Abstract
Somatic mutations of immunoglobulin genes characterize mature memory B cells, and intraclonal B-cell diversification is typically associated with expansion of B-cell clones with greater affinity for antigen (antigen drive). Evidence for a role of antigen in progression of intraclonal chronic lymphocytic leukemia (CLL) cell diversification in patients with mutated immunoglobulin genes has not been previously presented. We performed a single-cell analysis of immunoglobulin heavy and light chains in 6 patients with somatically mutated CLL-cell immunoglobulin genes and identified 2 patients with multiple related (oligoclonal) subgroups of CLL cells. We constructed genealogic trees of these oligoclonal CLL-cell subgroups and assessed the effects of immunoglobulin somatic mutations on the ratios of replacement and silent amino acid changes in the framework and antigen-binding regions (CDRs) of the immunoglobulin heavy and light chains from each oligoclonal CLL-cell population. In one subject, the amino acid changes were consistent with an antigen-driven progression of clonally related CLL-cell populations. In the other subject, intraclonal diversification was associated with immunoglobulin amino acid changes that would have likely lessened antigen affinity. Taken together, these studies support the hypothesis that in some CLL cases intraclonal diversification is dependent on antigen interactions with immunoglobulin receptors.
Introduction
Most lymphoid malignancies develop from B cells because B cells modify their genomic DNA throughout differentiation by V(D)J recombination, somatic mutation, and class switching.1,2 The analysis of immunoglobulin genes has been important to understanding the origins of B-cell malignancies.2 Acute lymphoid leukemias develop in less mature cells, whereas chronic leukemias tend to develop in more mature cells.1 Among leukemias and lymphomas that are derived from mature B cells, most have somatically mutated immunoglobulin chains; only mantle-zone lymphomas and some B-cell chronic lymphocytic leukemias (CLLs) have unmutated immunoglobulins.2 Ongoing somatic mutation is seen in lymphoplasmacytoid lymphoma, Burkitt lymphoma, diffuse large-cell lymphoma, mucosa-associated lymphoid tissue (MALT) lymphoma, and CLL.2,3 CLL can be either somatically mutated or unmutated1,4-6 ; therefore, the precise origin of these cells is still under investigation.4-10
Greater than 50% of CLL cases have somatically mutated immunoglobulin heavy chain genes, and these patients have a better prognosis than those with unmutated immunoglobulin genes.4,6 One hypothesis regarding the difference in CLL-cell mutation status is that mutation status reflects the maturation stage of the B cell during malignant transformation.4-7 This hypothesis postulates that unmutated CLL cases develop from pregerminal center B cells and mutated CLL cases develop from postgerminal center B cells. Another hypothesis postulates that unmutated CLL cases develop from B cells driven by T-independent antigens, whereas mutated CLL cases develop from B cells driven by T-dependent antigens.4-6 Finally, it has been suggested that CLL is derived from a memory B-cell population regardless of mutation status because the genetic profiles of the unmutated and mutated cases are similar.8,9
Earlier, CLL cells were considered to be static in their immunoglobulin variable gene expression and mutation load.5,11 Evidence for CLL-cell intraclonal diversification by ongoing somatic mutation,3,12-16 heavy chain receptor editing,17 and class switching18 suggests CLL cells can continue to differentiate. Prior studies of CLL-cell intraclonal diversification were performed using bulk CLL-cell populations and cloned heavy chain immunoglobulin gene variable regions.3,19 A single-cell analysis of mutational diversification was performed in one study and emphasized clonal analysis of immunoglobulin light chains.12 We used single-cell analysis of immunoglobulin heavy and light chain genes to investigate the possibility that CLL cells can undergo intraclonal diversification. We found evidence that CLL-cell intraclonal diversification with progressive somatic mutation was associated with evidence of antigen drive in some but not all cases of CLL.
Patients, materials, and methods
Patients with CLL
The diagnosis of CLL was established according to National Cancer Institute (NCI) Working Group criteria.20 Informed consent was obtained as part of a protocol approved by the Veterans Administration (VA) and Duke University Institutional Review Boards. We analyzed 6 patients with CLL characterized by CLL cells with mutated (≥ 2% difference from the most similar germ line gene) variable region immunoglobulin heavy chains (VH) from our cohort of patients from the Duke University and Durham VA Medical Centers. Patients had white blood cell (WBC) counts of more than 15 × 109/L (15 000/μL) and had not received CLL therapy in the past month. Patient characteristics are listed in Table 1. Rai staging was performed as previously described.20 Fluorescence in situ hybridization (FISH) analysis of CLL-cell chromosomes was performed using fluorescent probes to detect deletion of 13q14.3 (D13S319), 13q34, 17p13.1 (TP53), and 11q22.3 (ATM) and using a chromosomal enumeration probe (CEP) (12p11.1-q11) to detect trisomy 12 (Vysis, Des Plaines, IL). In contrast to the initial 5 subjects (011, 012, 038, 043, and 052), the sixth subject (151) was selected for analysis based on evidence of VH sequence heterogeneity during analysis of DNA from unsorted partially purified CLL cells from this subject. Retrospective analysis of the VH gene sequences from the unsorted partially purified CLL cells of subject 012 indicated VH sequence heterogeneity similar to that of subject 151.
CLL-cell isolation
Peripheral blood mononuclear cells (PBMCs) were purified from peripheral blood samples of patients with CLL using Ficoll-Hypaque density gradients.21 CLL cells were enriched by depletion of T cells and monocytes using RosetteSep Human B Cell Enrichment Cocktail (Stem Cell Technologies, Vancouver, BC), according to the manufacturer's instructions.
CLL-cell sorting
CLL cells from CLL subjects 012, 038, 011, 052, and 043 were phenotyped by flow cytometry using anti–CD19-PE-Texas Red, anti–CD5-TriColor, anti–Igκ-APC, anti–Igλ-Alexa488 (MH10620) (Caltag Laboratories, Burlingame, CA) and anti–CD27-PE (BD Pharmingen, San Diego, CA). CLL cells from subject 151 were phenotyped by flow cytometry using anti–CD19-APC-Cy7, anti–CD5-PE-Cy7, anti–Igλ-FITC, anti–CD27-PE (BD Pharmingen), and anti–Igκ-FITC (Caltag). Unfixed, fluorescently labeled CLL cells were identified based on cell-surface expression of CD19, CD5, and CD27 and either immunoglobulin κ or λ. CLL cells were sorted into Applied Biosystems (Foster City, CA) polymerase chain reaction (PCR) plates containing 20 μL 1 × Promega (Madison, WI) PCR buffer and with 20 ng rRNA (16S and 23S) (Roche, Indianapolis, IN). CLL cells were sorted in groups of 1 or 1000 cells per PCR well. For every 5 single cells or group of 1000 cells sorted, one well in the PCR plate with PCR buffer did not receive sorted cells and was used as a negative control. After sorting, samples were quick-frozen in an ethanol/dry ice bath and then stored at −80°C until further analysis.
Whole genome amplification
Sorted CLL cells were incubated with 0.3 mg/mL proteinase K (Roche) for 2 hours at 50°C followed by 10 minutes at 95°C. Then 40 μL of a mixture containing 1 × Promega PCR buffer, 0.17 mM dNTPs (Stratagene, La Jolla, CA), 2.5 mM MgCl2 (Qiagen, Valencia, CA), and 33.3 μM random 15 nucleotide oligomers (Operon, Huntsville, AL) was added to each tube. Whole genome amplification (WGA) was performed at 95°C for 2 minutes followed by the addition of 5 U Taq polymerase (Promega) at 80°C, heating at 37°C for 2 minutes, and then followed by 49 cycles of 94°C for 1 minute, 37°C for 2 minutes, and a 5% ramp to 55°C for 4 minutes.
PCR amplification of immunoglobulin heavy chain variable (VH) region DNA
Analyses of immunoglobulin VH regions were performed by amplification of WGA DNA using a nested PCR approach in which all external VH family primers were combined for the first round of PCR (PCRa), and the second round of PCR (PCRb) was performed using separate reactions containing the internal VH family primers. Immunoglobulin VH oligonucleotide primers were chosen based on those previously published22,23 and are listed in Table S1 (available on the Blood website; see the Supplemental Table link at the top of the online article). PCR conditions were optimized using PBMC DNA from healthy individuals, and this PBMC DNA was used during each PCR run as a positive control.
For PCRa, 5 μL WGA DNA was used in a 50-μL reaction. For PCRb, 2 μL PCRa product was used in a 50-μL reaction. PCR reactions contained 200 μM of each dNTP, 1 μM each of the appropriate primers, 10 mM Tris-HCl (pH 8.3) buffer, 50 mM KCl, 2 U Taq polymerase (HotStarTaq; Qiagen) and either MgCl2 or 10 μL Q buffer (Qiagen). The PCR primers and whether Q buffer and/or MgCl2 was used for each PCR reaction are listed in Table S1. The PCRa amplification was 95°C for 15 minutes followed by 25 cycles of 94°C for 30 seconds, 58°C for 1 minute, and 72°C for 1 minute, followed by a 7-minute elongation period at 72°C. The PCRb amplification was the same except that 35 cycles were used.
PCR amplification of immunoglobulin κ chain variable (Vκ) region DNA
Analyses of immunoglobulin Vκ regions were performed by amplification of WGA DNA using the same strategy as for heavy chains; the variable components of each reaction are listed in Table S1. Oligonucleotide primers were chosen based on those previously published.24
For Vκ region PCR amplification, the PCRa amplification was 95°C for 15 minutes followed by 25 cycles of 94°C for 30 seconds, 56°C for 1 minute, and 72°C for 1.5 minutes, followed by a 7-minute elongation period at 72°C. The Vκ region PCRb amplification was the same, except that 35 cycles were used and an annealing temperature of 60°C was used.
PCR amplification of immunoglobulin λ chain variable (Vλ) region DNA
Analyses of immunoglobulin Vλ regions were performed by amplification of WGA DNA using the same strategy as for heavy chains; the variable components of each reaction are listed in Table S1. Oligonucleotide primers were chosen based on those previously published.24
For Vλ region PCR amplification, the PCRa amplification was 95°C for 15 minutes followed by 25 cycles of 94°C for 30 seconds, 50°C for 1 minute, and 72°C for 1.5 minutes, followed by a 7-minute elongation period at 72°C. The Vλ region PCRb amplification was the same, except that 35 cycles were used and an annealing temperature of 64°C was used.
PCR amplification of β-globin DNA
A region of the β-globin gene was used as a positive control to confirm the presence of single sorted CLL cells in PCR plate wells and during sequencing to control for PCR-induced mutations. Analysis of β-globin gene DNA was performed by amplification of WGA DNA using the same strategy as for heavy chains. The PCR primers and the variable components of each reaction are listed in Table S1.
For β-globin gene amplification, the PCRa amplification was 95°C for 15 minutes followed by 25 cycles of 94°C for 30 seconds, 54°C for 1 minute, and 72°C for 1.5 minutes, followed by a 7-minute elongation period at 72°C. The PCRb amplification was the same except that an annealing temperature of 56°C was used.
DNA sequencing
PCR products were run on a 3% agarose gel, and appropriate-sized products were extracted and purified using GenElute spin columns (Sigma, St Louis, MO) according to the manufacturer's instructions. Purified products were sequenced directly on an automated DNA sequencer (Applied Biosystems) using the BigDye Terminator kit (Perkin Elmer, Boston, MA). Products were sequenced in the forward and reverse direction using the internal PCR primers.
RT-PCR analysis of immunoglobulin RNA
RNA was isolated from frozen pellets of CLL-enriched PBMCs using RNAlater-ICE and a RiboPure RNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA samples were treated with DNase (Ambion) during preparation to remove contaminating DNA. RNA (8 μL) was reverse transcribed in a 20-μL volume of buffer containing 10 μM random hexamer primers (IDT, Coralville, IA), 1 μL ImProm-II RT (Promega), Promega reverse transcriptase (RT) buffer, 2.5 mM MgCl2 (Promega), 0.5 mM of each dNTP (Stratagene), and 0.5 U RNasin (Stratagene). The RNA and primers were first incubated together at 70°C for 5 minutes and then added to the other mix components and incubated at 25°C for 5 minutes, 42°C for 1 hour, and 70°C for 15 minutes. RNA samples incubated in the same buffer without reverse transcriptase were used as controls to confirm the absence of amplification of genomic DNA. Following reverse transcription, cDNA was PCR amplified using the same nested protocol for amplification of WGA, except 2 μL of the RT-PCR reaction mixture was used instead of WGA. Immunoglobulin PCR products were analyzed and sequenced as described in “DNA sequencing.”
Analysis of immunoglobulin gene sequences
Sequences were compared with those in the online V BASE sequence directory25,26 or IgBlast.27 Forward and reverse sequences were aligned into a single resolved sequence, and then aligned with germ line sequences derived from DNA Plot on the V BASE26 directory or IgBlast27 using Sequencher 4.1 software (Gene Codes, Ann Arbor, MI). The sequences were compared with the germ line gene with the most homology, and the number of somatic mutations was determined by counting those from the beginning of FR1 to the end of FR3.
Statistical analysis
Antigen drive was computed by the method of Lossos et al28 using the web application available at http://www-stat.stanford.edu/immunoglobulin. The number of DNA mutations associated with amino acid changes (replacement) and the number of DNA mutations not associated with amino acid changes (silent) in either the framework regions (FRs) or complementarity-determining regions (CDRs) of the rearranged immunoglobulin genes were compared to determine whether the mutations were random or antigen driven. P values associated with variable region mutations were calculated using the web application of Lossos et al.28 Low P values for FR analyses signify an excess of silent mutations, and low P values for CDR analyses signify an excess of replacement mutations.
Results
Mutational analysis of rearranged Ig heavy and light chain genes from 6 patients with CLL with mutated immunoglobulin genes
An analysis of rearranged immunoglobulin heavy chain variable region DNA (VH) from 156 patients with CLL identified 100 subjects with somatically mutated immunoglobulin genes. Fresh blood samples from 6 of the subjects with somatically mutated immunoglobulin genes were sorted by flow cytometry into PCR plates in groups of 1 or 1000 cells and analyzed by PCR amplification and DNA sequencing of rearranged heavy and light chain immunoglobulin genes. Results from the 1000-cell analyses were used to delineate which heavy and light chains were rearranged in each subject (data not shown), and results from these analyses were used to guide the single-cell analyses. The results of the single-cell analyses are summarized in Table 2. Analysis of heavy and light chain immunoglobulin RNA from bulk CLL-cell populations was used to identify which immunoglobulin genes were expressed in CLL cells from each subject and are indicated in Table 2. Except for CLL subject 011, all subjects had evidence of multiple, related rearranged heavy and/or light chain immunoglobulin genes based on the presence of different somatic mutations in sequenced immunoglobulin genes. Two subjects with CLL (012 and 151) had multiple related rearranged VH chain genes and Vκ chain genes that contained partially shared mutations and formed oligoclonal subgroups. Taken together, our findings support recent studies indicating intraclonal diversification of CLL during the course of the disease.3,12,19
To estimate the effect of PCR-induced Taq polymerase errors on mutational frequency, we sequenced the amplified β-globin gene PCR products from all single cells (90 total cells) that underwent immunoglobulin sequencing from CLL subjects 012 and 151 (Table 2). Only 2 unique mutations in the β-globin gene were identified in this analysis. In addition, we confirmed the heavy chain and light chain sequences from a randomly selected set of 16 of the 43 cells from CLL subject 012 that underwent immunoglobulin sequencing by reamplifying WGA DNA from these 16 sorted single cells. Taken together, we believe that this makes Taq polymerase-induced errors an unlikely source of the mutations observed in our immunoglobulin sequence analyses.
For CLL subjects 012 and 038, we only identified one rearranged heavy chain gene and one rearranged light chain gene (Table 2). We were unable to identify a rearranged light chain gene in CLL subject 011. In contrast, CLL subjects 043, 052, and 151 had CLL cells with either more than one rearranged light chain gene (052 and 151) or more than one heavy and light chain gene (043). Immunoglobulin genes from all 6 subjects with CLL were productively rearranged except the VH6-01, Vk2 (A18), and VL4b immunoglobulin genes from CLL subject 043 and the VL8a immunoglobulin gene from subject 151 which had a somatic mutation resulting in a stop codon (Table 2). Analysis of CLL-cell RNA from subjects 011, 012, 038, and 151 identified immunoglobulin RNA transcripts corresponding to each of the rearranged immunoglobulin genes in these subjects (Table 2). Analysis of CLL-cell RNA from subject 043 identified transcripts corresponding to the VH2-05, VL3m, and VL4b rearrangements (Table 2). We also identified a Vk2 (A19) transcript in CLL cells from subject 043 that was likely missed in the genomic DNA analysis because of similarity with the Vk2 (A18) rearrangement. CLL cells from subject 043 expressed a λ immunoglobulin chain on their cell surface, suggesting that the VL3m rearrangement was the predominantly expressed light chain in this subject. In subject 052, we only detected immunoglobulin RNA corresponding to the VH3-11 and Vk3 (L2) rearrangements. In subject 151, we detected immunoglobulin RNA corresponding to the VH3-23, Vk2 (A19), and VL8a rearrangements. CLL cells from this subject expressed a κ immunoglobulin chain on their cell surface, suggesting that the Vk2 (A19) rearrangement was the predominantly expressed light chain in this subject.
Genealogic analysis of oligoclonal CLL-cell subgroups from subjects 012 and 151
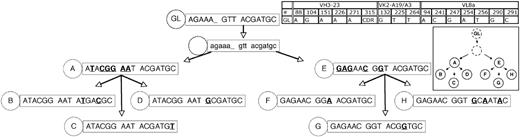
The immunoglobulin VH and Vκ gene sequences from CLL subjects 012 and 151 are shown in Figures 1 and 2. Genealogic trees of related CLL-cell subgroups from CLL subjects 012 and 151 are shown in Figures 3 and 4, respectively. The trees were constructed on the assumption that CLL-cell subgroups lower in each tree would sequentially accumulate somatic mutations and that CLL-cell subgroups lower in each tree would be unlikely to revert earlier mutations to the germ line configuration. Because a plausible genealogic relation between the oligoclonal subgroups of single cells analyzed from CLL subjects 012 and 151 could be constructed, this suggested a pattern of development of oligoclonal subgroups from single founder CLL cells.
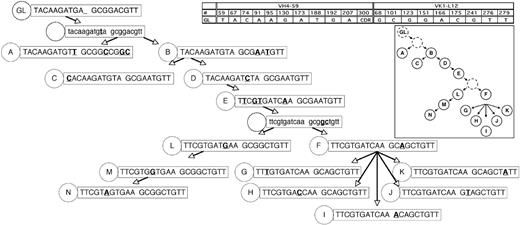
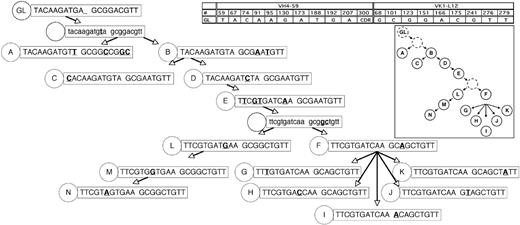
Rearranged immunoglobulin variable region heavy chain and light chain sequence analysis of 43 single sorted cells from CLL subject 012. Sequence alignment for 14 subgroups (A-N) of the VH4-59 (A) and Vk1-L12 (B) immunoglobulin genes sequenced from 43 single CLL cells from subject 012. Dashes signify identical bases, and dots represent the absence of bases compared with the germ line genes for VH4-59 and Vk1-L12. Replacement mutations are shown in uppercase, and silent mutations are shown in lowercase. The number of sequences or CLL cells in each group is indicated and is preceded by an x.
Rearranged immunoglobulin variable region heavy chain and light chain sequence analysis of 43 single sorted cells from CLL subject 012. Sequence alignment for 14 subgroups (A-N) of the VH4-59 (A) and Vk1-L12 (B) immunoglobulin genes sequenced from 43 single CLL cells from subject 012. Dashes signify identical bases, and dots represent the absence of bases compared with the germ line genes for VH4-59 and Vk1-L12. Replacement mutations are shown in uppercase, and silent mutations are shown in lowercase. The number of sequences or CLL cells in each group is indicated and is preceded by an x.
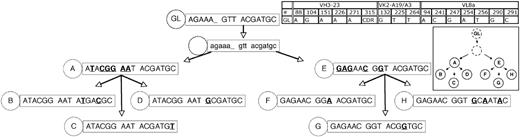
Rearranged immunoglobulin variable region heavy chain and light chain sequence analysis of 20 single sorted cells from CLL subject 151. Sequence alignment for 8 subgroups (A-H) of the VH3-23 (A), Vk2-A19/A3 (B), and VL8a (C) immunoglobulin genes sequenced from 20 single CLL cells from subject 151. Complete paired sequencing data from the VH3-23, Vk2-A19/A3, and VL8a immunoglobulin genes were available from only 20 single CLL cells. This accounts for the different numbers presented in this figure compared with Table 2. Dashes signify identical bases, and dots represent the absence of bases compared with the germ line genes for VH3-23, Vk2-A19, and VL8a. Replacement mutations are shown in uppercase, and silent mutations are shown in lowercase.
Rearranged immunoglobulin variable region heavy chain and light chain sequence analysis of 20 single sorted cells from CLL subject 151. Sequence alignment for 8 subgroups (A-H) of the VH3-23 (A), Vk2-A19/A3 (B), and VL8a (C) immunoglobulin genes sequenced from 20 single CLL cells from subject 151. Complete paired sequencing data from the VH3-23, Vk2-A19/A3, and VL8a immunoglobulin genes were available from only 20 single CLL cells. This accounts for the different numbers presented in this figure compared with Table 2. Dashes signify identical bases, and dots represent the absence of bases compared with the germ line genes for VH3-23, Vk2-A19, and VL8a. Replacement mutations are shown in uppercase, and silent mutations are shown in lowercase.
Genealogic analysis of CLL-cell subgroups from CLL subject 012. The 14 subgroups labeled A-N from CLL subject 012 (Figure 1) were sequentially arranged based on the assumption that descendant CLL-cell groups lower in the tree would have more somatic mutations than predecessor subgroups, that it would be unlikely for already mutated bases within the heavy and light chain immunoglobulin sequences to revert back to germ line sequences, and that the same mutation would not occur twice independently. In some instances, these assumptions could not be followed (subject 012, heavy chain at position 69 reverts back to germ line from stage D to E; subject 012, light chain at position 51 reverts back to germ line from stage E to F; patient 151, lambda light chain at position 31b, the same mutation occurs twice independently); therefore, efforts were made to minimize instances where these assumptions were violated. The germ line sequence (GL) is indicated at the top of the tree, and each subgroup is designated with its respective letter (A-N). The number of sequences or CLL cells in each group is listed in Figure 1. For the sequence data in the figure, only the bases which indicated the positions of partially shared and unique mutations are shown. The germ line base positions for each chain are indicated in the grid. Sequence changes from each previous subgroup are indicated in bold and underlined. For simplicity, shared mutations are not shown and are listed in Figure 1. Sequence data subgroups shown in lowercase letters and with a dashed circle label are intermediates that were not identified but whose existence was postulated.
Genealogic analysis of CLL-cell subgroups from CLL subject 012. The 14 subgroups labeled A-N from CLL subject 012 (Figure 1) were sequentially arranged based on the assumption that descendant CLL-cell groups lower in the tree would have more somatic mutations than predecessor subgroups, that it would be unlikely for already mutated bases within the heavy and light chain immunoglobulin sequences to revert back to germ line sequences, and that the same mutation would not occur twice independently. In some instances, these assumptions could not be followed (subject 012, heavy chain at position 69 reverts back to germ line from stage D to E; subject 012, light chain at position 51 reverts back to germ line from stage E to F; patient 151, lambda light chain at position 31b, the same mutation occurs twice independently); therefore, efforts were made to minimize instances where these assumptions were violated. The germ line sequence (GL) is indicated at the top of the tree, and each subgroup is designated with its respective letter (A-N). The number of sequences or CLL cells in each group is listed in Figure 1. For the sequence data in the figure, only the bases which indicated the positions of partially shared and unique mutations are shown. The germ line base positions for each chain are indicated in the grid. Sequence changes from each previous subgroup are indicated in bold and underlined. For simplicity, shared mutations are not shown and are listed in Figure 1. Sequence data subgroups shown in lowercase letters and with a dashed circle label are intermediates that were not identified but whose existence was postulated.
Genealogic analysis of CLL-cell subgroups from CLL subject 151. The 8 subgroups labeled A-H from CLL subject 151 (Figure 2) were sequentially arranged based on the assumptions listed in Figure 3. The number of sequences or CLL cells in each group is listed in Figure 2. The description and labeling of sequence data are described in Figure 3.
Genealogic analysis of CLL-cell subgroups from CLL subject 151. The 8 subgroups labeled A-H from CLL subject 151 (Figure 2) were sequentially arranged based on the assumptions listed in Figure 3. The number of sequences or CLL cells in each group is listed in Figure 2. The description and labeling of sequence data are described in Figure 3.
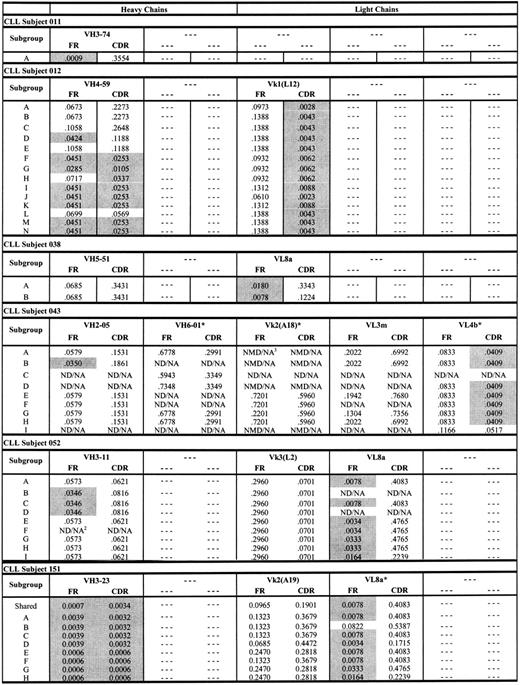
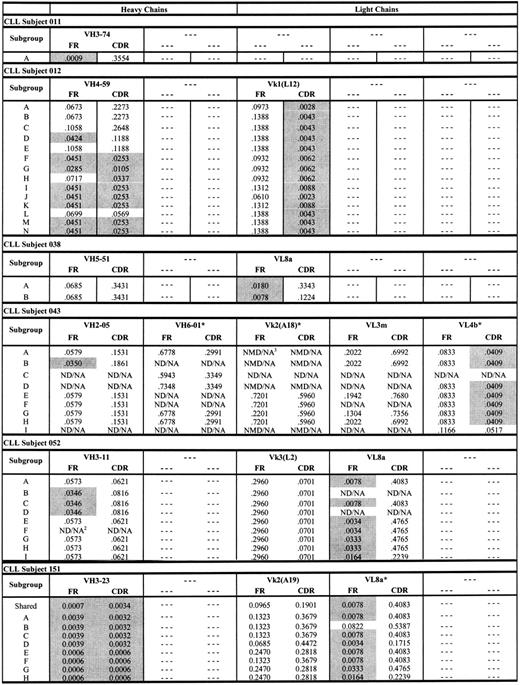
Antigen drive analysis of CLL subjects 012 and 151 and the other 4 subjects with CLL
The genealogic relation between CLL-cell subgroups from subjects 012 and 151 depicted in Figures 3 and 4 are consistent with previously described antigen-driven responses in normal B cells.29-31 To better understand whether these genealogies were consistent with antigen-driven responses, we used the method of Lossos et al28 to calculate P values for ratios of replacement and silent mutations (R/S) in the FR and CDR regions of each CLL-cell subgroup from subjects 012 and 151 (Figure 5). The criteria for evidence of antigen selection used by Lossos et al28 depends on lower R/S ratios in the FR regions and higher R/S ratios in the CDR regions. As shown in Figure 5, there was a general decrease in (ie, increased significance of) P values for the ratios of replacement to silent mutations in the FR and CDR regions of the heavy chain from CLL subject 012 for CLL-cell subgroups lower in the genealogic tree depicted in Figure 3. The P values for the ratios of replacement to silent mutations for CLL-cell subgroups of CLL subject 012 were all less than .05 for the light chain CDRs (Figure 5). For CLL subject 151, there were 2 large oligoclonal subgroups of CLL cells and evidence for 6 smaller subgroups of CLL cells (Figure 4). The P values for the heavy chain FR and CDR R/S ratios of the 2 large oligoclonal CLL-cell subgroups in subject 151 were less than .05. However, there were no decreases in the P values for the light chain FR and CDR R/S ratios in the smaller subgroups lower in the genealogic tree (Figure 5). Overall, the pattern of low and progressive decreases in the P values for the R/S ratios of immunoglobulin sequences from CLL-cell subgroups in subject 012 were consistent with an antigen-driven response. In contrast, the pattern of P value changes for the R/S immunoglobulin sequences from CLL-cell subgroups in subject 151 were consistent with proliferation of CLL subgroup clones with random immunoglobulin gene mutations.
P values for ratios of replacement and silent mutations (R/S) in the FR and CDR regions of each CLL-cell subgroup from subjects. Shaded P values are less than .05 and significant. ND/NA indicates not detected/not applicable; NMD/NA, no mutation detected/not applicable. *Nonproductive rearrangements.
P values for ratios of replacement and silent mutations (R/S) in the FR and CDR regions of each CLL-cell subgroup from subjects. Shaded P values are less than .05 and significant. ND/NA indicates not detected/not applicable; NMD/NA, no mutation detected/not applicable. *Nonproductive rearrangements.
Figure 5 also displays P values for the ratios of replacement to silent mutations for the other rearranged immunoglobulin genes from the other 4 subjects with CLL. Nearly all of these immunoglobulin chains exhibited evidence of antigen drive based on low P values for the ratios of replacement to silent mutations in the FR of each immunoglobulin gene. This was consistent with derivation of these CLL cells from a memory B-cell population. However, in contrast to subject 012 (and similar to subject 151), there was no clear evidence of progressive changes in the P values for the ratios of replacement to silent mutations in the immunoglobulin genes from subgroups of CLL cells from each of these 4 subjects.
Discussion
Prior studies presented evidence that CLL cells use a relatively restricted repertoire of immunoglobulin genes and postulated a role for antigen binding in the development and progression of CLL.5,32-36 More recent studies have highlighted the potential importance of antigen interaction with unmutated CLL cells in the development and progression of CLL.37-39 These recent studies primarily focused on cases of CLL with unmutated immunoglobulin genes and concluded that many cases of CLL derived from autoreactive B cells. In the study by Messmer et al,37 1 of every 5 patients with CLL with unmutated CLL cells fit into one of several sets of subjects with similar heavy chain immunoglobulins. The researchers speculated that immunoglobulins associated with these CLL-cell sets were likely directed to autoantigens or carbohydrates based on preferential use of VH4-34 immunoglobulin genes and the similarity of one VH1-69 CLL sequence to an anticardiolipin antibody. In the same way, the study by Ghiotto et al38 identified 5 cases of IgG+ CLL with immunoglobulin sequence similarities. The sequence similarities of these 5 IgG+ CLL cases were similar to other antibodies known to bind autoantigens such as dsDNA. Finally, in the study by Herve et al,39 immunoglobulins from unmutated CLL cells were produced and shown to bind autoantigens such as dsDNA. Importantly, in that study, immunoglobulin genes from mutated cases that were reverted to their germ line immunoglobulin genes were also shown to bind autoantigens. The researchers speculated that mutated CLL cases might originate from the same population of autoreactive B cells as unmutated CLL cases.
Although our study is not the first to demonstrate intraclonal diversification in CLL cells, we believe our study is the first to demonstrate evidence of intraclonal diversification in CLL cells associated with antigen drive. The ability to pair heavy and light chain immunoglobulin DNA analyses using a single-cell technique may have contributed to our ability to detect evidence of antigen drive in contrast to prior studies that used analysis of cloned immunoglobulin RNA from bulk populations of CLL cells.3,19 In the study by Gurrieri et al3 of patients with CLL with mutated immunoglobulin transcripts, 11 of 18 subjects with CLL analyzed had evidence of intraclonal diversification but did not show evidence of antigen drive. Likewise, analysis of only light chains in a single-cell analysis performed by Ruzickova et al12 did not show evidence of antigen drive but did speculate that CLL cells from this subject developed from B cells expressing a cold agglutinin antibody directed against a red-cell antigen. Had we only performed analyses of heavy chains like Gurrieri et al3 or only light chains like Ruzickova et al,12 we would have been unable to construct a genealogic tree from subject 012 that exhibited evidence of antigen drive.
A recent report by Bagnara et al19 demonstrated evidence for intraclonal diversification of oligoclonal subgroups in a patient with CLL. That study included a genealogic tree of related oligoclonal CLL-cell subgroups with evidence for antigen drive in FR regions but not CDR regions. Our analysis of antigen drive in the CDR regions of the clones depicted in the oligoclonal tree reported by Bagnara et al,19 using the method of Lossos et al,28 indicated that P values for descendant clones increased. Therefore, we believe that the oligoclonal tree described by Bagnara et al19 is more similar to that of subject 151 in our report. Taken together, this emphasizes the unique aspects of the CLL-cell subgroups from patient 012 but also supports the notion that intraclonal diversification of some mutated CLL cases (such as subject 151 or the subject described by Bagnara et al19 ) may be random or favor immunoglobulin gene mutations that lessen antigen affinity.39,40 More definitive evidence to support these hypotheses will require identification of antigens that bind immunoglobulins from these CLL cells and comparisons of immunoglobulin antigen avidity from different CLL-cell clones from each subject.
In the CLL patient with evidence of antigen drive (subject 012), we searched for similar immunoglobulin sequences in the NCBI databases to identify potential antigens that might bind CLL-cell immunoglobulins from this subject. These searches did not identify any potential antigens. A review of clinical records for subject 012 suggested 2 possible sources of antigen. This subject had a history of recurrent Staphylococcus aureus infections of the skin, and after his diagnosis of CLL this subject developed metastatic adenocarcinoma of unknown primary and subsequently died of the carcinoma. Other researchers have suggested that some cases of CLL may represent a response to infectious agents41,42 ; this makes a S aureus antigen an attractive potential target for the CLL-cell immunoglobulins of subject 012. Likewise, there is a high incidence of second neoplasms in patients with CLL.43-46 Although several researchers have suggested that the high incidence of second neoplasms in CLL is due to weakened immune surveillance or to a genetic predisposition to malignancy, it is possible that in some cases the development of CLL is triggered by the presence of clinically undetectable secondary neoplasms that serve as antigenic sources for expansion of CLL cells.
Because we did not identify subgroups of CLL cells from CLL subject 012 that lacked evidence of increases in antigen drive compared with predecessor CLL cells (Figures 1, 3, 5), we believe that expansion of CLL cells that recognize a particular antigen may be critical to progression of disease in CLL subject 012. There is increasing evidence that other B-cell malignancies may proliferate in response to antigenic stimuli. For example, early-stage MALT B-cell lymphomas expand in the presence of T cells reactive to Helicobacter pylori and undergo autoantigen-driven diversification and expansion.47 MALT lymphomas can be successfully treated with antibiotics that eliminate H pylori,47 and, as such, understanding the mechanisms and identifying the antigens associated with diversification of CLL cells in subject 012 is important. Identification of other CLL subjects who have mutated immunoglobulin genes and evidence of intraclonal diversification and continuing antigen drive would enhance understanding of the role of antigen in CLL pathogenesis. Such an analysis may also contribute to an understanding of the relation that antigen drive has to CLL prognosis. In patients with CLL with mutated immunoglobulin genes and evidence that their CLL cells derived from an antigen-driven B-cell population, Degan et al48 found that these subjects had a better prognosis than did patients with CLL with unmutated immunoglobulin genes or with mutated non–antigen-driven immunoglobulin genes. Therefore, in summary, we believe an understanding of antigenic triggers in CLL may permit targeted elimination of factors that promote CLL-cell development and progression.
Authorship
Contribution: A.D.V. designed and performed the research, collected and analyzed the data, and wrote the manuscript; J.B.W. designed the research, contributed patient samples, and collected and analyzed the data; B.E.B. and J.F.W. performed the research and collected data; J.P.G. contributed patient samples; J.O.M. contributed patient samples; G.K. designed the research and analyzed the data; B.K.G. performed the research and analyzed the data; M.C.L. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marc C. Levesque, Duke University Medical Center, Box 3266, Durham, NC 27710; e-mail: marc.levesque@duke.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Flow cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Core Facility, which is supported by National Institutes of Health award AI-051445.
This work was supported by grants from the Leukemia and Lymphoma Society, the National Cancer Institute, and by the Veterans Affairs Research Service.