Abstract
To identify genes that are commonly dysregulated in a murine model of acute promyelocytic leukemia (APL), we first defined gene expression patterns during normal murine myeloid development; serial gene expression profiling studies were performed with primary murine hematopoietic progenitors that were induced to undergo myeloid maturation in vitro with G-CSF. Many genes were reproducibly expressed in restricted developmental “windows,” suggesting a structured hierarchy of expression that is relevant for the induction of developmental fates and/or differentiated cell functions. We compared the normal myeloid developmental transcriptome with that of APL cells derived from mice expressing PML-RARα under control of the murine cathepsin G locus. While many promyelocyte-specific genes were highly expressed in all APL samples, 116 genes were reproducibly dysregulated in many independent APL samples, including Fos, Jun, Egr1, Tnf, and Vcam1. However, this set of commonly dysregulated genes was expressed normally in preleukemic, early myeloid cells from the same mouse model, suggesting that dysregulation occurs as a “downstream” event during disease progression. These studies suggest that the genetic events that lead to APL progression may converge on common pathways that are important for leukemia pathogenesis.
Introduction
For several years, investigators have used gene expression profiling to define genes and pathways that are dysregulated in acute myeloid leukemia (AML) cells.1-8 These efforts have been hampered by the difficulty in obtaining normal hematopoietic cells at the same stage of hematopoietic development for comparison purposes.9,10 While gene expression profiling studies have been able to correlate expression “signatures” with a number of translocations and other cytogenetic abnormalities,11-13 it is not yet clear whether these expression signatures represent the direct consequences of the translocations or the downstream events that these translocations set in motion.
In an attempt to define gene expression signatures at various times during myeloid development, Borregaard and colleagues (Bjerregaard et al14 and Theilgaard-Mönch et al15 ) have developed techniques for enriching human myeloid cells at early (ie, promyelocyte predominant), mid (ie, metamyelocyte predominant), and late (ie, neutrophil predominant) stages of myeloid development for expression analysis. Expression studies performed on these enriched samples were very useful, since the cells were minimally manipulated and not maintained in culture. These studies are possible with human myeloid cells because of the large number of starting cells that can be obtained from normal human marrow samples. In mice, however, studies of this kind are neither practical nor feasible. To overcome this problem, McLemore et al16 developed an in vitro culture system that uses primary hematopoietic progenitor cells as the starting material for the induction of myeloid differentiation in vitro with G-CSF. G-CSF induces a coordinated wave of myeloid differentiation in these cultures, so that morphologic promyelocytes are the predominant cell in culture on days 2 to 3, mid myeloid cells are most abundant on days 4 to 5, and terminally differentiated myeloid cells are enriched on days 6 to 7.17 While this system is different from the purification approach described for human myeloid cells,14,15 it does provide a method for obtaining highly enriched populations of murine myeloid cells at well-defined stages of differentiation. It also allows for the assessment of myeloid progenitors prior to the promyelocyte stage.
In this report, we use the murine myeloid differentiation system and array-based gene expression profiling to define the “transcriptome” at various stages of myeloid differentiation. The patterns of gene expression observed correlate closely with the patterns seen with enriched human myeloid populations. “Waves” of transcriptional activation and repression are apparent during myeloid differentiation, and are highly reproducible. Signature patterns of gene expression correspond with morphologic stages of myeloid maturation. To determine whether the murine myeloid transcriptome database could be used to define dysregulated genes in transformed promyelocytes, we performed gene expression profiling on acute promyelocytic leukemia (APL) cells derived from “knock-in” animals that express PML-RARα under the control of the murine cathepsin G locus,18 and compared this to the reference myeloid development data set. While transformed promyelocytes expressed many of the signature genes of promyelocytes,19 the expression profiles of APL cells were distinct. A number of well-characterized genes were consistently dysregulated in APL cells. The same genes were expressed normally in preleukemic early myeloid cells from the same mouse model, suggesting that these genes are not direct targets of PML-RARα, but rather distal events that are important for APL pathogenesis.
Materials and methods
In vitro myeloid differentiation
These experiments were performed exactly as described.17 Two experiments were performed with 2 independent groups of 10 to 15 C57Bl/6 donor mice that were 6 to 8 weeks of age. In brief, mice were treated with 150 mg 5-fluorouracil per kilogram intraperitoneally, and 48 hours later, marrow was harvested and plated in Dulbecco modified Eagle medium containing 20% fetal calf serum, 100 ng murine stem cell factor (SCF; R&D Systems, Minneapolis, MN) per milliliter, 6 ng murine interleukin-3 (R&D Systems) per milliliter, 50 ng murine FLT3 ligand (R&D Systems) per milliliter, and 10 ng human thrombopoietin (PeproTech, Rocky Hill, NJ) per milliliter. After 72 hours, mononuclear cells were purified by centrifugation on Histopaque-1077 (Sigma, St Louis, MO), and 100 000 Lineage−/Sca+ light-density cells were collected on a MoFlo high-speed cell sorter (DakoCytomation, Fort Collins, CO). Those cells were plated in Dulbecco modified Eagle medium with 20% fetal calf serum containing 100 ng SCF per milliliter and 100 ng human G-CSF (Amgen, Thousand Oaks, CA) per milliliter. Cells were harvested daily for total RNA collected with RNeasy (Qiagen, Valencia, CA), manual cell counts, and May-Grünwald-Giemsa staining of cytospin-prepared cells (Sigma).
Murine leukemia samples
Acute promyelocytic leukemia cells from the bone marrow and/or spleens of 22 independent mCG-PML-RARα knock-in mice were harvested and cryopreserved as previously described.20
RNA isolation and microarray processing
Total cellular RNA was purified using the Trizol reagent (Invitrogen, Carlsbad, CA), quantified using UV spectroscopy (Nanodrop Technologies), and qualitatively assessed using a BioAnalyzer 2100 and RNA NanoChip assay (Agilent Technologies, Palo Alto, CA). For some samples (as indicated), 100 ng total cellular RNA was linearly amplified, labeled, and hybridized to Affymetrix MOE430v2.0 GeneChip microarrays (Affymetrix, Santa Clara, CA) using standard protocols from the Siteman Cancer Center Multiplexed Gene Analysis Core Facility (for protocols, see http://pathimm.wustl.edu/∼mgacore/index.htm).
Analysis of array data from the murine myeloid differentiation sets
To perform interarray comparisons, the raw scan data from each microarray were scaled to a target intensity of 1500 using the Affymetrix GCOS 1.2 (MAS 5) statistical algorithm. Scaled data for each array were exported to the Siteman Cancer Center Bioinformatics Server (http://bioinformatics.wustl.edu), merged with updated gene annotation data for each probe set on the array, and downloaded for further data visualization and analysis. The completely annotated data set can be found at this URL. Further statistical analysis, hierarchic clustering, and data visualization were performed using DecisionSite for Functional Genomics software (Spotfire, Somerville, MA).
To compare data generated from 2 independent sets of myeloid differentiation assays (set 1 and set 2), the signal value for a probe set at a single time point was expressed as a percentage of the maximum value of that probe set across all time points within each set. Differentially expressed probe sets were then selected using 3 filters: (1) positive detection call (“P”) and more than 150 signal value in at least one sample; (2) a “significance” value of P below .05 (uncorrected for multiple comparisons) resulting from a t test between the percentage changes for both data sets at any 2 time points between days 0 and 7; (3) more than 2-fold changes in signal intensity between those 2 time points for both data sets. For heat map displays of data, either the percentage of maximal expression or z-score values were used, as indicated. In cases where multiple probe sets represented a single gene, the probe set with the most “present” calls and the highest average intensity was used as the representative probe set for the gene.
Identification of dysregulated genes in APL cells
Algorithms were designed to discover genes with altered expression patterns in APL cells when compared with normal promyelocytes. Each of the 4 independent analyses began with the complete set of probe sets from the Affymetrix MOE430v2 GeneChip array (45 037 probe sets after removal of the Affymetrix controls). Genes were then selected in sequence, all criteria were required to be met in both biological replicate samples of normal differentiated myeloid cells. The algorithm for set A was designed to isolate genes with the following pattern: high expression on day 0, low expression on day 2, high expression in APL. The following 3 criteria were for set A: (1) select genes whose maximum expression is on day 0 and with expression signals more than 5000 units; (2) select genes with decreasing expression signals from day 0 to day 7 (genes with the most negative slopes); and (3) select genes with expression in the APL samples consistently equal to or greater than in both normal myeloid cell replicates. The algorithm for set B was designed to isolate genes with the following pattern: high expression on day 7, low expression on day 2, high expression in APL. The following 3 criteria were for set B: (1) as in set A, except that the reference is day 7; (2) select genes with increasing expression signals from day 0 to day 7 (genes with the most positive slopes); and (3) as in set A. The algorithm for set C was designed to isolate genes with the following pattern: high expression on day 2, lower expression on all other days, low expression in APL. The following 2 criteria were for set C: (1) select genes whose expression peaks on day 2 and with expression signals more than 5000 units; and (2) select genes with expression in the APL samples consistently lower than in both normal myeloid cell replicates (ratio of APL/day 2 < 0.2). The algorithm for set D was designed to isolate genes with the following pattern: low expression on day 2, relatively low expression on all other days, high expression in APL. The following 3 criteria were for set D: (1) select genes with consistently high expression in the APL samples (> 5000 units); (2) select genes with expression on day 2 in both normal myeloid cell replicates lower than in the APL samples (ratio of day 2/APL < 0.2); and (3) subtract genes that are highly expressed on other days (slope near 0).
Gene ontology, biological process, and pathway analysis
Gene ontology analysis was performed using the DecisionSite Gene Ontology browser, the Gene Ontology (GO) database,21 and Affymetrix annotation files (www.affymetrix.com). Biological process analysis was performed with the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (https://panther.appliedbiosystems.com). Pathway analysis was performed with PathwayArchitect software (Stratagene, La Jolla, CA).
Real-time quantitative reverse transcription–polymerase chain reaction (qRT-PCR) analysis
cDNA was made from nonamplified RNA using the Taqman reverse-transcription reagent (Applied Biosystems, Foster City, CA). Real-time PCR assays were performed using SYBR green PCR master mix (Applied Biosystems) with 400-nM concentrations of each oligonucleotide primer. The primers used for the amplification of each cDNA were as follows: Fos: 5′-GGACAGCCTTTCCTACTACCATTC-3′ (forward) and 5′-CACTAGAGACGGACAGATCTGC-3′ (reverse); Jun: 5′-CTGAACTGCATAGCCAGAACACG-3′ (forward) and 5-GCAGGCTGGCGCTGTAGCCAC-3 (reverse); Egr-1: 5′-GAACAACCCTATGAGCACCTGAC-3 (forward) and 5-AAGCGGCCAGTATAGGTGATG G-3 (reverse); Tnf: 5′-CGCTCTTCTGTCTACTGAACTTCG-3′ (forward) and 5′-GACGTGGGCTACAGGCTTGTC-3′ (reverse); Vcam-1: 5′-GTGAAGGGATTAACGAGGCTGG-3′ (forward) and 5′-GTGCAGGAGATAATGACGGTGTC-3 (reverse); and glyceraldehyde-3-phosphate dehydrogenase (Gapdh): 5′-GTGATGGGTGTGAACCACGAG-3′ (forward) and 5′-TGGCAGTGATGGCATGGACTG-3′ (reverse).
The raw expression values for Fos, Jun, Egr-1, Tnf, and Vcam-1 mRNA abundance were normalized to Gapdh mRNA abundance. All assays were performed in duplicate.
Results
Defining the normal murine myeloid developmental transcriptome
To characterize the transcriptional programs that govern myeloid differentiation, we performed RNA expression profiling analyses of cells derived from a G-CSF–dependent in vitro myeloid differentiation system, as previously described.16,17 These enriched progenitors undergo a coordinated wave of myeloid maturation, as demonstrated in Figure 1A-B. Cells were harvested on day 0 through day 7 for gene expression microarray analysis. Two completely independent sets of data from 2 different sets of 10 to 15 C57Bl/6 donor mice were created and compared in all studies. While small differences were present between the 2 independent experiments, overall patterns of gene expression were highly reproducible between the data sets (Figures 1, 2, and 5, and data not shown).
G-CSF–driven myeloid differentiation from day 0 to day 7. (A) Morphology of G-CSF–treated cells from day 0 to day 7. Cytospins were made with cells harvested on days 0 to 7, and differentials were performed after May-Grunwald-Giemsa staining. (Sigma, St. Louis, MO). Cells were visualized using a Nikon Microphot-SA microscope (Nikon, Tokyo, Japan) with a 100×/1.40 oil objective. Images were captured using a Colorview II camera (Soft Imaging System, Lakewood, CO), and images were produced with analySIS software (Soft Imaging System). The images were not edited. (B) Differential counts for early mononuclear cells, early myeloid cells (promyelocytes), mid myeloid cells (myelocytes and metamyelocytes), and late myeloid cells (bands and neutrophils) from day 0 to day 7. Two hundred cell differentials were performed for each day. (C) Changes in the expression of known early, mid, and late myeloid genes during G-CSF–driven myeloid differentiation in 2 independent gene expression analyses (set 1 and set 2). “Early” genes include CD34, fms-related tyrosine kinase 3 (Flt3), Kit, and Sca1; “mid” genes include cathepsin G (Ctsg), myeloperoxidase (Mpo), neutrophil elastase (Ela2), and proteinase 3 (Prtn3); “late” genes include CD11b (Itgam), formyl peptide receptor 1 (Fpr1), lysozyme (Lyzs), and Mmp9. Signal intensity values (normalized to whole array target intensity of 1500 for all probe sets) are displayed on the y-axis. The inset in the “early” panel displays expression values on a linear scale instead of a log scale.
G-CSF–driven myeloid differentiation from day 0 to day 7. (A) Morphology of G-CSF–treated cells from day 0 to day 7. Cytospins were made with cells harvested on days 0 to 7, and differentials were performed after May-Grunwald-Giemsa staining. (Sigma, St. Louis, MO). Cells were visualized using a Nikon Microphot-SA microscope (Nikon, Tokyo, Japan) with a 100×/1.40 oil objective. Images were captured using a Colorview II camera (Soft Imaging System, Lakewood, CO), and images were produced with analySIS software (Soft Imaging System). The images were not edited. (B) Differential counts for early mononuclear cells, early myeloid cells (promyelocytes), mid myeloid cells (myelocytes and metamyelocytes), and late myeloid cells (bands and neutrophils) from day 0 to day 7. Two hundred cell differentials were performed for each day. (C) Changes in the expression of known early, mid, and late myeloid genes during G-CSF–driven myeloid differentiation in 2 independent gene expression analyses (set 1 and set 2). “Early” genes include CD34, fms-related tyrosine kinase 3 (Flt3), Kit, and Sca1; “mid” genes include cathepsin G (Ctsg), myeloperoxidase (Mpo), neutrophil elastase (Ela2), and proteinase 3 (Prtn3); “late” genes include CD11b (Itgam), formyl peptide receptor 1 (Fpr1), lysozyme (Lyzs), and Mmp9. Signal intensity values (normalized to whole array target intensity of 1500 for all probe sets) are displayed on the y-axis. The inset in the “early” panel displays expression values on a linear scale instead of a log scale.
Microarray analysis of myeloid differentiation. (A) Heat map of the 27 127 “expressed” probe sets (with MAS 5 “present” call and > 150 signal units for at least one day) in experimental set 1, normalized to the percentage of the maximal value of each probe set from days 0 to 7. (B) Definition of the most highly expressed probe sets on each day of differentiation (100 for each day) chosen by z-score analysis. A heat map displaying z-score values for each of the selected probe sets on all days of differentiation are shown. (C) The top 100 probe sets for each day (as displayed in B) were replotted with the data from experimental set 2. Day-1 data were not available for this set. (D) Graphic representation of the absolute signal values from the top 100 probe sets on each day of myeloid differentiation from set 1, as defined in panel B. (E) Biological process classification of the genes shown in panel B.
Microarray analysis of myeloid differentiation. (A) Heat map of the 27 127 “expressed” probe sets (with MAS 5 “present” call and > 150 signal units for at least one day) in experimental set 1, normalized to the percentage of the maximal value of each probe set from days 0 to 7. (B) Definition of the most highly expressed probe sets on each day of differentiation (100 for each day) chosen by z-score analysis. A heat map displaying z-score values for each of the selected probe sets on all days of differentiation are shown. (C) The top 100 probe sets for each day (as displayed in B) were replotted with the data from experimental set 2. Day-1 data were not available for this set. (D) Graphic representation of the absolute signal values from the top 100 probe sets on each day of myeloid differentiation from set 1, as defined in panel B. (E) Biological process classification of the genes shown in panel B.
To determine whether G-CSF–driven myeloid maturation parallels gene expression patterns from human myeloid precursors, we examined the expression of a number of genes whose expression patterns have been characterized in enriched human myeloid precursors. Several genes that are known to be expressed in early hematopoietic progenitors (CD34, Flt3, Kit, and Sca1) were expressed at their highest levels at days 0 to 1 in both experiments. Azurophil granule–specific genes, which are expressed at their highest levels in promyelocytes, were highly induced between days 0 and 2 (Ctsg, Mpo, and Prtn3) or days 0 and 4 (neutrophil elastase [NE; E1a2]. Finally, genes associated with terminal myeloid differentiation (CD11b [Itgam], Fpr1, Lyzs, and Mmp9) demonstrated peak expression at days 6 to 7. Data from both myeloid maturation sets are shown in Figure 1C; the patterns are highly similar in both sets. We used the data from set 1 for discovery, and validated all results with set 2 (which is missing data from day 1 due to inadequate numbers of cells harvested on that day).
We used several analytical approaches to define differentially expressed genes within the myeloid differentiation data sets. We first defined the 27 127 “expressed” probe sets (MAS 5 “present” call, and normalized signal value of > 150 in at least one sample) from all 8 arrays in experimental set 1. As shown in Figure 2A (a heat map of the 27 127 expressed probe sets from set 1, scaled to the percentage of the maximal value for each probe set between days 0-7), this analysis revealed that a number of genes are differentially expressed as a function of time in culture (for this and several other designated heat maps, data were normalized to percent of maximal expression, so that small alterations in absolute expression levels would not be overemphasized22 ; an alternative method for showing variation among probe sets [the z-score] is designed to expand differences among probe sets to similar “scales,” no matter how much absolute variation occurs). While the hybridization signals from many probe sets do not change during myeloid differentiation, many others display patterns suggesting that expression is strongly regulated.
We subsequently focused on 8225 of the differentially regulated probe sets that had at least one signal value of more than 1000 (an arbitrarily chosen value designed to focus the analysis on probe sets with relatively high expression levels at some point during myeloid development). We defined the 100 probe sets that were most characteristically expressed on a single day of myeloid differentiation using z-score statistics. These data are displayed in Figure 2B, which demonstrates that these signature genes are generally expressed at peak values for a very limited time during myeloid differentiation. Analysis of the expression values of these probe sets from experimental set 2 revealed that these expression patterns are similar and reproducible (Figure 2C). We also plotted the actual expression values from the 100 daily signature genes for each day in Figure 2D. Striking changes in the absolute expression levels of many genes are clearly evident, validating the methods used to define these signature probe sets. Biological process analysis by the PANTHER classification revealed the percentage of gene “hits” with respect to several fundamental biological processes (Figure 2E). Heat maps demonstrating patterns of expression for many of these annotated genes are presented as Figures S1–S3, available on the Blood website (see the Supplemental Materials link at the top of the online article). A number of genes previously associated with AML pathogenesis23-25 were also developmentally regulated during myeloid maturation (Figure S4; Table S1).
Using the normal myeloid transcriptome to define genes that are dysregulated in acute promyelocytic leukemia cells
Our laboratory previously developed 2 mouse models of APL18,26 ; in this study, we used the model where the bcr-1 isoform of PML-RARα cDNA was knocked into the 5′ untranslated region of the murine cathepsin G gene (mCG-PML-RARα),18 since the penetrance of APL in this model is nearly 100%.18,27 Most bone marrow– and spleen-derived APL cells display aberrant coexpression of CD34 (and/or c-Kit) and Gr-1 on the cell surface.20,28 All but one of the APL samples used for profiling analysis in this study were highly enriched for this malignant population (Table S2). For many years, our laboratory has banked splenic cells from overtly leukemic mice as a source of material for flow analysis and genetic analysis (since the spleens are very large, as many as 109 cells can be banked from each animal). To determine whether leukemic spleen cells can serve as an accurate surrogate for leukemic bone marrow cells in gene expression studies, we compared the expression profiles of the spleen cells and bone marrow cells from 4 leukemic mCG-PML-RARα mice (Table S2; APL samples 19-22). The percentage of CD34+/Gr-1+ cells in the bone marrow and spleen samples was similar for all pairs for which data were available (Table S2). We compared the expression profiles of these APL samples with that of normal bone marrow and spleen samples (Figure 3A). Unsupervised hierarchical clustering analysis of these 14 data sets revealed that all APL samples cluster separately from the wild-type spleen and marrow samples, as expected. Each APL sample had a unique and reproducible expression signature revealed by the clustering analysis, probably reflecting the heterogeneity of mutations that contribute to APL progression in these tumors.29 Of importance, the spleen and bone marrow samples from each individual tumor clustered together; the profiles of APL cells from the 2 different tissue sources are therefore highly concordant. The expression profiles obtained from RNA derived from cryopreserved versus unmanipulated spleen cells were not significantly different (data not shown). We therefore used cryopreserved leukemic spleen samples as the source of APL cells for all subsequent studies in this report.
The expression phenotype of murine APL cells. (A) Unsupervised hierarchic clustering analysis of RNA array data (32 420 probe sets) from the spleens and bone marrows of 4 independent mCG-PML-RARα mice with APL, 4 normal murine bone marrow samples, and 2 normal murine spleen samples. Expression data on the heat map are displayed as z-scores. (B) Expression levels of 12 genes associated with early, mid, and late myeloid differentiation (during murine G-CSF–induced differentiation, days 0-1: blue bars; days 2-3: pink bars; days 4-7: blue-green bars), and in 6 murine APL samples (red indicates averages for 6 samples; brackets, SEMs). (Inset) Expression levels of PU.1 on an expanded scale. (C) Heat map of expression data from set 1, and 6 independent APL samples. Probe sets (4244) with expression values peaking on day 2 were selected for analysis. Expression is shown as a ratio of each sample to the reference sample (day 2). Data from the same differentiation experiment (set 1) were displayed in panels B-C.
The expression phenotype of murine APL cells. (A) Unsupervised hierarchic clustering analysis of RNA array data (32 420 probe sets) from the spleens and bone marrows of 4 independent mCG-PML-RARα mice with APL, 4 normal murine bone marrow samples, and 2 normal murine spleen samples. Expression data on the heat map are displayed as z-scores. (B) Expression levels of 12 genes associated with early, mid, and late myeloid differentiation (during murine G-CSF–induced differentiation, days 0-1: blue bars; days 2-3: pink bars; days 4-7: blue-green bars), and in 6 murine APL samples (red indicates averages for 6 samples; brackets, SEMs). (Inset) Expression levels of PU.1 on an expanded scale. (C) Heat map of expression data from set 1, and 6 independent APL samples. Probe sets (4244) with expression values peaking on day 2 were selected for analysis. Expression is shown as a ratio of each sample to the reference sample (day 2). Data from the same differentiation experiment (set 1) were displayed in panels B-C.
To identify genes that are dysregulated in APL samples, we compared the expression patterns of normal myeloid cells at different stages of development with that of APL cells. The profiling studies performed with the G-CSF–differentiated bone marrow cells were all amplified before they were labeled, since the number of cells available for analysis was very small. To compare gene expression data from murine APL tumors with these data sets, we analyzed amplified RNA derived from the spleens of 6 independent mice that were clinically ill from the effects of widespread leukemia. The characteristics of these tumors (APL1-6) are described in Table S2.
Despite some variability in the percentages of CD34+/Gr-1+ cells in these samples (42%-92%), the expression profiles of these 6 mouse tumors all exhibited a distinct promyelocyte signature, in that all tumors expressed high levels of most promyelocyte-specific genes (Figure 3B). c-Kit is relatively overexpressed in APL cells compared with normal promyelocytes (ie, it is dysregulated in APL cells), a finding that has been previously described.28 Genes that are normally activated during the promyelocyte stage of development (Mpo, Ctsg, and NE [Ela2]) were highly expressed in all of the murine APL samples; levels of Prtn3 were somewhat lower in APL cells than in enriched promyelocytes. PU.1 (Sfpi1), a transcription factor that is known to be down-regulated by PML-RARα,29,30 is expressed at lower levels in APL samples than in enriched promyelocytes, as expected (Figure 3B, inset).
To begin to define the repertoire of dysregulated genes in APL cells, we identified all probe sets that displayed maximal signals on day 2 of myeloid differentiation, and compared the expression of these probe sets with that of the 6 APL samples. As shown in Figure 3C, a group of 4244 probe sets was maximally expressed on day 2 for both myeloid differentiation data sets. The expression of most of these genes increases during early myeloid development and declines on later days. The expression levels for these probe sets in the 6 APL samples revealed that the majority of genes were consistently expressed at similar or higher levels in the APL samples. In contrast, approximately one third were expressed at lower levels in all 6 samples. These data suggest that there is a set of consistently dysregulated genes in transformed promyelocytes.
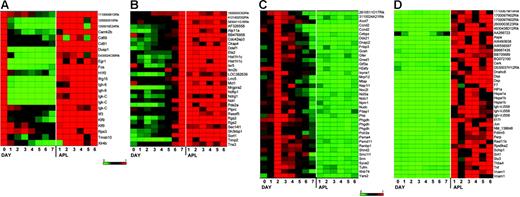
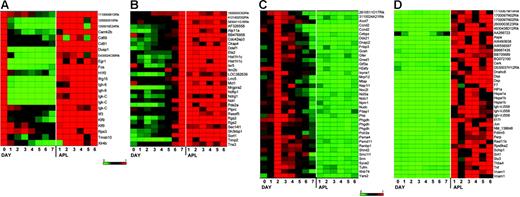
To identify the specific genes that are consistently dysregulated (referred to as the “dysregulome” for the remainder of the paper), we defined 4 independent sets of genes that displayed altered expression patterns in APL cells when compared with normal enriched promyelocytes (day-2 cells). These data are shown in Figure 4, displayed as heat maps normalized to the expression level on the day during myeloid development when expression was maximal (data used to create these figures is shown in Table S3). In Figure 4A, we defined genes that are normally expressed on day 0 but down-regulated during myeloid development (set A); these genes are persistently expressed in APL cells, and are therefore dysregulated. In Figure 4B, we defined genes that were maximally expressed on day 7 of normal myeloid development (with low-level expression on day 2 in most cases), but inappropriately expressed in most APL samples (set B). In Figure 4C, we defined genes that were normally expressed at high levels in day-2 cells, but were not expressed in APL samples (set C). In Figure 4D, we defined genes that were highly expressed in APL samples, but not expressed in day-2 cells (set D; details of the algorithms used to identify these genes are provided in “Materials and methods”). Only one of the genes identified in the APL dysregulome was previously shown to be regulated by PML-RARα expression in PR-9 cells (Tmsb10).31,32
A subset of genes is dysregulated in most murine APL samples. Expression profiles from GeneChip analyses designed to identify genes that were dysregulated in APL leukemic cells when compared with G-CSF–differentiated normal myeloid cells. APL expression was compared with 3 time points in the differentiation assay to capture the least and most differentiated myeloid cells (day 0, day 7) and the time point most enriched for promyelocytes (day 2). See “Materials and methods” for the algorithms used for each analysis (sets A-D). Heat maps show gene expression as a ratio of each sample to the reference sample in a continuous color range (green to red) of 0 to 1 (A, day 0; B, day 7; C, day 2; D, average of 6 APL samples). The same differentiation experiment (from set 1) is shown in each heat map. (A) Nineteen genes were highly expressed on day 0 and in APL cells, and had much lower expression on days 1 to 7 (set A). (B) Twenty-nine genes were highly expressed on day 7 and in APL cells, and had much lower expression on days 0 to 6 (set B). (C) Thirty-six genes were maximally expressed on day 2, had much lower expression on the other days, and in APL had less than 0.2 times the expression on day 2 (set C). (D) Thirty-two genes were highly expressed in APL cells and had much lower expression on day 2 (set D).
A subset of genes is dysregulated in most murine APL samples. Expression profiles from GeneChip analyses designed to identify genes that were dysregulated in APL leukemic cells when compared with G-CSF–differentiated normal myeloid cells. APL expression was compared with 3 time points in the differentiation assay to capture the least and most differentiated myeloid cells (day 0, day 7) and the time point most enriched for promyelocytes (day 2). See “Materials and methods” for the algorithms used for each analysis (sets A-D). Heat maps show gene expression as a ratio of each sample to the reference sample in a continuous color range (green to red) of 0 to 1 (A, day 0; B, day 7; C, day 2; D, average of 6 APL samples). The same differentiation experiment (from set 1) is shown in each heat map. (A) Nineteen genes were highly expressed on day 0 and in APL cells, and had much lower expression on days 1 to 7 (set A). (B) Twenty-nine genes were highly expressed on day 7 and in APL cells, and had much lower expression on days 0 to 6 (set B). (C) Thirty-six genes were maximally expressed on day 2, had much lower expression on the other days, and in APL had less than 0.2 times the expression on day 2 (set C). (D) Thirty-two genes were highly expressed in APL cells and had much lower expression on day 2 (set D).
Classification and validation of the commonly dysregulated genes
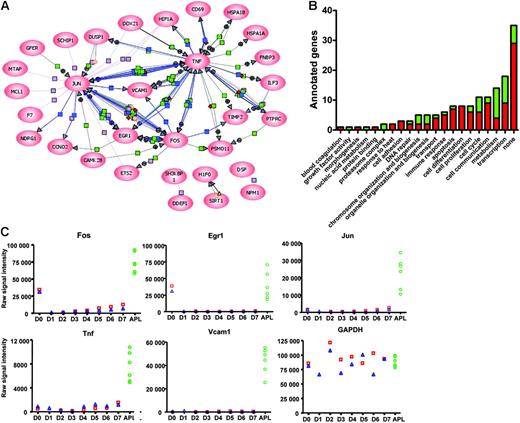
To understand whether functional relationships exist among the frequently dysregulated genes identified in Figure 4, we attempted to organize them into common pathways. Pathway analysis with PathwayArchitect software (Stratagene) allowed us to classify 31 of the 116 dysregulated genes into well-recognized pathways. From this analysis, it is clear that many genes are dysregulated as a consequence of the expression of the transcription factors Fos, Jun, and/or Egr1; Tnf and Vcam1 overexpression also accounts for additional downstream changes (Figure 5A). Further, GO annotations of all dysregulated genes revealed that genes involved in transcription, metabolism, cell communication, cell proliferation, cell cycle control, and the control of apoptosis are overrepresented in the APL dysregulome (Figure 5B).
Organization of dysregulated genes into common pathways. (A) One-hundred-and-sixteen dysregulated genes (sets A-D) were analyzed in PathwayArchitect (Stratagene) to determine a relevance interaction network. Thirty-one genes with a high confidence index of interactions were included in the pathway layout graph. Connecting lines between gene symbols indicate interactions; different types of interactions are denoted by symbols on the lines. Green square indicates regulation; purple square, binding; blue square, expression; orange circle, protein modification; red diamond, metabolism; green circle, promoter binding; yellow triangle, transport; + in gray circle, positive effect; and − in gray circle, negative effect. (B) GO annotations of the murine APL dysregulome genes. Red indicates genes with increased expression in APL cells with respect to enriched promyelocytes; green, genes with decreased expression. (C) Two grouping variable category graphs show raw probe set signal intensity values for the common pathway genes shown in panel A (and also Gapdh) for days 0 to 7 of the myeloid differentiation assay, and for 6 APL samples.
Organization of dysregulated genes into common pathways. (A) One-hundred-and-sixteen dysregulated genes (sets A-D) were analyzed in PathwayArchitect (Stratagene) to determine a relevance interaction network. Thirty-one genes with a high confidence index of interactions were included in the pathway layout graph. Connecting lines between gene symbols indicate interactions; different types of interactions are denoted by symbols on the lines. Green square indicates regulation; purple square, binding; blue square, expression; orange circle, protein modification; red diamond, metabolism; green circle, promoter binding; yellow triangle, transport; + in gray circle, positive effect; and − in gray circle, negative effect. (B) GO annotations of the murine APL dysregulome genes. Red indicates genes with increased expression in APL cells with respect to enriched promyelocytes; green, genes with decreased expression. (C) Two grouping variable category graphs show raw probe set signal intensity values for the common pathway genes shown in panel A (and also Gapdh) for days 0 to 7 of the myeloid differentiation assay, and for 6 APL samples.
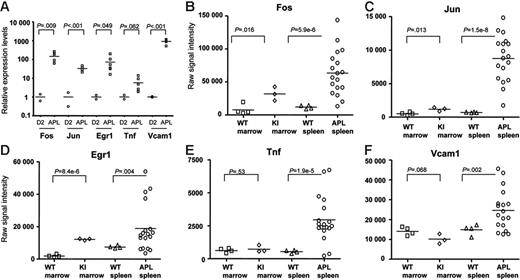
To validate the expression changes identified in the array analysis, we examined the expression of the common pathway genes (ie, Fos, Jun, Egr1, Tnf, and Vcam1) in both of the myeloid differentiation sets, and compared levels of expression with the 6 amplified APL samples described above (Figure 5C). Patterns of expression of all genes were highly concordant between the 2 myeloid differentiation sets, and were strikingly dysregulated in all 6 APL samples. We further validated these findings with qPCR using nonamplified RNA samples. We evaluated RNA from 2 independent day-2 samples from the myeloid differentiation studies, and the same 6 APL samples used above (Figure 6A). All qPCR data were normalized to Gapdh, since its expression level does not significantly change during myeloid development (Figure 5C). Fos mRNA levels were, on average, 147-fold higher in APL cells than in day-2 cells, Jun levels were 33-fold higher, Egr-1 levels were 72-fold higher, Tnf levels were 6-fold higher, and Vcam1 levels were 918-fold higher. These data therefore validate the differences in expression levels defined for these genes with microarray analysis.
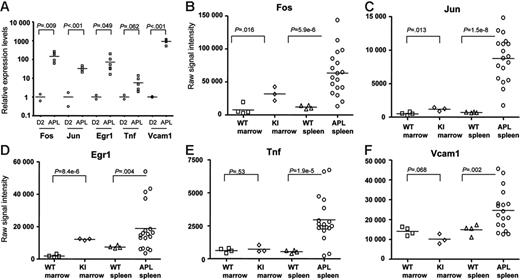
Common pathway gene expression patterns in leukemic and nonleukemic hematopoietic tissues. (A) qRT-PCR analysis of Fos, Jun, Egr-1, Tnf, and Vcam1 expression levels was performed using primers specific for each cDNA. All data were normalized to Gapdh, since the expression of this gene does not change during myeloid differentiation (Figure 5C). Day-2 samples were obtained from 2 independent myeloid differentiation experiments. The 6 APL samples used were the same as those used in the studies shown in Figures 4–5. All RNA samples used for this study were nonamplified. (B-F) Common pathway gene expression patterns in whole bone marrow and spleen samples using nonamplified RNA- and array-based expression profiling. Two grouping variable category graphs show raw probe set signal intensity values for the common pathway genes identified in Figure 5. Unmanipulated bone marrow and spleen samples were obtained from either wild-type mice (WT), or from mCG-PML-RARα mice (KI), and were cryopreserved prior to analysis. APL samples were obtained from the cryopreserved spleens of 18 overtly leukemic mCG-PML-RARα mice, including the 6 APL samples in Figures 3C, 4, and 5 (Table S2, APL samples 1-18).
Common pathway gene expression patterns in leukemic and nonleukemic hematopoietic tissues. (A) qRT-PCR analysis of Fos, Jun, Egr-1, Tnf, and Vcam1 expression levels was performed using primers specific for each cDNA. All data were normalized to Gapdh, since the expression of this gene does not change during myeloid differentiation (Figure 5C). Day-2 samples were obtained from 2 independent myeloid differentiation experiments. The 6 APL samples used were the same as those used in the studies shown in Figures 4–5. All RNA samples used for this study were nonamplified. (B-F) Common pathway gene expression patterns in whole bone marrow and spleen samples using nonamplified RNA- and array-based expression profiling. Two grouping variable category graphs show raw probe set signal intensity values for the common pathway genes identified in Figure 5. Unmanipulated bone marrow and spleen samples were obtained from either wild-type mice (WT), or from mCG-PML-RARα mice (KI), and were cryopreserved prior to analysis. APL samples were obtained from the cryopreserved spleens of 18 overtly leukemic mCG-PML-RARα mice, including the 6 APL samples in Figures 3C, 4, and 5 (Table S2, APL samples 1-18).
We also repeated array studies with 18 APL samples using nonamplified RNA (this set included the original 6 APL samples), and compared these samples with 3 normal murine bone marrow RNA samples, 4 bone marrow RNA samples from young (6-8 weeks), preleukemic mCG-PML-RARα mice, and 4 normal murine spleen samples (Figure 6B-F). Microarray expression data are displayed for the 5 central pathway genes but are representative of most of the genes in the APL dysregulome (data not shown). Levels of expression of all 5 genes are markedly higher in APL spleens than in normal bone marrow or spleen samples. The abundance of Fos, Jun, and Egr-1 mRNAs (but not Tnf or VCAM1) is significantly higher in the bone marrows of young, preleukemic mCG-PML-RARα mice than in wild-type mice. Since each of these genes is expressed at higher levels in very early myeloid cells (Figures 4–5), there are 2 potential explanations for these findings. First, since the hematopoietic compartments of young mCG-PML-RARα mice are known to be enriched for early myeloid cells,17,18 the increased expression levels may be due to an expanded population of early myeloid cells that normally express higher levels of these genes. Alternatively, PML-RARα might activate these genes directly by acting as a novel gain-of-function transcription factor. To distinguish between these possibilities, we enriched early myeloid populations from young WT versus mCG-PML-RARα mice and performed expression profiling analysis, as described in the next section.
Normal expression of the dysregulome genes in preleukemic promyelocytes
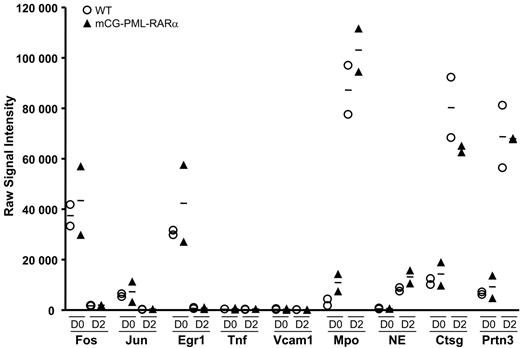
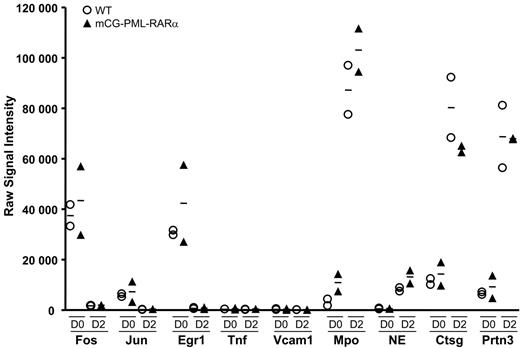
Since virtually all mCG-PML-RARα mice are destined to develop APL,18,27 young mice can be thought of as “preleukemic.” Myeloid cells from young mice can therefore be used to determine whether genes are dysregulated by PML-RARα before the mice become overtly leukemic. Accordingly, we harvested and pooled bone marrow cells from 2 additional groups of 10 young (6-8 week old) wild-type mice, and 2 groups of 10 young, preleukemic mCG-PML-RARα mice (in the C57Bl/6 background) and performed the G-CSF myeloid differentiation assay, exactly as described.17 Cells were harvested at days 0, 2, and 7 for RNA amplification and expression profiling analysis. The mutant mCG-PML-RARα allele is activated concurrently with the endogenous cathepsin G allele between days 0 and 217 ; endogenous cathepsin G is persistently expressed until day 7 (Figures 1C and 3B). The expression patterns of the central pathway genes (which are representative of all the dysregulome genes, data not shown), and also of the highly expressed azurophil granule genes of promyelocytes, are shown in Figure 7. The WT and mCG-PML-RARα samples all displayed similar patterns of expression on days 0 and 2. Fos, Jun, and Egr-1 mRNA abundance declined similarly between days 0 and 2 for both wild-type and both mCG-PML-RARα samples; these patterns persisted at day 7 (data not shown). Tnf and Vcam1 were minimally expressed in all 4 samples on all days. The expression levels of the remaining 111 dysregulome genes in the wild-type and mCG-PML-RARα samples were also carefully examined. Hspa1b was expressed 3.1-fold higher in the day-0 mCG-PML-RARα samples compared with the WT samples (P < .05); expression was not different on day 2. Perp was expressed 2.8-fold higher in the day-0 mCG-PML-RARα samples (P < .02), but was unchanged on day 2. Finally, Ccnd2 expression was reduced 55% in mCG-PML-RARα samples on day 0 (P < .03), but was unchanged on day 2. None of the other dysregulome genes exhibited significant changes in expression on either day (a global analysis of these arrays did yield a small number of reproducible changes in gene expression in cells from mCG-PML-RARα mice that will be reported separately). The promyelocyte-specific genes all demonstrated substantial increases in mRNA abundance between days 0 and 2 for both wild-type and both mCG-PML-RARα samples, indicating that myeloid differentiation was fully activated for all pools. These data show that the APL dysregulome genes are generally expressed at normal levels in the preleukemic myeloid cells of mCG-PML-RARα mice, suggesting that dysregulation of these genes occurs at a later stage in the pathogenesis of APL.
Expression patterns of common pathway genes in preleukemic early myeloid cells. Two grouping variable category graphs display raw signal intensity values for the common pathway genes, and for several abundantly expressed azurophil granule genes. Myeloid differentiation studies were performed as shown in Figure 1. Two independent pools of wild-type mice and 2 independent pools of mCG-PML-RARα mice were used in the study. Cells were harvested on days 0 and 2 of differentiation, and array-based expression analysis was performed. The data for the common pathway genes are representative of nearly all genes in the APL dysregulome.
Expression patterns of common pathway genes in preleukemic early myeloid cells. Two grouping variable category graphs display raw signal intensity values for the common pathway genes, and for several abundantly expressed azurophil granule genes. Myeloid differentiation studies were performed as shown in Figure 1. Two independent pools of wild-type mice and 2 independent pools of mCG-PML-RARα mice were used in the study. Cells were harvested on days 0 and 2 of differentiation, and array-based expression analysis was performed. The data for the common pathway genes are representative of nearly all genes in the APL dysregulome.
Discussion
In this report, we used microarray-based gene expression profiling to define genes that are developmentally regulated during G-CSF–induced myeloid maturation of primary murine hematopoietic progenitor cells. The patterns of expression observed were similar to that of enriched human myeloid cells at various stages of differentiation, suggesting that G-CSF–induced maturation in vitro mimics normal myeloid maturation in vivo. The database generated with this experiment was used to define genes that are dysregulated in murine acute promyelocytic leukemia cells. We identified a number of genes that were highly expressed in murine APL cells, but not normally expressed in enriched promyelocytes. Some of these genes were normally expressed at highest levels in very early hematopoietic progenitors, some were normally expressed in late myeloid cells, and some were not expressed at any stage of normal hematopoietic development. We also identified a small group of genes that are normally expressed in enriched promyelocytes, but not in murine APL cells. Many of the dysregulated genes had previously been linked to oncogenic transformation in AML cells and other tissues. Our data strongly suggest that a well-defined cohort of genes is consistently dysregulated in most APL tumors (the APL “dysregulome”), despite the fact that different progression events can contribute to APL pathogenesis.19
One question raised by the identification of an APL dysregulome is whether the genes identified represent direct targets of PML-RARα, or whether they represent the consequences of downstream events that were initiated by PML-RARα (but executed by other genes or pathways). The consistency and reproducibility of the dysregulated gene set among many APL samples might suggest that many of these genes are direct targets of PML-RARα. However, since the central pathway genes (and other dysregulome genes) are not overexpressed in preleukemic, enriched promyelocytes (Figure 7), this is probably not the case. More likely, the genes identified in the dysregulome are activated by additional “progression events” that occur in early myeloid cells that have an altered proliferative fate that is directly due to PML-RARα expression.17 It is still possible that persistent PML-RARα expression may have indirect effects on gene expression; for example, a recent study revealed that the Fos gene is activated by PML-RARα as a consequence of an alteration in the chromatin structure near the Fos gene, making it more accessible to transactivation by other transcription factors.33 The activation of Fos expression in APL cells may therefore represent an indirect—but specific—effect of PML-RARα on gene expression.
We have attempted to organize the dysregulated genes into pathways to better understand their potential pathophysiological relevance for APL. Pathway analysis revealed that many of the identified genes are dysregulated as a consequence of the activation of the transcription factors Fos, Jun, and/or Egr1 (Figure 5A). These genes have indeed been implicated in the pathogenesis of many tumors, including AML, and are intimately connected to the control of cell proliferation and/or apoptosis34-39 ; further, Egr1 is important for the development of the macrophage lineage.40,41 Similarly, the dysregulation of Tnf in murine APL cells may also alter the expression of a number of other target genes. Tnf is sometimes expressed by AML cells,42,43 and may promote APL development and/or progression by a variety of mechanisms, including the activation of Vcam1. Vcam1, primarily through its interaction with very late antigen 4 (VLA4; α4 β1 integrin), provides a key signal regulating the interaction of hematopoietic cells with stromal cells in the bone marrow (see Hall and Gibson for a review44 ). Vcam1 signaling has been implicated in the regulation of hematopoietic cell migration, growth, and survival. Although this gene was originally thought to be expressed primarily on stromal cells, a recent study showed that Vcam1 also is expressed on subsets of hematopoietic progenitors and myeloid cells.45 In this study, we found that Vcam1 is expressed at minimal levels during all stages of in vitro myeloid differentiation, but it is highly expressed in APL samples. Its dysregulation in APL cells may promote homotypic interactions between APL cells, which express beta integrin receptors. These interactions may alter the ability of APL cells to interact normally with stromal support cells in the bone marrow, and could potentially alter normal patterns of cellular egress from the marrow and/or responses to chemotherapy.43
We identified several additional dysregulated genes that had previously been implicated in AML or cancer pathogenesis. Examples included genes associated with antiapoptotic activities, including the Bcl2 family member, Mcl1, which is normally expressed in early hematopoietic progenitor cells, and down-regulated during normal myeloid development.46 The overexpression of the heat shock proteins may also be relevant for protecting APL cells from death; overexpression of heat shock proteins is an adverse risk factor for AML.47 Hif1α has been implicated in the pathogenesis of many tumors (including AML), and was expressed in all murine APL samples tested.48-50 Mtap1a, a tumor suppressor implicated in several solid tumor syndromes,51-55 was normally expressed in murine promyelocytes, but was expressed at very low levels in all APL samples tested. Much additional work will be required to fully understand the relationship between these dysregulated genes, and their respective roles in the pathogenesis of this disorder. Similar studies with purified human myeloid precursors and M3 AML cells are in progress.
We have used G-CSF–induced myeloid differentiation to describe genes that are highly regulated during G-CSF–driven murine myeloid differentiation in vitro. With these data sets, which are now publicly available (http://bioinformatics.wustl.edu), the expression patterns of genes during myeloid differentiation can be assessed, and the dysregulated genes of other murine AML model systems can be identified. This system has been useful for defining commonly dysregulated genes in murine APL cells and understanding the timing of activation of these genes during disease progression.
Authorship
Contribution: W.Y. and T.J.L. designed research; W.Y. performed research; D.C.L., M.A.W., and J.F.D. contributed vital new reagents or analytical tools; W.Y., M.S.H., and J.F.D. collected data; W.Y., J.E.P., and T.J.L. analyzed data; W.Y., J.E.P., and T.J.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy J. Ley, Washington University, Division of Oncology, Campus Box 8007, 660 South Euclid Ave, St Louis, MO 63110; e-mail: tley@im.wustl.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by grants NIH RO1 CA083962 and NIH PO1 CA0101937.
The authors thank the Hi Speed Flow Core, the Multiplexed Gene Analysis Core, the Tissue Procurement Core, and the Bioinformatics Core of the Siteman Cancer Center for their invaluable support. Nancy Reidelberger provided expert editorial assistance. Mieke Hoock and Kelly Schrimpf provided excellent animal husbandry. Dr Geoffrey Uy provided invaluable assistance with APL banking studies, and he and Dr Matthew Walter critically read the paper.