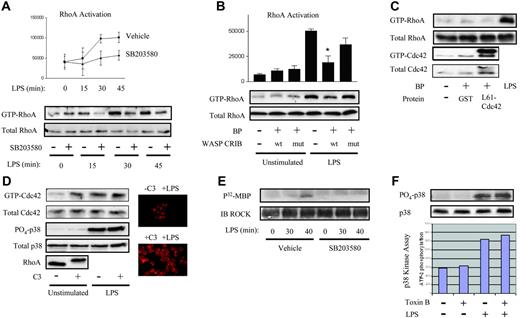
To address this issue, we tested for LPS-induced activation of RhoA in human PMN lysates, using a Rhotekin-Rho Binding Domain (RBD)–GST-Sepharose pulldown.26 Active RhoA was detected in the resting PMN, and LPS induced further accumulation of GTP-RhoA in a time-dependent fashion, first noted by 15 to 30 minutes (Figure 1A). Activation of RhoA was p38 dependent, as revealed by the use of SB203580, a p38 inhibitor. We have previously reported p38-dependent activation of the Rho GTPase Cdc42 in the human PMN within 10 to 15 minutes of LPS exposure.30 As this is earlier than RhoA activation (Figure 1A), and Cdc42 has been reported to be upstream of RhoA,31 we next questioned whether Cdc42 lies upstream of RhoA in the PMN's LPS signaling cascade. No cell-permeant inhibitors specific for Cdc42 are available. Hence, we used BioPORTER reagent to protein-transduce the Cdc42 binding domain of the Cdc42 effector, Wiskott Aldrich syndrome protein (WASP-CRIB), a method previously reported specifically to inhibit Cdc42 in the human PMN by sequestering GTP-Cdc42.24 As a negative control, we transduced a mutant, non–Cdc42-binding WASP-CRIB. As shown in Figure 1B, LPS-induced RhoA activation was inhibited in PMNs pretreated with wild-type but not mutant WASP-CRIB protein, confirming that Cdc42 lies upstream of RhoA in the LPS signaling pathway. As we have previously reported basal activation of Cdc42 in the resting human PMN,30 the lack of any effect of WASP-CRIB on GTP-RhoA expression in the resting PMN (Figure 1B) suggests that Cdc42-independent mechanisms are responsible for basal RhoA activation. In support of this, transduced constitutively active L61Cdc42 was not sufficient to activate RhoA in the resting PMN (Figure 1C).
LPS activates RhoA through a p38- and Cdc42-dependent pathway. (A) Human PMNs were preincubated with 10 μM SB203580 or 0.1% DMSO vehicle, treated with LPS for the indicated times, and lysates assayed for RhoA activation.26 Active RhoA was quantified by densitometry and plotted (P < .05 by 2-way ANOVA for SB203580 versus vehicle). (B) PMNs were transduced using BioPORTER (BP) reagent with wild-type WASP-CRIB or mutant non-Cdc42 binding WASP-CRIB,24 LPS-exposed (20 minutes), and then assayed for RhoA activation. Active RhoA was quantified by densitometry and plotted (*P < .001 for LPS/wt WASP-CRIB versus LPS/−). (A-B) Error bars represent SE. (C) PMNs were untreated or transduced with either GST (control) or L61Cdc42 and then assayed for RhoA and Cdc42 activation. LPS was used as a positive control. (D) PMNs were treated with buffer or C3 transferase (20 μg/mL, 37°C, 4 hours) and assayed for Cdc42 activation.27 Lysates were also probed with rabbit anti-Cdc42, -PO4-p38, p38, and -RhoA antibodies. C3- and buffer-treated PMNs were treated with LPS (37°C, 45 minutes), stained with rhodamine-phalloidin, and imaged under × 60 objective.30 Image intensity was enhanced on C3-treated PMNs to facilitate comparisons of morphology. (E) Human PMNs treated as in panel A were lysed, and the activity of immunoprecipitated ROCKα was tested.28 (F) PMNs were pretreated with 500 ng/mL Clostridium difficile toxin B or buffer (90 minutes), exposed to LPS (30 minutes), lysed, and immunoblotted for PO4-p38 and total p38. Under the same conditions, p38 kinase activity was assayed.8 All panels are representative of 3 or more independent experiments.
LPS activates RhoA through a p38- and Cdc42-dependent pathway. (A) Human PMNs were preincubated with 10 μM SB203580 or 0.1% DMSO vehicle, treated with LPS for the indicated times, and lysates assayed for RhoA activation.26 Active RhoA was quantified by densitometry and plotted (P < .05 by 2-way ANOVA for SB203580 versus vehicle). (B) PMNs were transduced using BioPORTER (BP) reagent with wild-type WASP-CRIB or mutant non-Cdc42 binding WASP-CRIB,24 LPS-exposed (20 minutes), and then assayed for RhoA activation. Active RhoA was quantified by densitometry and plotted (*P < .001 for LPS/wt WASP-CRIB versus LPS/−). (A-B) Error bars represent SE. (C) PMNs were untreated or transduced with either GST (control) or L61Cdc42 and then assayed for RhoA and Cdc42 activation. LPS was used as a positive control. (D) PMNs were treated with buffer or C3 transferase (20 μg/mL, 37°C, 4 hours) and assayed for Cdc42 activation.27 Lysates were also probed with rabbit anti-Cdc42, -PO4-p38, p38, and -RhoA antibodies. C3- and buffer-treated PMNs were treated with LPS (37°C, 45 minutes), stained with rhodamine-phalloidin, and imaged under × 60 objective.30 Image intensity was enhanced on C3-treated PMNs to facilitate comparisons of morphology. (E) Human PMNs treated as in panel A were lysed, and the activity of immunoprecipitated ROCKα was tested.28 (F) PMNs were pretreated with 500 ng/mL Clostridium difficile toxin B or buffer (90 minutes), exposed to LPS (30 minutes), lysed, and immunoblotted for PO4-p38 and total p38. Under the same conditions, p38 kinase activity was assayed.8 All panels are representative of 3 or more independent experiments.
RhoA, by contrast, inhibited basal activation of Cdc42 in the resting PMN. In the resting cell, C3 transferase, a specific Rho inhibitor that ADP-ribosylates RhoA on Asn41, leading to its gel-retardation and degradation32 (Figure 1D) and thereby modifies the actin cytoskeleton (Figure 1D), was sufficient to induce modest Cdc42 activation (Figure 1D), as assessed by a p21-binding domain pulldown.27 By contrast, C3 had no effect on either basal or LPS-induced p38 activation (Figure 1D). Taken together with the preceding findings, this suggests that (1) Cdc42 mediates LPS-induced, but not spontaneous activation of RhoA; (2) Cdc42 is not sufficient for RhoA activation in the resting PMN; and (3) RhoA suppresses Cdc42 in the resting PMN.
Finally, to confirm further the downstream significance of RhoA activation, we tested for activation of a prototypical Rho-specific effector, ROCKα. ROCKα activation was of particular interest because this kinase has been associated with cytokine expression in other cell types.33,34 To test for ROCKα activation in the LPS-exposed PMN, and to place it downstream of p38, like its activator, RhoA (Figure 1A), we performed an in vitro ROCKα kinase assay28 on LPS-exposed PMNs, both untreated and pretreated with SB203580. As depicted in Figure 1E, ROCKα activity was first detected 40 minutes following LPS exposure, temporally consistent with our observation of RhoA activation at 15 to 30 minutes of LPS exposure (Figure 1A). Moreover, ROCKα activation was sensitive to p38 inhibition, as also shown for RhoA (Figure 1A) and previously by us for Cdc42 in the LPS-exposed human PMN.30
In summary, these data suggest contrasting suppressive versus stimulatory roles for RhoA in the resting and LPS-exposed human PMN: (1) inhibition of Cdc42 by RhoA in the resting state and (2) a pathway leading from p38 to Cdc42 to RhoA to ROCKα in the LPS-stimulated condition.
LPS-induced p38 activation and Rho GTPases
In contrast to our findings, existing reports of Rho GTPases in other cell types place these molecular switches upstream of MAP kinases.35,36 Nevertheless, neither of 2 surrogate assays of p38 activation, phosphospecific immunoblotting and in vitro kinase assay, revealed diminution of p38 activation in the presence of a pan-Rho GTPase inhibitor, C difficile toxin B16 (Figure 1F), at the inhibitor concentration tested. Similar results were noted with C3 transferase (Figure 1D; data not shown). Alternatively, RhoA has also been reported to be functionally upstream of ERK by regulating its nuclear/cytoplasmic translocation,37 an effect that might conceivably be important to transcriptional and translational regulation. Immunoblotting revealed no effect of either toxin B or Y27632, a ROCK inhibitor,38 on nuclear localization of either PO4-p38 or total p38 (data not shown). Thus, in contrast to other cell types, Rho GTPases and ROCK, as tested by the inhibitors toxin B and Y27632 respectively, do not appear to regulate either the activity or nuclear/cytoplasmic localization of p38 in the LPS-stimulated human PMN.
Cdc42 regulates TNFα transcript and protein expression
We have previously reported that p38 MAPK regulates LPS-induced TNFα transcript and protein expression in the human PMN.8 Cdc42 has been implicated in nuclear translocation of transcription factors and posttranscriptional regulation.39,40 Having placed Cdc42 downstream of p38 in the LPS signaling cascade,30 we next queried what regulatory effect, taken in isolation, that Cdc42 exercises on TNFα induction. As shown in Figure 2A, wild-type WASP-CRIB inhibited LPS induction of TNFα protein, whereas the inactive mutant had no effect. No effect of WASP-CRIB was seen on TNFα transcript abundance (Figure 2B). Transduced constitutively active L61Cdc42 was, nevertheless, sufficient in the resting PMN not only to induce modest expression of TNFα protein but also to induce a marked increase in TNFα transcripts (Figure 2C-D). These data identify an additional role for Cdc42 in regulation of TNFα transcripts. Moreover, they highlight fundamental differences between Cdc42 function in the resting and LPS-stimulated human PMN, as observed with Cdc42 regulation of RhoA activation (Figure 1B-C). Because L61Cdc42 does not activate RhoA (Figure 1C), TNFα gene induction by Cdc42 in the resting PMN is likely RhoA independent.
Cdc42 regulates TNFα transcript and protein. (A-B) PMNs were left untreated or transduced with wild-type WASP-CRIB or mutant non–Cdc42-binding WASP-CRIB (10 μg/mL, 25°C, 2 hours) using BioPORTER (BP). Cells were then left untreated or exposed to LPS (2 hours). Supernatants were analyzed by ELISA for TNFα protein (A) and lysates by RT-PCR for TNFα and GAPDH transcripts (B) (*P < .05 compared with buffer/LPS). The results shown are representative of 4 experiments. (C-D) PMNs were left untreated or transduced with GST or L61Cdc42 (10 μg/mL, 25°C, 2 hours) using BP. Cells were then left untreated or exposed to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein (C) and lysates by RT-PCR for TNFα and GAPDH (D). (*P < .001 compared with untreated and GST treated). The gel shown is representative of 3 experiments. (E) PMNs were untreated or transduced with GST or L61Cdc42 (4 hours) and then exposed to buffer or LPS (25 minutes). Cell sonicates were assayed by ELISA for p65 activation (*P < .05) in the presence or absence of competitor (wt) and noncompetitor (mutant) oligonucleotides, and (F) cytoplasmic fractions were immunoblotted for IκBα. A representative immunoblot is shown. PCR gels are representative of 3 independent experiments. (A, C, E) Error bars represent SE.
Cdc42 regulates TNFα transcript and protein. (A-B) PMNs were left untreated or transduced with wild-type WASP-CRIB or mutant non–Cdc42-binding WASP-CRIB (10 μg/mL, 25°C, 2 hours) using BioPORTER (BP). Cells were then left untreated or exposed to LPS (2 hours). Supernatants were analyzed by ELISA for TNFα protein (A) and lysates by RT-PCR for TNFα and GAPDH transcripts (B) (*P < .05 compared with buffer/LPS). The results shown are representative of 4 experiments. (C-D) PMNs were left untreated or transduced with GST or L61Cdc42 (10 μg/mL, 25°C, 2 hours) using BP. Cells were then left untreated or exposed to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein (C) and lysates by RT-PCR for TNFα and GAPDH (D). (*P < .001 compared with untreated and GST treated). The gel shown is representative of 3 experiments. (E) PMNs were untreated or transduced with GST or L61Cdc42 (4 hours) and then exposed to buffer or LPS (25 minutes). Cell sonicates were assayed by ELISA for p65 activation (*P < .05) in the presence or absence of competitor (wt) and noncompetitor (mutant) oligonucleotides, and (F) cytoplasmic fractions were immunoblotted for IκBα. A representative immunoblot is shown. PCR gels are representative of 3 independent experiments. (A, C, E) Error bars represent SE.
Having identified a regulatory role for Cdc42 in TNFα transcript expression in the resting PMN, we next sought to characterize further the underlying mechanism. TNFα transcription is strongly NF-κB dependent.41 We are unaware of any reports associating Cdc42 with NF-κB activation in the PMN. To address this, we assayed the effect of L61Cdc42 on binding of the p65 subunit of NF-κB to its cognate DNA sequence. Using a sandwich ELISA of p65 binding to immobilized κB oligonucleotide sequence (p65 TransAM; Active Motif), we observed that L61Cdc42 was sufficient to induce p65 activation in the PMN (Figure 2E). Because proteasomal degradation of cytoplasmic IκBα is a critical mechanistic step in canonical NF-κB activation,42 we also evaluated IκBα expression in cytoplasmic fractions of L61Cdc42-transduced PMNs. L61Cdc42 but not GST control induced IκBα degradation (Figure 2F). These results suggest that Cdc42 is sufficient to activate NF-κB in the resting human PMN, and, moreover, that Cdc42 may induce TNFα gene expression through a transcriptional mechanism involving NF-κB. Consistent with our results showing no effect of WASP-CRIB on LPS-induced TNFα gene expression, no consistent effect of WASP-CRIB was seen on LPS-induced IκBα degradation (data not shown).
RhoA suppresses TNFα and NF-κB–dependent chemokines in the resting human neutrophil
We next sought to define the independent role of RhoA in regulation of TNFα expression. Given our findings of contrasting roles for RhoA in the resting versus LPS-stimulated state, we evaluated the effect of Rho inhibition under both conditions. Unexpectedly, RhoA inhibition by C3 transferase was sufficient to induce TNFα protein (Figure 3A) and mRNA (Figure 3B) in resting cells. In corroboration, Rhotekin-RBD, a RhoA-sequestering agent,14,43 which depleted active RhoA in the PMN as indicated by surrogate inactivation of ROCKα (Figure 3C), was sufficient to induce expression of both TNFα protein and transcripts (Figure 3D). C3 was similarly sufficient to induce NF-κB–dependent chemokines44,45 (IL-8, a CXC chemokine, and MIP-1β, a CC chemokine) (Figure 3B), as was Rhotekin-RBD (data not shown). Like LPS, but unlike poly-IC, C3 was not sufficient to induce IFN-β, an antiviral cytokine that depends on both NF-κB and interferon regulatory factor-3 for induction 46 (Figure 3B). Of note, C3 and all other recombinant proteins used in the present study were confirmed by Limulus amebocyte lysate assay to have less than 50 pg/mL (< 0.5 EU) of contaminating LPS (data not shown). Pretreatment with polymyxin B (1 μg/mL, 30 minutes) completely abolished TNFα induction by 10 ng/mL LPS but had a negligible effect on C3-induced TNFα induction (data not shown). Moreover, boiling completely ablated TNFα induction by C3 but did not affect TNFα induction by 100 ng/mL LPS or peptidoglycan (data not shown). Collectively, these findings suggest that it is the enzymatic activity of C3 on Rho16 and not contaminants (ie, LPS, peptidoglycan, lipoproteins) that account for the observed induction of TNFα by C3. In summary, RhoA inhibition by 2 different approaches was sufficient to induce 3 NF-κB–dependent cytokines in the resting human PMN: TNFα, IL-8, and MIP-1β. By inference, these data indicate that active RhoA suppresses proinflammatory cytokine production in the resting PMN.
RhoA negatively regulates TNFα induction in the PMN. (A) PMNs were left untreated or treated with 20 μg/mL C3 transferase (37°C, 4 hours) and then exposed either to media or to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein. (*P < .05). (B) PMNs were untreated or treated with C3 transferase, exposed to buffer or LPS (1 hour) and then analyzed by RT-PCR. PMNs were also exposed to either poly-inosine/cytidine (pIC) or LPS (4 hours) and analyzed for IFN-β and GAPDH expression. The gels shown are representative of 3 experiments. (C) PMNs were left untreated or transduced with BSA or Rhotekin-RBD (25°C, 2 hours) using BioPORTER (BP). ROCKα was immunoprecipitated, and its kinase activity was assayed.28 (D) PMNs were left untreated or transduced with BSA or Rhotekin-RBD (10 μg/mL, 25°C, 2 hours) using BP. Cells were then either left untreated or exposed to LPS (2 hours). Supernatants were analyzed by ELISA for TNFα protein (*P < .05 compared with buffer/unstimulated) and analyzed by RT-PCR. The gel shown is representative of 3 experiments. (E) PMNs were left untreated or exposed to 0.1% DMSO vehicle or 5 μg/mL cytochalasin B and analyzed by RT-PCR. The gel shown is representative of 4 experiments. (F) PMNs were either left untreated or transduced with wild-type or mutant WASP-CRIB and then exposed to C3 transferase (20 μg/mL, 4 hours). Lysates were analyzed by RT-PCR for TNFα and GAPDH. The gel shown is representative of 3 separate experiments. (G) PMNs were treated with buffer or C3 as in panel B and then with buffer or LPS (25 minutes). Cell sonicates were assayed for p65 activation (*P < .05 compared with NS/no oligo), and cytoplasmic fractions were immunoblotted for anti-IκBα. A representative immunoblot of 3 is shown. PCR gels are representative of 4 independent experiments. (A, D, G) Error bars represent SE.
RhoA negatively regulates TNFα induction in the PMN. (A) PMNs were left untreated or treated with 20 μg/mL C3 transferase (37°C, 4 hours) and then exposed either to media or to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein. (*P < .05). (B) PMNs were untreated or treated with C3 transferase, exposed to buffer or LPS (1 hour) and then analyzed by RT-PCR. PMNs were also exposed to either poly-inosine/cytidine (pIC) or LPS (4 hours) and analyzed for IFN-β and GAPDH expression. The gels shown are representative of 3 experiments. (C) PMNs were left untreated or transduced with BSA or Rhotekin-RBD (25°C, 2 hours) using BioPORTER (BP). ROCKα was immunoprecipitated, and its kinase activity was assayed.28 (D) PMNs were left untreated or transduced with BSA or Rhotekin-RBD (10 μg/mL, 25°C, 2 hours) using BP. Cells were then either left untreated or exposed to LPS (2 hours). Supernatants were analyzed by ELISA for TNFα protein (*P < .05 compared with buffer/unstimulated) and analyzed by RT-PCR. The gel shown is representative of 3 experiments. (E) PMNs were left untreated or exposed to 0.1% DMSO vehicle or 5 μg/mL cytochalasin B and analyzed by RT-PCR. The gel shown is representative of 4 experiments. (F) PMNs were either left untreated or transduced with wild-type or mutant WASP-CRIB and then exposed to C3 transferase (20 μg/mL, 4 hours). Lysates were analyzed by RT-PCR for TNFα and GAPDH. The gel shown is representative of 3 separate experiments. (G) PMNs were treated with buffer or C3 as in panel B and then with buffer or LPS (25 minutes). Cell sonicates were assayed for p65 activation (*P < .05 compared with NS/no oligo), and cytoplasmic fractions were immunoblotted for anti-IκBα. A representative immunoblot of 3 is shown. PCR gels are representative of 4 independent experiments. (A, D, G) Error bars represent SE.
Inhibition of RhoA has been described to disrupt the actin cytoskeleton (Figure 1D)47,48 and cytoskeletal remodeling to modulate signal transduction.49 Moreover, cytochalasins, which are actin depolymerizing agents, have long been known to “prime” leukocytes for certain proinflammatory functions, such as release of O
Because C3 transferase activated Cdc42, and both C3 and constitutively active Cdc42 induced TNFα, we next questioned whether induction of TNFα by C3 is Cdc42 dependent. To address this, we left PMNs untreated or transduced them with either wild-type or mutant WASP-CRIB and subsequently exposed them to C3 transferase. RT-PCR revealed that C3 induction of TNFα was modestly inhibited by pretreatment with wild-type but not mutant WASP-CRIB (Figure 3F). These results suggest that RhoA suppresses TNFα induction in the resting PMN through suppression of Cdc42 but do not rule out the existence of Cdc42-independent mechanisms of TNFα suppression by RhoA.
RhoA suppresses NF-κB activation in the resting PMN
NF-κB is a critical proinflammatory transcription factor whose activation has been tightly linked to TNFα, IL-8, and MIP-1β induction by an extensive body of literature.41,44,45 There are no reports, to our knowledge, connecting RhoA and NF-κB in the PMN. Our group has previously reported p38-dependent activation of NF-κB in the LPS-stimulated human PMN.8 Hence, we sought to establish a regulatory role for RhoA in NF-κB activation in the human PMN. Human PMNs were preincubated with buffer or C3 (20 μg/mL, 4 hours, 37°C) and then left untreated or treated with LPS (25 minutes). C3 activated p65 to a magnitude comparable to LPS and induced greater IκBα degradation than that observed with LPS (Figure 3G). Significant p65 expression was detected in the nucleus of the resting PMN (data not shown; Ear et al11 ), with little additional nuclear translocation observed with LPS and no significant translocation with C3 (data not shown). Taken together, these findings indicate that inhibition of RhoA is sufficient to activate NF-κB, and that, hence, by inference, active RhoA suppresses NF-κB in the resting human PMN. This suppression occurs at or upstream of IκBα degradation and does not appear to involve cytoplasmic sequestration of p65. Because NF-κB activation has been tightly linked to TNFα gene induction,41 these findings provide a tentative transcriptional mechanism for the increase in TNFα mRNA observed with RhoA inhibition (Figure 3B).
We next queried whether RhoA might physically associate with p65, and whether such an association might be sensitive to RhoA activation state. To address this question, we exposed PMNs to a time course of LPS and then performed pulldowns on lysates, using Sepharose-conjugated GST fusions of wt RhoA and constitutively active L63RhoA. As depicted in Figure 4A, p65 coprecipitated with wt RhoA, but minimal coprecipitation was observed with L63RhoA.
Regulatory roles of RhoA and ROCK. (A) PMNs were exposed to the indicated time course of LPS. Lysates were then precleared with 15 μg GST-Sepharose (1 hour) and incubated (2 hours) with 15 μg indicated Sepharose conjugates of GST fusion protein. Precipitates were then eluted, electrophoresed on a 10% SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted with anti-p65. Nitrocellulose membranes were stained with Ponceau S to confirm protein loading. Blots are representative of 3 independent experiments. (B-C) PMNs were either left untreated or pretreated with Y27632 (10 μM, 60 minutes) and then exposed to buffer or to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein (B) and lysates by RT-PCR for TNFα and GAPDH (C)(*P < .05 compared with LPS alone). The gel shown is representative of 3 separate experiments. (B) Error bars represent SE.
Regulatory roles of RhoA and ROCK. (A) PMNs were exposed to the indicated time course of LPS. Lysates were then precleared with 15 μg GST-Sepharose (1 hour) and incubated (2 hours) with 15 μg indicated Sepharose conjugates of GST fusion protein. Precipitates were then eluted, electrophoresed on a 10% SDS-PAGE gel, transferred to nitrocellulose, and immunoblotted with anti-p65. Nitrocellulose membranes were stained with Ponceau S to confirm protein loading. Blots are representative of 3 independent experiments. (B-C) PMNs were either left untreated or pretreated with Y27632 (10 μM, 60 minutes) and then exposed to buffer or to LPS (2 hours). Supernatants were tested by ELISA for TNFα protein (B) and lysates by RT-PCR for TNFα and GAPDH (C)(*P < .05 compared with LPS alone). The gel shown is representative of 3 separate experiments. (B) Error bars represent SE.
ROCK, a RhoA effector, positively regulates LPS-induced TNFα expression
The marked activation of NF-κB and induction of TNFα seen after C3 pretreatment in resting PMNs made it difficult for us to draw firm conclusions about the regulatory role of RhoA in subsequent LPS induction of these processes (Figure 3A-B,G). Hence, we tested the regulatory role of specific RhoA effectors. ROCK, one of several effectors of RhoA,54 has been implicated in both transcription and protein synthesis.55 Having confirmed activation of ROCKα by LPS in the human PMN, placing it downstream of p38 (Figure 1E) and RhoA (Figure 3C), we next tested the regulatory role of ROCK in LPS-induced TNFα expression with the use of Y27632, a ROCK inhibitor. As shown in Figure 4, Y27632 was not sufficient to induce TNFα in the resting PMN, indicating that RhoA suppression of TNFα expression in the resting cell is mediated either by RhoA itself or by a RhoA effector other than ROCKα. Of interest, Y27632 reduced LPS-induced TNFα protein but not gene expression, identifying an additional TNFα protein-promoting role for RhoA in the context of the LPS cascade counter to its TNFα-suppressive role in the resting cell.
Discussion
The prototypical acute-response cytokine TNFα is produced by multiple different cell types and plays a variety of roles, beneficial and deleterious, in inflammation and host defense.1,2 Perhaps as a consequence, TNFα induction is precisely controlled,3-5 with important differences among cell types.5,7,8,56 Fundamental differences exist between RhoA signaling in the human PMN and transfectable cell lines, such as HEK293s.57 Hence, these lines cannot be used to model RhoA signaling in the human PMN. Moreover, the human PMN differs in several important ways from other leukocytes, likely because of its unique role as a short-lived, acute-response effector cell.15,21 An improved understanding of the specific regulatory mechanisms underlying TNFα induction in the PMN provides an opportunity for better defining the controls underlying the “resting” and “activated” states of the PMN itself.
Rho GTPases have been extensively studied in the PMN, wherein they have been classically described to mediate cytoskeletal-related proinflammatory functions.17 In the present study, we applied protein transduction tools to the human PMN to describe novel roles for RhoA and Cdc42 in TNFα induction that differ between the resting and LPS-stimulated state (Figure 5). Inhibition of RhoA by 2 independent approaches was sufficient to induce TNFα transcript and protein in the resting PMN, whereas specific inhibition of the RhoA effector, ROCKα, inhibited LPS-induced TNFα protein expression. These findings indicate that, downstream of RhoA, at least 2 antagonistic TNFα-regulatory pathways must exist: (1) a ROCK-independent pathway that tonically inhibits TNFα transcript and protein expression and (2) a ROCK-dependent pathway that promotes LPS-induced TNFα protein expression (Figure 5). Our observation that RhoA inhibition is sufficient to activate NF-κB (Figure 3G) suggests that the former, ROCK-independent pathway, likely operates, at least in part, through suppressing NF-κB–regulated gene transcription. Our results obtained with L61Cdc42 and WASP-CRIB transduction suggest, moreover, that RhoA suppresses cytokine induction in the resting PMN, at least in part, through inhibiting Cdc42. Consistent with our findings, antagonistic effects have previously been reported between alternate Rho effectors,18,19 and apparent contrary effects of RhoA in resting versus stimulated cells have been explained by stimulus-dependent redirection of signaling among RhoA effectors.58
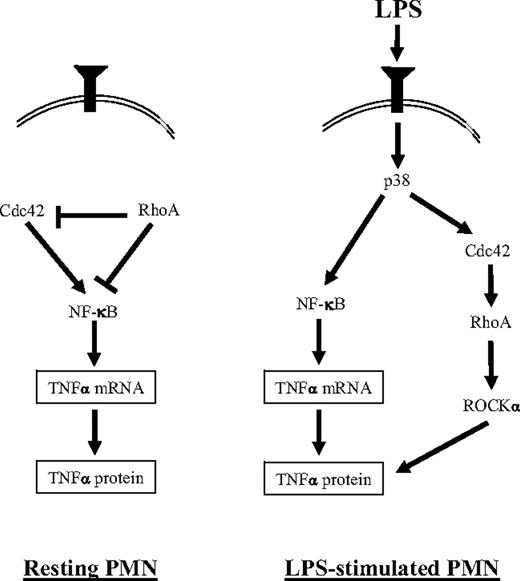
A proposed scheme for regulation of TNFα induction in the LPS-stimulated human PMN. (Left) In the resting cell, RhoA suppresses NF-κB activation and TNFα expression. This may occur through its inhibition of Cdc42 and possibly also through a Cdc42-independent mechanism. RhoA activity in the resting PMN is p38- and Cdc42-independent. (Right) In the LPS-stimulated PMN, we have previously reported that p38 is activated8 and promotes the activation of NF-κB8 and Cdc42.30 LPS-activated Cdc42 regulates RhoA activation. Downstream of RhoA, ROCKα is activated and promotes TNFα protein expression. LPS-activated p38 and Cdc42 may regulate TNFα protein through ROCKα, but ROCKα-independent pathways are also possible (not shown).
A proposed scheme for regulation of TNFα induction in the LPS-stimulated human PMN. (Left) In the resting cell, RhoA suppresses NF-κB activation and TNFα expression. This may occur through its inhibition of Cdc42 and possibly also through a Cdc42-independent mechanism. RhoA activity in the resting PMN is p38- and Cdc42-independent. (Right) In the LPS-stimulated PMN, we have previously reported that p38 is activated8 and promotes the activation of NF-κB8 and Cdc42.30 LPS-activated Cdc42 regulates RhoA activation. Downstream of RhoA, ROCKα is activated and promotes TNFα protein expression. LPS-activated p38 and Cdc42 may regulate TNFα protein through ROCKα, but ROCKα-independent pathways are also possible (not shown).
We propose a working model (Figure 5) to synthesize our present findings in the context of previous literature. In the resting cell, inhibition of p38 and Cdc42 do not recapitulate the positive effects on TNFα expression observed with inhibition of RhoA, likely because RhoA activity in the resting cell is p38- and Cdc42-independent (Figures 1A-B and 5). This differs from the LPS-stimulated state, wherein RhoA activity is both p38 and Cdc42 dependent (Figure 1A-B). These findings suggest that a condition (eg, an activated molecule) characteristic of the LPS-stimulated, but not resting PMN, may be required for p38 and Cdc42 to regulate (ie, be “upstream” of) RhoA activity. In the context of LPS stimulation, inhibition of p38 and Cdc42 both reduce TNFα protein8 (Figure 2), possibly through regulation of ROCK downstream (Figure 5). Although the predominant regulatory effect of p38 on TNFα may be through this Cdc42-mediated pathway, our previous finding of subtle, transient effects of p38 on LPS-induced TNFα transcripts and on NF-κB activation8 (neither of which were observed with Cdc42 inhibition in the present study) suggests that p38 may also regulate TNFα through Cdc42- and RhoA-independent means (Figure 5), for example, through MK2, as has been reported.59-61 Nevertheless, our data do not exclude a role for Rho GTPases in LPS-induced NF-κB activation in the human PMN.
In vivo, the human PMN is unlikely to encounter LPS as a solitary stimulus in isolation from cytokines or other pathogen-associated molecular patterns. We speculate that the multiple-level, counter-regulatory controls that we have observed may allow for integration of extracellular signals at hierarchical signaling hubs that permit for contextual control over TNFα production, as well as for coordination of TNFα production with other Rho GTPase-dependent functions. As uncontrolled TNFα release may well be maladaptive,1 tonic activation of RhoA by stimuli such as adhesion26 may serve as a natural “brake” on induction of cytokines in PMNs localized to sites of inflammation. In this respect, the PMN differs significantly from other cells, such as endothelial and monocytic cells, in which RhoA plays an NF-κB– and TNFα-promoting role.13,15,20,22
Limitations of the present study should be noted. We cannot be certain that transduced L61Cdc42 has all of the necessary features of endogenous Cdc42 for RhoA activation or that we sampled the correct time point for RhoA activation following L61Cdc42 transduction. Because we did not confirm toxin B stoichiometry or assay all aspects of p38 activation, we cannot be fully certain that p38 activation is Rho independent in all respects. How closely the ex vivo isolated PMN models the in vivo resting PMN was not addressed. The RhoA effector(s) operative in the ROCK-independent suppressive mechanism in the resting PMN was not identified in the present study and will be the focus of future efforts. Of interest, examples exist in the literature of both cooperative and antagonistic interactions among RhoA effectors,18,19,62 of coregulatory proteins such as CNK1 that direct signaling specificity downstream of RhoA,63 and of proteins that inhibit specific RhoA effectors.64 Hence, the potential exists for sophisticated cellular control over signaling specificity downstream of RhoA. Moreover, the existence of multiple independent RhoA effectors provides precedence for distinct, and even contrary, roles for RhoA under different states of cellular activation, as seen in the present study.
Finally, our data may suggest an additional mechanism of clostridial pathogenesis: “inappropriate” elaboration of proinflammatory cytokines. Nevertheless, the potential for dynamic RhoA inhibition in biologic systems exists not only through the action of bacterial toxins but also through host endogenous proteins. For example, cadherin engagement,65 serine phosphorylation,66 tenascin-C,67 and c-AMP–dependent protein kinase68 are reported to inhibit RhoA and the Rnd proteins to inhibit RhoA and ROCK.69,70 Such reports raise the intriguing possibility that TNFα induction in the human PMN may, in fact, in some situations be the consequence of dynamic signals inhibiting RhoA or its effectors. Extrapolating to the therapeutic front, a potential role for Rho GTPase inhibitors, such as hydroxy-methylglutaryl coenzyme A reductase inhibitors, has been reported in sepsis and acute pulmonary inflammation.71 However, our findings, taken together with those of others,12,71 suggest that systemic treatment with RhoA inhibitors may carry some cell-specific, untoward proinflammatory effects warranting further evaluation.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael B. Fessler, National Institute of Environmental Health Sciences, 111 T. W. Alexander Dr., PO Box 12233, MD D2-01, Research Triangle Park, NC 27709; e-mail: fesslerm@niehs.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Klaus Hahn for providing the WASP-CRIB constructs, and Yu Hong Liu for assistance with recombinant protein expression.
This work was supported by the American Heart Association (grant 0275035N) (M.B.F.) and the National Institutes of Health (grant 5R01HL061407-08) (G.S.W.), (grant 5P01HL68743-04) (J.A.N.), and (grant HL67179) (P.G.A.).