Abstract
Persistent activation of Stat5 is frequently found in hematologic neoplasms. Studies conducted with constitutively active Stat5 mutants (Stat51*6 and cS5F) have shown that deregulated Stat5 activity promotes leukemogenesis. To investigate the oncogenic properties of these mutants, we used cS5F-expressing bone marrow cells which induce a multilineage leukemia when transplanted into recipient mice. Here, we show by immunocytochemistry that cS5F is localized mainly in the cytoplasmic compartment of leukemic cells, suggesting that the transforming nature of cS5F may be associated with a cytoplasmic function. In support of this hypothesis, we found that cS5F forms a complex with the p85 subunit of the phosphatidylinositol 3-kinase (PI3-K) and the scaffolding adapter Gab2 in leukemic bone marrow cells, resulting in the activation of Akt/PKB, a crucial downstream target of PI3-K. By using transducible TAT-Gab2 or TAT-Akt recombinant proteins, we were able to demonstrate that activation of the PI3-kinase/Akt pathway by cS5F molecules through Gab2 is essential for induction of cell growth. We also found that persistently phosphorylated Stat5 in primary cells from patients with myeloid leukemias has a cytoplasmic localization. These data suggest that oncogenic Stat5 proteins exert dual transforming capabilities not only as transcriptional activators but also as cytoplasmic signaling effectors.
Introduction
Stat5A and Stat5B transcription factors are important mediators of cytokine-induced cell survival and proliferation.1 There is a large body of evidence indicating that they play crucial roles in hematopoiesis in humans and mice. Stat5a/b−/− mice have multiple hematopoietic defects which affect the proliferation and/or survival of both lymphoid and myeloid lineages.2-7 In addition, Stat5 proteins regulate the growth of hematopoietic progenitor cells, and a recent report has suggested that Stat5 might be involved in self-renewal of human CD34+ progenitor cells.8,9
Deregulation of the Jak-Stat signaling pathway, particularly Stat3 and Stat5, was reported in many different types of cancer, including hematopoietic neoplasms.10,11 Persistent activation of these transcription factors is frequently found in many tumor cells, most probably as a consequence of deregulated tyrosine kinase activity. Importantly, Stat5 is a common target for different oncoproteins with tyrosine kinase activity such as Tel-Jak2, Bcr-Abl, the mutated forms of Flt3 and c-Kit, and the Jak2V617F mutant which has been recently characterized in various myeloproliferative disorders.12-16 Furthermore, it was shown that Stat5 plays a critical role in Bcr-Abl– and Tel-Jak2–induced leukemia.17-19 The most direct evidence that constitutive activation of Stat5 is an important causative event in cell transformation came from the analysis of the Stat5 mutants, Stat5A1*6, Stat5B1*6, and cS5F. These proteins with mutations at residues H299→R and S711/716→F (Stat5A1*6) or with the single mutation S711→F (cS5F) possess constitutive tyrosine phosphorylation and DNA binding activity and are capable of transforming cell lines to growth factor independence.20 Stat5A1*6 can also induce a fatal myeloproliferative disease in mice, whereas cS5F has been shown to induce a multilineage leukemia and to restore the defective phenotype of Stat5a/b-deficient mice, closely mimicking wt Stat5 function.20,21
Comprehensive analysis of gene-deleted mice as well as dominant-negative approaches shed light onto the mechanisms by which Stat5 controls cell growth and survival of hematopoietic cells. In particular, Stat5 has been shown to regulate expression of genes involved in cell survival and cell-cycle progression, such as Bcl-xL, A1 and D type cyclins, as well as genes encoding cytokines or growth factors, such as Osm or Igf-1, and the proto-oncogene Pim-1.2,22-26 Thus, persistent transcriptional activation of these genes by Stat51*6 or cS5F transcription factors may be the sole consequence for the oncogenic properties of these mutants, a hypothesis that awaits experimental evidence. Several studies suggested that Stat5 proteins may control cellular processes independently of their nuclear functions, through interactions with various signaling molecules. Accordingly, the tyrosine-phosphorylated forms of Stat5 were shown to interact with the SH2-SH3–containing adapter CrkL, the scaffolding adapter Gab, and the PI3-K.27-29 We previously reported that activation of the PI3-K–signaling cascade plays an important role in Stat51*6-induced cell growth and survival via the scaffolding adapter Gab2.30,31 It is well established that PI3-K function requires the activation of the serine threonine kinase Akt.32 Akt regulates the activity of a number of substrates involved in cell apoptosis or proliferation such as Bad, Forkhead, NFκB, and GSk3β.33,34 Akt is the cellular homologue of the v-akt oncogene, and there is strong evidence that inappropriate activation of the PI3-K/Akt pathway contributes to the development of cancers.35 Altogether, these different data argue that oncogenic activation of Stat5 triggers its association with PI3-kinase and that activation of this pathway may be involved in the leukemic potential of constitutively active Stat5 molecules. By using bone marrow (BM) cells from mice that received a transplant of cS5F-expressing cells, we demonstrated for the first time that cS5F is essentially localized in the cytoplasm of primary leukemic cells. We also found that cS5F forms a signaling complex with the PI3-K and the scaffolding adapter Gab2 which results in the activation of Akt in primary leukemic cells. We were able to show that Gab2 and Akt play a critical role in cS5F-induced cell growth through the use of transducible TAT fusion proteins. In addition, we demonstrated that persistent Stat5 phosphorylation is detected mainly in the cytoplasm of primary cells from patients with chronic myeloid leukemia (CML) and patients with acute myeloid leukemia (AML). Collectively, these data indicate that oncogenic activation of Stat5 affects its cellular localization and function and that activation of the PI3-kinase/Akt pathway via an interaction with p85 and Gab2 is implicated in Stat5-induced leukemia.
Patients, materials, and methods
Animals, primary cell isolation, retroviral infection
Bone marrow (BM) was harvested from hind limbs of 6-week-old male mice (C57/B6 × Sv129j F1). Freshly isolated BM cells were preactivated for 48 hours in medium containing IL-3 (25 ng/mL), IL-6 (50 ng/mL), and SCF (10 ng/mL). Cells were then cocultured on irradiated (1.5 Gy) semiconfluent ecotropic producer cell lines for 48 hours in the presence of 6 μg/mL Polybrene. Generation of retroviral packaging cell lines carrying the cS5F mutant or the GFPv was described elsewhere.2 Lethally irradiated wt female (1 Gy) recipients were then reconstituted with the transduced BM cells by tail vein injection (4 × 106 cells/mouse). Mice that received a transplant were checked for disease onset through blood analysis.
Patients
Four patients with acute myeloid leukemia (AML; FAB-subtype: M2, M3, M6, M7), 3 patients with chronic myeloid leukemia (CML; chronic phase, cp, n = 1; myeloid blast phase, pb, n = 1; megakaryoblast crisis, n = 1), and 1 control patient (no hematologic neoplasm, normal bone marrow) were examined. Diagnosis and classification of cases were performed according to established criteria.36 This study was approved by the institutional review board (Medical University of Vienna) and conducted in accordance with the declaration of Helsinki. Informed consent was obtained before bone marrow biopsies were taken.
Cell culture, plasmids, and reagents
BM cells from mice that received a transplant of cS5F were grown for 24 hours in RPMI 1640 with 10% fetal calf serum (FCS) and SCF (10 ng/mL; Valbiotech, Paris, France), and GFPv BM cells were grown in the same medium supplemented with IL-3 (10 ng/mL). The next day, GFP+ cells were sorted by flow cytometry and cultured in their respective medium for 6 weeks.
Ku-812 cells were grown in RPMI 1640 medium (Life Technologies, Cergy-Pontoise, France) containing 10% FCS, 2 mM L-glutamine (10 U/mL), and penicillin/streptomycin (10 μg/mL) at 37°C with 5% CO2. The LY294002 PI3-K inhibitor and imatinib mesylate were purchased from Sigma (St Quentin Fallavier, France) and Novartis (Basel, Switzerland), respectively.
The coding regions of Akt and Akt (K179→M), Gab2, and Gab2-3YF were amplified by polymerase chain reaction (PCR) and cloned at NcoI and EcoRI sites of the bacterial expression vector pTAT-HA (for the Akt cDNAs) or at the KpnI-XhoI sites (for the Gab2 cDNAs).
Fluorescence-activated cell sorting (FACS) analysis
Cells were incubated 20 minutes with the following phycoerythrin-conjugated antibodies: CD117, CD34, Mac-1, Gr-1, Ter-119, Sca-1, CD19, and Thy1.2 (BD Biosciences, le Pont de Claix, France) and analyzed by flow cytometry (Elite; Becton Dickinson, Le Pont de Claix, France).
Immunohistochemistry and immunocytochemistry
Immunohistochemistry was performed on sections prepared from paraffin-embedded formalin-fixed BM specimens using the indirect immunoperoxidase staining technique as described.37,38 Endogenous peroxidase was blocked by methanol/H2O2. Prior to staining with anti–P-Y-Stat5a/b antibody, sections were pretreated by microwave oven. Sections were stained with an anti–P-Y-Stat5a/b antibody AX-1 (Advantex Bioreagents, Conroe TX) (1.2 μg/mL) diluted in 0.05 M Tris-buffered saline (TBS, pH 7.5) plus 1% BSA for 20 hours. Then slides were washed and incubated with biotinylated goat anti–mouse IgG for 30 minutes, washed, and exposed to streptavidin-peroxidase complex. AEC was used as chromogen. Slides were counterstained in Mayer Hemalun. For immunocytochemical analysis, cells were spun on cytospin slides prior to staining with the AX1 antibody. DAB was used as chromogen. Slides were counterstained with Mayer Hemalun. Acquisition of micrographs was performed by an Olympus DP11 camera connected to an Olympus microscope equipped with 40×/0.85 (Figure 7A,C) and 100×/1.35 (Figure 3A) Uplan-Apo objective lenses (Olympus, Hamburg, Germany). Images were acquired with Photoshop CS2 software version 9.0 (Adobe Systems, San Jose, CA) and were processed with PowerPoint software (Microsoft, Redmond, WA).
Purification of TAT fusion proteins
Purification of TAT fusion proteins was performed as previously described39 with the following modifications: BL21 (DE3) pLysS bacteria (Stratagene, Amsterdam, The Netherlands) expressing the TAT fusion proteins were cultured in LB broth medium containing 50 μg/mL ampicillin and 34 μg/mL chloramphenicol at 37°C. Protein expression was induced by addition of 1 mM IPTG at 37°C during 4 hours. Cells were harvested and sonicated in buffer A (8 M urea, 20 mM HEPES [pH = 8.0], and 500 mM NaCl). Lysates were clarified by centrifugation at 15 600g for 4 minutes at 4°C. The supernatants containing the recombinant TAT fusion proteins were equilibrated in 30 mM imidazole and then applied to Ni-NTa agarose columns (Qiagen, Courtabeuf, France). Columns were washed with 6 bed volumes of buffer B (4 M urea, 20 mM HEPES [pH 8.0], 500 mM NaCl, and 30 mM imidazole). After washing, bound proteins were eluted with buffer B containing 250 mM imidazole. Eluates containing the purified proteins were dialyzed against PBS or (0.9% NaCl) with 4 buffer changes overnight at 4°C. The purity and concentration of TAT fusion proteins was assessed by Coomassie blue–stained SDS–polyacrylamide gel electrophoresis (PAGE) using standard concentrations of BSA. Stability and biologic activity of TAT fusion proteins were determined in Ba/F3 cells expressing the constitutively active form Stat51*6 (See Figures S1–S6, available on the Blood website; see the Figure link at the top of the online article).
In vitro proliferation studies
Cells were incubated as triplicates in flat-bottom 24-well plates at a density of 2 × 104 cells/well in medium supplemented with the different TAT fusion proteins at the indicated concentration for 16, 24, or 48 hours. The percentage of living cells was evaluated using the trypan blue dye exclusion assay.
Subcellular fractionation
Cells were lysed in hypotonic buffer (20 mM HEPES, 10 mM KCl, 1 mM EDTA, 0.2% NP40, 10% glycerol, 5 μg/mL aprotinin, 5 μg/mL leupeptin, 1 mM phenylmethylsulfonide fluoril, and 1 mM Na2VO4) and centrifuged for 5 minutes at 800g (Eppendorf centrifuge). Supernatants (cytoplasmic fraction) were frozen at −70°C. Pelleted nuclei were resuspended in hypertonic buffer (Hypotonic buffer plus 350 mM NaCl), and proteins extracts were prepared by constant agitation during 30 minutes at 4°C. Debris was removed by centrifugation, and nuclear extracts were frozen at −70°C.
Immunoprecipitations, Western blotting, and antibodies
Cells were lysed in Laemmli buffer (62.5 mM Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol). Equal amounts of each protein sample were separated by electrophoresis on SDS-PAGE and blotted onto nitrocellulose membrane (Bio-Rad, Munich, Germany). Blots were incubated as indicated with antibodies raised against the following proteins: Actin, Akt, Gab2, Raf-1, and Lamin C (Santa Cruz Biotechnology, Santa Cruz, CA), p27kip,1 Stat5 (Transduction Laboratories, Lexington, KY), Bim (Affinity BioReagents, Golden, CO), HA (Roche, Basel, Switzerland), P-Stat5 or P-AktSer473 (Cell Signaling Technology, Boston, MA), and topoisomerase I. The blots were developed with the enhanced chemiluminescence (ECL) system (Amersham Pharmacia Biotech, Orsay, France) using specific peroxidase-conjugated anti-IgG Abs. Immunoprecipitation experiments were performed as previously described.27
Results
Mice that received a transplant of cS5F as a model to characterize the transforming properties of Stat5
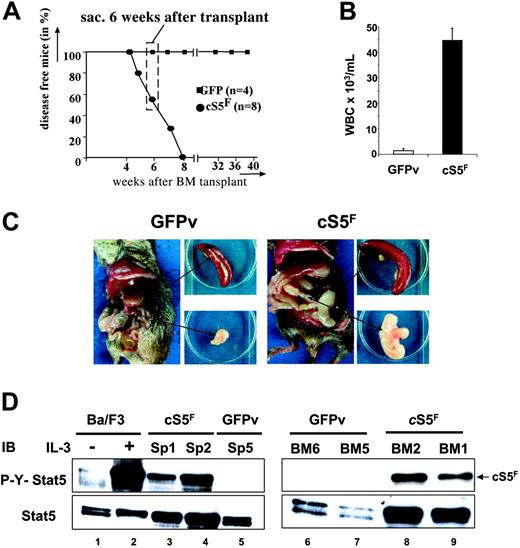
Mice that received a transplant of cS5F have been previously reported to develop a multilineage leukemia as a result of enhanced and sustained Stat5 tetramer DNA binding complexes.21 It remained questionable how cS5F proteins transduce signals to transform hematopoietic cells and if the predominant function can be attributed only to the transcriptional activity of the oncogenic mutant. In the present model, BM cells were infected with a retrovirus carrying the cS5F mutant or the GFP vector (GFPv) alone, analyzed for infection efficiency by FACS and injected into lethally irradiated wt mice. Eight mice that received a transplant of cS5F and 4 mice that received a transplant of GFPv were analyzed. All 8 cS5F mice were fully reconstituted 4 weeks after transplantation. They developed multilineage leukemia after 4 weeks, whereas no disease was seen in the 4 control mice (Figure 1A). None of the cS5F-expressing mice survived more than 8 weeks after transplantation, whereas all control mice remained disease free. Thus, in our experiments we chose to analyze diseased mice that received a transplant of cS5F at 6 weeks after transplantation (Figure 1A). Blood from these animals always contained a remarkable increase in white blood cell counts as compared with controls that was associated with splenomegaly and mesenterial lymphadenopathy (Figure 1B-C). Persistent activation of Stat5 was demonstrated by Western blotting in extracts from cS5F-transformed splenic (SP) and BM cells using anti–phospho-Y694-Stat5 (P-Y-Stat5) antisera (Figure 1D, lanes 3, 4, 8, 9). Extracts from unstimulated or IL-3–stimulated Ba/F3 cells were also loaded as control (lanes 1 and 2). By contrast no phosphorylation was detected in the splenocytes or BM cells of control GFPv mice (lanes 5, 6, 7).
Leukemogenic mice that received a transplant of cS5F as a model to characterize the transforming properties of Stat5. (A) Kaplan-Meier plot of mice that received a transplant of cS5F (n = 8) versus mice that received a transplant of GFPv (n = 4). Mice that received a transplant of cS5F died within 7 weeks after transplantation, while control GFPv mice remain disease free (followed for more then 10 months after transplantation). Thus, experimental mice that received a transplant of cS5F were killed and analyzed 6 weeks after transplantation for all following experiments (dashed box). (B) White blood cell counts from mice that received a transplant of cS5F (n = 8) and from GFPv mice (n = 4). Error bars represent SE. (C) Typical organic alterations of mice that received a transplant of cS5F and control GFPv mice that were killed at 6 weeks after transplantation. Leukemic mice that received a transplant of cS5F have a unique pathologic appearance with massively enlarged mesenterial lymph nodes (lymphadenopathy) and splenomegaly. (D) Splenic- and BM-cell extracts from 2 mice that received a transplant of cS5F (Sp1, Sp2, BM1, BM2) or from control GFPv mice (Sp5, BM5, BM6) were analyzed by Western blotting with the indicated antibodies. Cell lysates from Ba/F3 cells unstimulated or stimulated with IL-3 for 30 minutes were also included as controls.
Leukemogenic mice that received a transplant of cS5F as a model to characterize the transforming properties of Stat5. (A) Kaplan-Meier plot of mice that received a transplant of cS5F (n = 8) versus mice that received a transplant of GFPv (n = 4). Mice that received a transplant of cS5F died within 7 weeks after transplantation, while control GFPv mice remain disease free (followed for more then 10 months after transplantation). Thus, experimental mice that received a transplant of cS5F were killed and analyzed 6 weeks after transplantation for all following experiments (dashed box). (B) White blood cell counts from mice that received a transplant of cS5F (n = 8) and from GFPv mice (n = 4). Error bars represent SE. (C) Typical organic alterations of mice that received a transplant of cS5F and control GFPv mice that were killed at 6 weeks after transplantation. Leukemic mice that received a transplant of cS5F have a unique pathologic appearance with massively enlarged mesenterial lymph nodes (lymphadenopathy) and splenomegaly. (D) Splenic- and BM-cell extracts from 2 mice that received a transplant of cS5F (Sp1, Sp2, BM1, BM2) or from control GFPv mice (Sp5, BM5, BM6) were analyzed by Western blotting with the indicated antibodies. Cell lysates from Ba/F3 cells unstimulated or stimulated with IL-3 for 30 minutes were also included as controls.
In vitro proliferation of cS5F-expressing BM cells
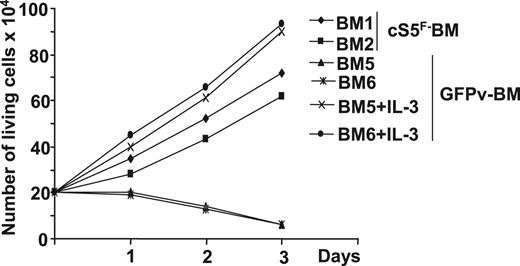
Proliferation and survival of BM progenitors are influenced by various hematopoietic growth factors, including stem-cell factor (SCF) and IL-3. Because persistent Stat5 signaling was shown to relieve the growth factor dependence of cell lines, we then analyzed the growth factor requirement of cS5F-expressing BM cells. GFP+ BM cells of 2 leukemic mice that received a transplant of cS5F or of 2 control GFPv mice were isolated and cultured in a medium containing SCF. In these conditions, cS5F-GFP+ BM cells (BM1 and BM2) were able to grow, whereas control GFP+ BM cells (BM5 and BM6) did not grow (Figure 2). Addition of IL-3 restored the proliferating capacities of control GFP+ BM cells. However, we observed that the presence of SCF was absolutely required for the growth of these cS5F-transformed primary cells, indicating that expression of this mutant partially relieved their cytokine (IL-3) dependence. Note, the growth rate of GFPv BM cells cultured in presence of SCF, and IL-3 was always higher than the growth rate of cS5F BM cells cultured in the presence of SCF alone (Table 1).
In vitro proliferation of cS5F-transformed bone marrow cells. Sorted GFP+ BM cells from 2 cS5F-grafted mice (BM1 and BM2) or from 2 control GFPv mice (BM5 and BM6) were cultured in the presence of SCF (10 ng/mL). In addition to SCF, IL-3 (10 ng/mL) was added to BM5- and BM6-cell cultures. Cells were counted daily using the trypan blue dye exclusion method.
In vitro proliferation of cS5F-transformed bone marrow cells. Sorted GFP+ BM cells from 2 cS5F-grafted mice (BM1 and BM2) or from 2 control GFPv mice (BM5 and BM6) were cultured in the presence of SCF (10 ng/mL). In addition to SCF, IL-3 (10 ng/mL) was added to BM5- and BM6-cell cultures. Cells were counted daily using the trypan blue dye exclusion method.
cS5F promotes the growth of hematopoietic progenitor cells in vitro
Freshly isolated BM cells from animals that received a transplant of cS5F (n = 4) were subjected to lineage markers and GFP expression by FACS analysis (Table 1). The lineage markers included Ter119 (erythroid); Gr-1/Mac-1 (myeloid); CD19 and Thy-1.2 (B and T lineages); and combinations of CD34, Sca-1, and CD117 (c-Kit), typical markers for immature multipotent cells or hematopoetic stem cells (HSCs). The majority of cS5F-GFP+ BM cells was Gr-1– and Mac-1–positive myeloid cells and around 20% of GFP+ BM cells were of B- or T-lymphoid origin. A fraction of cS5F-GFP+ BM cells also expressed markers typical for primitive multipotent cells. Following 1 week of culture, the myeloid- and lymphoid-specific markers disappeared from the cS5F-GFP+ BM cells. By contrast, GFP+ cells expressing the immature markers CD34, Sca-1, and CD117 were amplified, indicating that cS5F promotes the growth of immature progenitor cells. These data support the recent findings that the multilineage cell expansion observed in these mice arose from immature cells transformed by cS5F.21
Cytoplasmic localization of cS5F in transformed BM cells
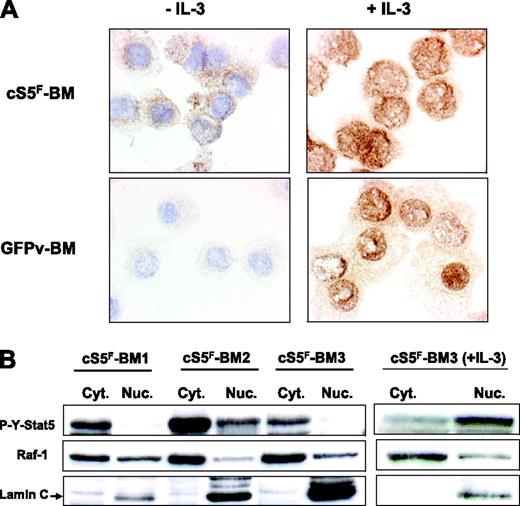
It was previously shown that phosphorylated forms of Stat5 interact with various cytoplasmic signaling effectors.27-29 We therefore addressed the question whether cS5F may have a cytoplasmic function. We first determined the localization of cS5F in transformed BM cells by immunocytochemistry with an anti–phospho-Y694-Stat5 antibody before and after stimulation with IL-3. Results clearly showed (Figure 3A) that constitutively active cS5F mutant is mainly localized in the cytoplasm of BM cells, whereas it is found in the nucleus after stimulation with IL-3. We also analyzed the localization of P-Y-Stat5 in control GFPv BM cells stimulated or not with IL-3. No phosphorylated Stat5 was detected in IL-3–deprived cells, whereas IL-3 treatment of GFPv cells induced the appearance of nuclear P-Y-Stat5. To confirm the specific cytoplasmic localization of cS5F, we prepared cytoplasmic and nuclear extracts of cS5F BM cells from 3 different leukemic mice, and the presence of P-Y-Stat5 in both fractions was analyzed by Western blotting with an anti–P-Y-Stat5 antibody (Figure 3B). P-Y-Stat5 was detected in the cytoplasmic fraction of cS5F BM cells, whereas it was mainly found in the nucleus after stimulation with IL-3. The purity of the extracts was controlled by Western blot analysis for the cytoplasmic ser/thr kinase Raf-1 and the nuclear protein Lamin C. Altogether, these results support the idea that cS5F has a cytoplasmic signaling function that might play an important role in cS5F-induced leukemia.
Cytoplasmic localization of cS5F in bone marrow cells. (A) Immunocytochemical detection of phosphorylated Stat5 in cS5F-expressing BM cells and control GFPv BM cells stimulated or not with IL-3. In each case, cells were spun on cytospin slides and analyzed by indirect immunocytochemistry using an anti–P-Y-Stat5 antibody (AX1). (B) The localization of cS5F was also determined by Western blot analysis using cytoplasmic and nuclear extracts of unstimulated or IL-3–stimulated cS5F BM cells from 3 different mice that received a transplant.
Cytoplasmic localization of cS5F in bone marrow cells. (A) Immunocytochemical detection of phosphorylated Stat5 in cS5F-expressing BM cells and control GFPv BM cells stimulated or not with IL-3. In each case, cells were spun on cytospin slides and analyzed by indirect immunocytochemistry using an anti–P-Y-Stat5 antibody (AX1). (B) The localization of cS5F was also determined by Western blot analysis using cytoplasmic and nuclear extracts of unstimulated or IL-3–stimulated cS5F BM cells from 3 different mice that received a transplant.
Critical role of the Gab2/PI3-K/Akt pathway in cS5F-mediated cell transformation
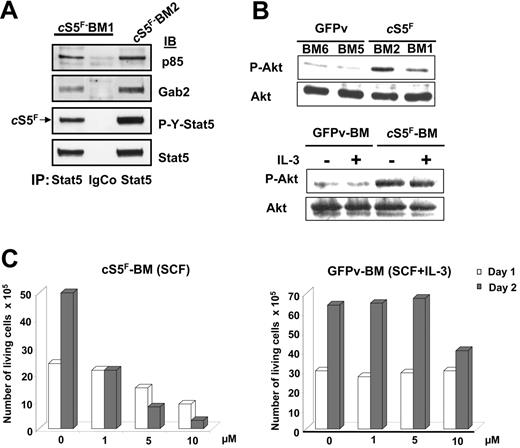
We have previously demonstrated that tyrosine-phosphorylated Stat5 is able to interact with both the PI3-K regulatory subunit p85 and the Gab2-scaffolding adapter in Ba/F3 cells.30,31 We therefore determined whether cS5F also interacts with PI3-K and Gab2 in the primary leukemic cells from diseased mice. For this purpose, cell lysates obtained from 2 different cS5F-expressing BM cells (BM1 and BM2) were immunoprecipitated with anti-Stat5 or isotype-matched control antibodies. The coimmunoprecipitations of the p85 regulatory subunit of PI3-K and Gab2 were next evaluated by immunoblotting (Figure 4A). Results showed that both p85 and Gab2 were associated with cS5F in primary leukemic-cell extracts. BM-cell lysates of mice that received a transplant with cS5F or control GFPv mice were next analyzed by immunoblotting with anti–Pser473-Akt and anti-Akt antibodies (Figure 4B, top). A weak phosphorylation of Akt was found in GFPv BM cells, whereas the level of phosphorylated Akt was much higher in cS5F-expressing BM cells, indicating that Akt phosphorylation was significantly induced by cS5F in these primary leukemic cells. IL-3 treatment of GFPv BM cells and cS5F BM cells did not further increase the level of phosphorylated Akt (Figure 4B, lower panel) but retained the capacity to activate Stat5 (see Figure S5A, available on the Blood website; click on the Supplemental Figures link at the top of the online article).
cS5F interacts with Gab2 and PI3-K and induces Akt activation in transformed bone marrow cells. (A) Stat5 was immunoprecipitated from 2 cS5F-expressing BM-cell lysates (BM1 and BM2). Subsequently, the content in p85 and Gab2 proteins was evaluated in the Stat5 immunoprecipitates by Western blotting using the indicated antisera. Cell lysates were also immunoprecipitated with an isotypic control IgG antibody. (B) BM-cell lysates from 2 mice that received a transplant of cS5F or from 2 control GFPv mice (top) and lysates from GFPv BM and cS5F BM cells stimulated or not with IL-3 (10 ng/mL) for 30 minutes (bottom) were analyzed by Western blotting with the indicated antibodies. (C) cS5F-expressing BM and control GFPv BM cells cultured in the presence of SCF and SCF + IL-3, respectively, were incubated or not with different concentrations of LY294002 for 48 hours. The number of living cells was determined daily using the trypan blue dye exclusion method. Results are the mean of 3 independent experiments.
cS5F interacts with Gab2 and PI3-K and induces Akt activation in transformed bone marrow cells. (A) Stat5 was immunoprecipitated from 2 cS5F-expressing BM-cell lysates (BM1 and BM2). Subsequently, the content in p85 and Gab2 proteins was evaluated in the Stat5 immunoprecipitates by Western blotting using the indicated antisera. Cell lysates were also immunoprecipitated with an isotypic control IgG antibody. (B) BM-cell lysates from 2 mice that received a transplant of cS5F or from 2 control GFPv mice (top) and lysates from GFPv BM and cS5F BM cells stimulated or not with IL-3 (10 ng/mL) for 30 minutes (bottom) were analyzed by Western blotting with the indicated antibodies. (C) cS5F-expressing BM and control GFPv BM cells cultured in the presence of SCF and SCF + IL-3, respectively, were incubated or not with different concentrations of LY294002 for 48 hours. The number of living cells was determined daily using the trypan blue dye exclusion method. Results are the mean of 3 independent experiments.
We next determined whether activation of the PI3-K is required for cS5F-induced cell growth and survival. For this purpose, we used the pharmacologic PI3-K inhibitor LY294002. cS5F BM cells cultured in the presence of SCF or control GFPv BM cells grown in the presence of SCF and IL-3 were treated with increasing concentrations (ranging from 1 to 10 μM) of LY294002 (Figure 4C). Growth arrest of the cS5F-expressing BM cells was observed with 1 μM LY294002 after 48 hours of culture, whereas 10 μM was required to inhibit the growth of GFPv BM cells, indicating that cS5F BM cells were more sensitive to the inhibition of PI3-K than were control cells. Collectively, our results suggest that cS5F molecules form a signaling complex with Gab2 and PI3-K to promote the phosphorylation of Akt.
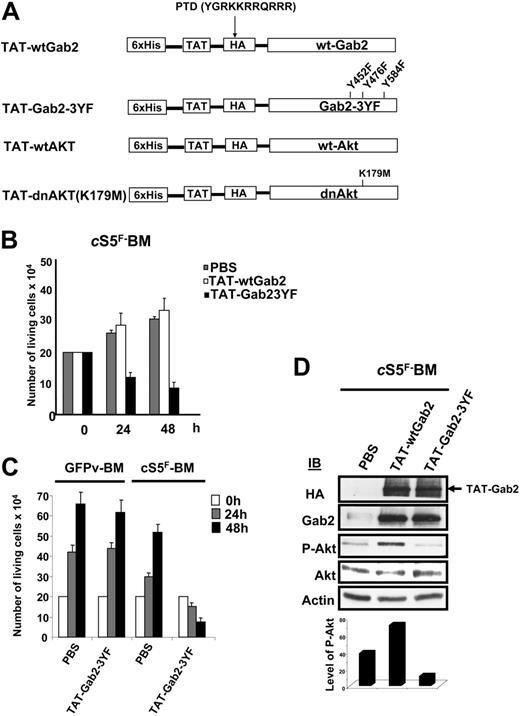
Recombinant TAT fusion proteins that interfere with PI3-K signaling block cS5F-induced cell growth and Akt phosphorylation
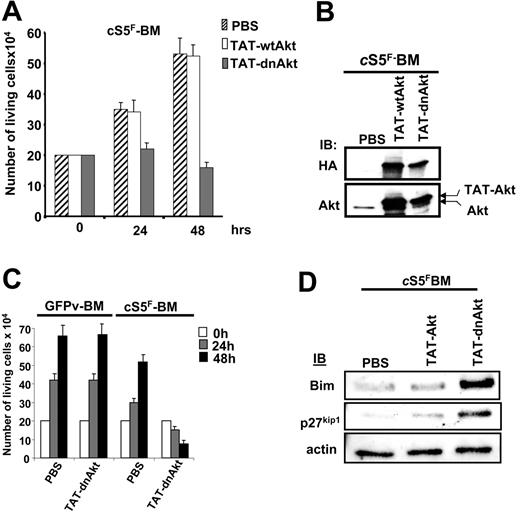
To investigate the role of Gab2 and Akt in the transforming properties of cS5F, we generated different recombinant TAT-Gab2 or TAT-Akt fusion proteins (Figure 5A). The use of TAT fusion proteins is an appealing approach to transduce cell lines or primary cells.39,40 Adding them to the culture medium requires, however, biochemical testing for each individual fusion protein purification which had to be done freshly for each individual experiment. We first analyzed the role of Gab2 in cS5F-induced cell growth, and, for this, we generated a TAT-wtGab2 and a TAT-Gab2-3YF deficient in PI3-kinase binding in which the 3 tyrosine residues (Y,452, Y476, and Y584) were changed to phenylalanine (Figure 5A). It was shown that mutations of these 3 tyrosine residues inhibit activation of the PI3-K/Akt pathway in cytokine-stimulated cells.41 TAT-wtGab2 and TAT-Gab2-3YF proteins were purified and quantified as described in “Patients, materials, and methods” (data not shown). cS5F BM cells from 3 different mice that received a transplant were transduced or not with the TAT-Gab2 fusion proteins (100 nM) for 24 and 48 hours, and the number of living cells was estimated by the trypan blue dye exclusion method (Figure 5B). Transduction of TAT-Gab2-3YF protein abolished the growth of the cells, whereas transduction of TAT-wtGab2 slightly increased their proliferation. Transduction efficiency of the different TAT-Gab2 proteins was evaluated in cS5F-expressing BM cells by Western blot analysis with anti-HA and anti-Gab2 antibodies (Figure 5D). Equal transduction of both proteins was observed. To verify the specific effect of the TAT-Gab2-3YF fusion protein, control GFPv BM cells grown in the presence of SCF and IL-3 or cS5F BM cells were transduced with the TAT-Gab2-3YF protein (100 nM), and cell growth was determined next (Figure 5C). The TAT-Gab2-3YF protein clearly inhibited the proliferation of cS5F BM cells, whereas control GFPv BM cells remained insensitive. Western blot analysis with anti-HA and anti-Gab2 antibodies showed that both types of cells were equally and efficiently transduced (data not shown). We also determined whether TAT-Gab2 proteins could alter the PI3-K activity in cS5F BM cells by analyzing the phosphorylation level of Akt. Although phosphorylation of Akt was clearly enhanced following transduction with TAT-wtGab2, it sharply decreased Akt activation in TAT-Gab2-3YF–treated cells (Figure 5D). Similarly, a wild-type (wt) or a dominant-negative (dn) form of Akt containing a point mutation that inactivates its kinase activity (K179→M) were fused to the protein transduction domain of the TAT HIV protein (Figure 5A). Resulting TAT-Akt fusion proteins were produced, purified, and quantified exactly like TAT-Gab2 fusion proteins (see Figure S1). The biologic effects of TAT-Akt proteins were then evaluated on cS5F-transformed BM cells. Primary cells were incubated with the different TAT-Akt fusions, and cell growth was determined in a time course experiment. Transduction of TAT-dnAkt completely inhibited the proliferation of cS5F BM cells, whereas TAT-wtAkt was without any detectable effects on cell growth (Figure 6A). Similar results were obtained after transduction of the TAT-Akt fusion proteins in Ba/F3 cells (Figure S2). Western blot analysis with anti-HA and anti-Akt antibodies showed that both TAT-Akt fusion proteins were equally transduced in cS5F-expressing BM cells (Figure 6B). We also analyzed the effect of the TAT-dnAkt fusion protein on the growth of GFPv-BM cells (Figure 6C). Although GFPv BM cells and cS5F BM cells were equally and efficiently transduced (data not shown), results clearly showed that transduction of the TAT-dnAkt fusion protein had no effect on GFPv BM cell growth, arguing again that cS5F BM cells but not GFPv BM cells were highly sensitive to inhibition of the PI3-kinase/Akt pathway. It was previously shown that Akt modulates the phosphorylation of the Forkhead family members.33 Inhibition of Akt activity promotes the nuclear translocation and transcriptional activity of these proteins. Accordingly, we found that transduction of TAT-dnAkt but not TAT-wtAkt inhibited FKHR (FoxO1A) phosphorylation (see Figure S3). Bim and p27kip1 are among the target genes of the Forkhead transcription factors, and they execute critical roles in apoptosis or inhibition of cell-cycle progression.42,43 We therefore determined whether TAT-dnAkt affected the transcriptional activity of Forkhead transcription factors and analyzed expression of Bim and p27kip1 by immunoblotting in cS5F-expressing BM cells transduced with the different TAT-Akt proteins (Figure 6D). Expression of both Bim and p27kip1 was up-regulated in cells transduced with the TAT-dnAkt but not with the TAT-wtAkt. These data clearly indicate that TAT-dnAkt is capable to specifically inhibit the phosphorylation of FKHR in primary transformed cS5F-expressing BM cells, thereby inducing its transcriptional target genes. These results also indicate that Akt plays an important regulatory role in the cS5F-induced cell growth and survival by down-modulating Bim and p27Kip1 molecules.
Effects of recombinant TAT-Gab2 fusion proteins on cS5F-induced cell growth and Akt phosphorylation. (A) Schematic representation of TAT-Akt and TAT-Gab2 constructs. The different cDNAs were introduced into the bacterial expression vector pTAT-HA. Resulting recombinant Akt (wt and dn) and Gab2 (wt and Gab23YF) proteins were fused in their N-terminal part to a 6 ×His-Tag followed by the protein transduction domain (PTD) of the TAT protein and a HA tag sequence. (B) cS5F BM cells were transduced or not with 100 nM of the different TAT-Gab2 proteins during 48 hours, and the number of living cells was determined daily using the trypan blue dye assay. Results are the mean of 3 independent experiments performed with 2 independent cS5F BM-cell cultures. (C) GFPv BM cells and cS5F BM cells were transduced with 100 nM TAT-Gab23YF fusion protein, and growth of the cells was determined daily. Results are the mean of 3 independent experiments. (D) After extensive washes, lysates from transduced cS5F BM cells were prepared and analyzed by Western blotting with the indicated antibodies. Densitometric analysis was also performed to determine the ratio of P-Akt/Akt in the different samples (bottom).
Effects of recombinant TAT-Gab2 fusion proteins on cS5F-induced cell growth and Akt phosphorylation. (A) Schematic representation of TAT-Akt and TAT-Gab2 constructs. The different cDNAs were introduced into the bacterial expression vector pTAT-HA. Resulting recombinant Akt (wt and dn) and Gab2 (wt and Gab23YF) proteins were fused in their N-terminal part to a 6 ×His-Tag followed by the protein transduction domain (PTD) of the TAT protein and a HA tag sequence. (B) cS5F BM cells were transduced or not with 100 nM of the different TAT-Gab2 proteins during 48 hours, and the number of living cells was determined daily using the trypan blue dye assay. Results are the mean of 3 independent experiments performed with 2 independent cS5F BM-cell cultures. (C) GFPv BM cells and cS5F BM cells were transduced with 100 nM TAT-Gab23YF fusion protein, and growth of the cells was determined daily. Results are the mean of 3 independent experiments. (D) After extensive washes, lysates from transduced cS5F BM cells were prepared and analyzed by Western blotting with the indicated antibodies. Densitometric analysis was also performed to determine the ratio of P-Akt/Akt in the different samples (bottom).
Specific inhibition of cS5F-induced cell growth by the recombinant TAT-dnAkt protein. (A) cS5F BM cells were grown in the presence of PBS, TAT-wtAkt, or TAT-dnAkt fusion proteins (100 nM), and the cells were counted daily using the trypan blue dye assay. Results are the mean of 3 independent experiments. (B) cS5F BM cells were transduced with the different TAT-Akt fusion proteins during 24 hours, and the presence of the recombinant proteins was detected by immunoblotting with the indicated antibodies. (C) GFPv BM cells and cS5F BM cells were transduced with 100 nM TAT-dnAkt fusion protein, and the number of living cells was enumerated daily. Results are the mean of 3 independent experiments. (D) Expression of Bim and p27kip1 was evaluated by Western blotting in cS5F-expressing BM cells transduced or not with recombinant TAT-Akt.
Specific inhibition of cS5F-induced cell growth by the recombinant TAT-dnAkt protein. (A) cS5F BM cells were grown in the presence of PBS, TAT-wtAkt, or TAT-dnAkt fusion proteins (100 nM), and the cells were counted daily using the trypan blue dye assay. Results are the mean of 3 independent experiments. (B) cS5F BM cells were transduced with the different TAT-Akt fusion proteins during 24 hours, and the presence of the recombinant proteins was detected by immunoblotting with the indicated antibodies. (C) GFPv BM cells and cS5F BM cells were transduced with 100 nM TAT-dnAkt fusion protein, and the number of living cells was enumerated daily. Results are the mean of 3 independent experiments. (D) Expression of Bim and p27kip1 was evaluated by Western blotting in cS5F-expressing BM cells transduced or not with recombinant TAT-Akt.
Constitutively phosphorylated Stat5 proteins are present in the cytoplasm of human neoplastic cells from patients with chronic myeloid leukemia (CML) and patients with acute myeloid leukemia (AML)
It was previously shown that Stat5 is constitutively active in leukemic cells from patients with CML and patients with AML.10 We then analyzed by immunohistochemistry the localization of P-Y-Stat5 in neoplastic cells from patients with CML at different phases of the disease progression (Figure 7A). In the normal bone marrow (BM), P-Y-Stat5 was detectable in the cytoplasm of megakaryocytes and some immature myeloid cells, whereas mature granulocytes and mature erythrocytes did not contain substantial amounts of P-Y-Stat5. In chronic-phase CML (CML-cp), an increase in P-Y-Stat5–positive myeloid progenitors was found when compared with normal BM. Blast cells in patients with CML blast phase (CML-pb) were also found to react with the anti–P-Y-Stat5 antibody. In both cases, P-Y-Stat5 was predominantly detected in the cytoplasm of the leukemic cells. The most abundant staining reaction was seen in a patient with CML with megakaryoblast crisis. To confirm the cytoplasmic localization of P-Y-Stat5, cytoplasmic and nuclear fractions of cells from a patient with CML were analyzed by Western blotting using an anti–P-Y-Stat5 antibody. As shown Figure 7B, P-Y-Stat5 was exclusively localized in the cytoplasm of these leukemic cells. The purity of the cellular fractions was controlled by a Western analysis for the cytoplasmic Raf-1 kinase and the nuclear Topoisomerase I p97/p70 proteins. Bone marrow sections obtained from patients with AML-M2, -M3, -M6, and -M7 were also stained with anti–P-Y-Stat5 antibody (Figure 7C). A clear staining of blast cells was found in those with AML-M2 and AML-M3, and in the patient with AML-M7, in whom blasts cells contained huge amounts of P-Y-Stat5. By contrast, erythroid progenitors were found to be P-Y-Stat5-low cells in one patient (AML-M6) closely reflecting the situation in the normal BM. Collectively, these results suggested that oncogenic activation of Stat5 promotes the cytoplasmic localization of these transcription factors in myeloid leukemias.
Cytoplasmic localization of phosphorylated Stat5 in myeloid leukemia. (A) Immunohistochemical detection of P-Y-Stat5 in neoplastic cells from patients with CML. P-Y-Stat5 immunostaining was realized on normal bone marrow (BM), on neoplastic cells from CML in chronic phase (CML-cp), in blast phase (CML-pb), and in megakaryoblast crisis. (B) Cytoplasmic and nuclear extracts were prepared from leukemic cells of a patient with CML and analyzed by Western blot with the indicated antibodies. (C) Immunohistochemical detection of P-Y-Stat5 in AML blasts. P-Y-Stat5 immunostaining was performed with bone marrow sections from patients AML-M2, -M3, -M6, and -M7.
Cytoplasmic localization of phosphorylated Stat5 in myeloid leukemia. (A) Immunohistochemical detection of P-Y-Stat5 in neoplastic cells from patients with CML. P-Y-Stat5 immunostaining was realized on normal bone marrow (BM), on neoplastic cells from CML in chronic phase (CML-cp), in blast phase (CML-pb), and in megakaryoblast crisis. (B) Cytoplasmic and nuclear extracts were prepared from leukemic cells of a patient with CML and analyzed by Western blot with the indicated antibodies. (C) Immunohistochemical detection of P-Y-Stat5 in AML blasts. P-Y-Stat5 immunostaining was performed with bone marrow sections from patients AML-M2, -M3, -M6, and -M7.
Discussion
Beside their physiologic role in hematopoietic-cell development and function, deregulated Stat5 activities promote leukemogenesis.10 Engineered Stat5 mutants with constitutive activities are a good model to mimic persistent tyrosine kinase activation and to investigate the role of deregulated Stat5 activity in cancers that are susceptible to tyrosine kinase oncogenes. Mutant tyrosine kinases activate distinct signaling pathways (eg, Ras/Map kinase, PI3-K/Akt, NFκB) that play a role in cell growth and survival.44-47 It is therefore crucial to determine the precise contribution of Stat5 molecules in the transforming process. Here, we have started to analyze cytoplasmic functions of Stat5 that bridge to PI3-K signaling both in a murine transplantation system with oncogenic Stat5 or in cells from patients with CML and patients with AML associated with a constitutive Stat5 activity. To address signaling properties of activated cytoplasmic Stat5 proteins, we used mainly a murine transplantation model of Stat5-induced leukemia.21 Mice that received a transplant with BM cells expressing cS5F exhibited a multilineage hematopoietic-cell expansion that lead to severe leukemia within 4 weeks after transplantation. We found that contrary to normal hematopoietic progenitors, the in vitro growth and survival of cS5F-transformed BM cells did not require IL-3, but SCF was essential for self-renewal of immature cell populations. The possibility of an autocrine loop of IL-3 secretion was excluded by the lack of alteration of cell growth in the presence of neutralizing anti–IL-3 antibodies (data not shown). These findings are reminiscent of the loss of IL-3 dependence of Stat5A1*6-expressing Ba/F3 cells.20
The present study demonstrates for the first time that a major fraction of constitutively active Stat5 molecules is cytoplasmic under steady state culture conditions in the presence of growth factors such as SCF or in leukemic patient samples. The cytoplasmic localization of P-Y-Stat5 is not restricted to CML and is also found in AML. We lack a full understanding of the cytoplasmic localization of P-Y-Stat5 in leukemic cells, but active Stat5 proteins are rapidly shuttling between nucleus and cytoplasm.48 It is therefore conceivable that persistent phosphorylation of Stat5 may affect its subcellular localization. Alternatively, it is possible that translocation signals provided by activated cytokine receptors or signals that induce activation of tyrosine phosphatases are lacking in cS5F BM cells, thereby inducing accumulation of cS5F in the cytoplasm. Studies with kinetic experiments of Stat5 protein shuttling in and out of the nucleus on cytokine action in normal versus transformed cell pairs would reveal important insights. Irrespective, persistent transcriptional activity of Stat5 seems not to be the dominant form of oncogenic Stat5 molecules in myeloid-cell types. Our results support the concept that a large part of oncogenic Stat5 activity might not only involve a nuclear function but also a cytoplasmic function. The data are consistent with previous work showing that phosphorylated forms of Stat5 act as signaling effectors by interacting with various signaling molecules such as Crkl, Gab2, and PI3-kinase.27-31 Accordingly, we also found that expression of cS5F in BM cells induced activation of the PI3-K/Akt pathway via an interaction with the scaffolding adapter Gab2 and p85, the regulatory subunit of the PI3-K. It will be difficult in future experiments to create mutants of Stat5 that do not interact with PI3-K signaling, because tyrosine phosphorylation of Stat5 is absolutely required for Gab2 and p85 interaction.31 Importantly, the PI3-K inhibitor LY294002 completely blocked the growth of cS5F BM cells, but PI3-K activation is also achieved efficiently through activation of the c-Kit receptor and SCF was always present in the primary murine-cell cultures. Thus, activation of the PI3-K/Akt pathway was required for cS5F-mediated cell transformation. We also addressed in detail how activation of the Gab2/PI3-kinase/Akt pathway is involved in the cS5F-induced cell transformation. Activation of the PI3-K by tyrosine kinases following activation by extracellular stimuli or after oncogenic challenge occurs via distinct mechanisms. One of them implies the binding of p85 to the scaffolding adapters Gab1/2.49 In such a scenario, tyrosine phosphorylation of Gab proteins allows recruitment of the p85 subunit via its SH2 domain and this leads to the allosteric activation of the catalytic subunit. Data from our laboratory are in line with this model and we demonstrate here by means of a recombinant and transducible TAT-Gab2-3YF fusion protein, a deficient p85 binding mutant, that Gab2 is absolutely necessary for cS5F-induced cell growth and Akt phosphorylation. We also analyzed the downstream effects of Akt on cS5F-induced cell growth by using transducible recombinant TAT fusion proteins fused to a wt or a dominant-negative form of Akt. Akt has been shown to directly phosphorylate and inactivate members of the Forkhead family which includes AFX, FKHR, and FKHRL1.33,34 Accordingly, we found that transduction of the mutant TAT-dnAkt protein inhibits the phosphorylation of FKHR and up-regulates expression of Bim and p27kip1 proteins in transformed bone marrow cells inhibiting cell growth (see also Figure S3). In regard to the transformation process induced via cS5F activity, the overall increase in Akt phosphorylation contributes to growth and survival advantages in the absence of cytokines. Therefore, signaling via the Akt kinase is an essential step in Stat5-induced leukemogenesis.
Persistent Stat5 and PI3-K activation were observed in cells expressing various oncogenic tyrosine kinases such as Bcr-Abl, and it was shown that both pathways contribute to the transformation process controlled by this oncoprotein.13,47 Interestingly enough, Bcr-Abl is unable to transform and to activate the PI3/Akt pathway in primary myeloid cells isolated from gab2−/− mice, indicating that Gab2 signaling via the PI3-K is an important step in Bcr-Abl–induced leukemogenesis.50 Collectively, these data support our novel findings that formation of a cytoplasmic signaling complex among Stat5, Gab2, and PI3-K might be one of the key steps in cS5F-evoked multilineage leukemia. A recent report documented lentivirus-mediated RNA interference to inhibit Stat5 or Gab2 protein expression, leading to diminished in vitro colony formation of CML colony-forming units (CFUs) pinpointing to an essential role of both proteins for Bcr-Abl transformation in line with knock-out studies of both molecules.17,51
Immunohistochemistry or nuclear versus cytoplasmic fractionation proved that persistent Stat5 activity is mainly present in the cytoplasm of leukemic cells isolated from patients with CML or from primary cS5F-transformed cells. In addition, we also found that phosphorylated Stat5 proteins interact with PI3-kinase and Gab2 in human Bcr-Abl–expressing cell lines such as K56230 and Ku812 (data not shown). Thus, Stat5 proteins might also play an integral part as cytoplasmic signaling effectors via its association with Gab2 and PI3-K in the development of human CML. It is possible that oncogenic activation of Stat5 may affect the ratio of nonphosphorylated/phosphorylated forms of Stat5 in both cellular compartments which could enhance the capacity of these molecules to interact with other signaling pathways, most importantly the PI3-K/Akt pathway. However, the cytoplasmic function of Stat5 is not the sole mechanism by which these proteins promote leukemogenesis.
Some previously identified target genes of Stat5 have been shown to contribute to the development of cancer. DNA binding and transcriptional activities play an important role in the transforming properties of Stat5. We have shown that tetramer formation is required for cS5F-induced leukemia.21 Formation of Stat5 tetramers results in DNA binding to low-affinity Stat5 binding sites and putative Stat5 tetramer target genes include CIS, SPI2.1, CD25, D type cyclins, OSM, or IGF-1.21,52-54 The transduction of persistently activated forms of Stat5 that are tetramerization deficient do not cause leukemia when transplanted in mice.21 Nonetheless, we were able to stain for localization of tetramerization-deficient Stat5 mutants that are persistently active from transfected Ba/F3 cells followed by FACS sorting and immunocytochemistry. cS5F and the tetramer-deficient mutants cS5FW37A and cS5FΔ136 proteins were found to have a predominant cytoplasmic localization under IL-3–deprived conditions in Ba/F3 cells, again IL-3 stimulation caused nuclear translocation and increased tyrosine phosphorylation of the mutants (see Figure S4; data not shown). Because the tetramer-deficient but constitutively active Stat5 mutants are not able to induce cell transformation, this would argue again that the cytoplasmic role of cS5F is also not sufficient to transform cells. The data parallel the predominant cytoplasmic activation on persistent activation of mutant Stat5 in primary mouse cells and the findings from CML or AML patient samples. Our current hypothesis is that both cytoplasmic and nuclear functions are linked to the full transforming activity of cS5F molecules or persistently active Stat5 proteins in leukemia.
In conclusion, both the transcriptional activity and the capacity to regulate the PI3-K/Akt activity are required for the full transforming properties through persistent Stat5 protein activation. Oncogenic activation of Stat5 revealed a surprising cytoplasmic localization and function. This report sheds light on the crucial role of the Gab2/PI3-K/Akt pathway to contribute to cancer progression through persistent Stat5 signaling.
Authorship
Contribution: N.H., C.P., M.K., K.S., B.K., R.N., A.R., I.D., C.S., and M.B. designed and performed the research; V.G.-G. designed the research and wrote the paper; H.B. contributed analytical tools and analyzed the data; K.L. analyzed data and wrote the paper; P.V., R.M., and F.G. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
R.M. and F.G. contributed equally to this study.
Correspondence: Fabrice Gouilleux, Institut National de la Santé et de la Recherche Médicale (EMI 351), Faculté de Médecine, 3 rue des Louvels, 80036 Amiens, France; e-mail: fabrice.gouilleux@sa.u-picardie.fr.
The online version of this article contains a data supplement
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Dr Gu (Harvard Institutes of Medicine) and Dr Dowdy (University of California) for the kind gift of the Gab2 or pTAT-HA vectors and Aline Régnier for her excellent technical support.
This work was supported by Association de la Recherche contre le Cancer, Ligue contre le Cancer (Comité du Nord-Pas de Calais), Conseil Régional de Picardie, Fondation de France and Cent pour Sang la Vie, Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich, FWF (grant P17205-B14) (P.V.) and (grant SFB F28) (R.M.), the French and Algerian Ministry for Research and Technology (N.H.), and ARERS Verre-Espoir (C.P.).