Abstract
Inhibition of BCR-ABL tyrosine kinase with imatinib represents a major breakthrough in the treatment of patients with chronic myeloid leukemia (CML). However, resistance to imatinib develops frequently, particularly in late-stage disease. To identify new cellular BCR-ABL downstream targets, we analyzed differences in global protein expression in BCR-ABL–positive K562 cells treated with or without imatinib in vitro. Among the 19 proteins found to be differentially expressed, we detected the down-regulation of eukaryotic initiation factor 5A (eIF5A), a protein essential for cell proliferation. eIF5A represents the only known eukaryotic protein activated by posttranslational hypusination. Hypusination inhibitors (HIs) alone exerted an antiproliferative effect on BCR-ABL–positive and –negative leukemia cell lines in vitro. However, the synergistic dose-response relationship found for the combination of imatinib and HI was restricted to Bcr-Abl–positive cells. Furthermore, this synergistic effect was confirmed by cytotoxicity assays, cell-cycle analysis, and CFSE labeling of primary CD34+ CML cells. Specificity of this effect could be demonstrated by cotreatment of K562 cells with imatinib and siRNA against eIF5. In conclusion, through a comparative proteomics approach and further functional analysis, we identified the inhibition of eIF5A hypusination as a promising new approach for combination therapy in BCR-ABL–positive leukemias.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder characterized by Philadelphia chromosome translocation t,(9,22), which generates the BCR-ABL fusion gene and oncoprotein, representing a constitutively activated tyrosine kinase. The selective tyrosine kinase inhibitor imatinib (formerly STI571 [Gleevec; Novartis, Basel, Switzerland]) blocks kinase activity by occupying the ATP-binding site of the Abl tyrosine kinases Bcr-Abl, c-Abl, v-ABL, and ABL-related gene (ARG), platelet-derived growth factor receptors (PDGFs) α and β, and the receptor for human stem cell factor (SCF) c-kit.1 Based on numerous studies in CML, including those of patients in early chronic phase (CP),2,3 accelerated phase (AP),4 and myeloid blast crisis (BC),5 imatinib is considered the new criterion for conventional treatment of CML.
Unfortunately, resistance to imatinib occurs frequently during AP and BC, resulting in remissions usually lasting only 6 to 12 months. Apart from second-generation Bcr-Abl inhibitors that have recently been described,6,7 identifying novel direct or indirect downstream targets of Bcr-Abl could contribute significantly to the development of new synergistic treatment strategies in CML. Bcr-Abl regulates diverse signaling pathways involving Ras, phosphatidylinositol 3-kinase (PI3-K), protein kinase C (PKC), Jak-STAT, and NF-κB.8 Whereas consequences of Bcr-Abl inhibition have mostly been investigated at the RNA level,9-12 little is known about changes in global protein expression.13,14 The introduction of standardized 2-dimensional gel electrophoresis methods using immobilized ampholyte pH gradients, combined with mass spectrometry and bioinformatics-based protein database searches, has permitted screening of tumor-related global protein changes on a genomewide scale.15-18
In the present study, we investigated the effects of imatinib on the protein expression profiles of BCR-ABL–positive cells. Among others, we found eIF5A, a protein involved in proliferation control, to be down-regulated with imatinib treatment. Interestingly, eIF5A is subject to a type of posttranslational modification (PTM) called hypusination, representing the transfer of an amino-butyl residue to lysine.19 Several pharmacologic substances are able to inhibit hypusination, rendering eIF5A a potentially promising target protein for single-agent and combined-treatment strategies.20,21 Therefore, we characterized the interaction between imatinib and hypusination inhibitors (HIs) in leukemic cell lines and primary CD34+ cells of CML patients at diagnosis. These results might have implications for the design of novel synergistic therapeutic strategies for patients with Bcr-Abl–positive leukemia and other imatinib-responsive diseases.
Materials and methods
Reagents
Imatinib (kindly provided by E. Buchdunger, Novartis, Basel, Switzerland) stock solution (10 mg/mL; in DMSO/H2O 1:1) was stored at −20°C. Ciclopirox (CPX) and N1 -guanyl-1,7-diaminoheptane (GC7) were obtained from Sigma (Steinheim, Germany) and from Biosearch Technology (Novato, CA), respectively.
Cell culture techniques
K562, HL-60, and BA/F3 cells were obtained from DSMZ (Bielefeld, Germany). BA/F3 p210, T315I, M351T, and E255K cells were kindly provided by C.L. Sawyers (University of California at Los Angeles). All cell lines were cultured in RPMI 1640 medium (Gibco-BRL, Invitrogen, Paisley, United Kingdom) containing 10% fetal calf serum (FCS) (Biochrom KG, Berlin, Germany) and 1 ng/mL recombinant murine interleukin-3 (IL-3) as indicated. Cells were incubated at 37°C in a humidified atmosphere with 5% CO2.
Purification of stem and progenitor cells
Fresh leukapheresis, peripheral blood, or bone marrow samples from CML patients were collected with informed consent. The study was approved by the ethics review board of the medical faculty of the University of Tübingen, Germany. CD34+ cells were selected using a Midi-MACS CD34 Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany), as described.22
Two-dimensional gel electrophoresis
Protein samples were isolated from 107 K562 cells, yielding approximately 1000 μg protein. Cells were lysed in sample buffer (9 M urea, 4% CHAPS, 0.5% Resolyte [BDH Biochemicals, Poole, United Kingdom], 10 μg/mL bromophenol blue) followed by centrifugation at 12 000g for 5 minutes. Protein concentrations were determined by the Bradford method.23 Isoelectric focusing was performed as described.24 Samples were applied to IPG strips (pH 4-7, 18 cm; Amersham Biosciences, Freiburg, Germany) by in-gel rehydration. After isoelectric focusing on Multiphor II (Pharmacia, Uppsala, Sweden), IPG strips were equilibrated for 2 × 15 minutes in 6 M urea, 4% SDS, 50 mM Tris-HCl, pH 8.8, containing 1% DTT for the first or 4.8% iodoacetamide for the second period of equilibration. Strips were placed on vertical SDS-PAGE gels and overlaid with 0.6% agarose. SDS-PAGE was carried out with the IsoDalt system (Amersham Biosciences), with 1.5-mm–thick gels and a 15% acrylamide concentration. Two-dimensional gels were stained overnight with colloidal Coomassie, followed by destaining for 1 day. Digitized images were analyzed with PDQuest 7.2 software (Bio-Rad, Hercules, CA). After spot detection, a match set of gels from treated and untreated cells was built. Individual protein spot quantity was normalized based on total density in the gel image and was expressed as ppm. Quantitative analysis was performed using the Student t test between gels of treated and untreated cells. The confidence level was 95%.
Mass spectrometry
Protein spots of interest were excised from the gel and were washed with Millipore-purified water and with 50% acetonitrile/water. In-gel protein digestion was performed as previously described with minor modifications.25 After excised gel spots were dried, trypsin (sequencing grade; Promega, Mannheim, Germany) was added to each sample. Tryptic protein fragments were extracted from the gel matrix with 5% formic acid and with 50% acetonitrile/5% formic acid. Extracts were pooled and concentrated in a speed vac concentrator. After purification with ZipTips (C18-ZipTip; Millipore, Bedford, MA), aliquots were deposited on a spot of α-cyano-4-hydroxycinnamic acid/nitrocellulose and were analyzed with a Reflex III MALDI-TOF mass spectrometer (Bruker Daltonic, Bremen, Germany) equipped with an N2 337-nm laser. All measurements were performed in the positive-ion reflection mode at an accelerating voltage of 23 kV and delayed-pulsed ion extraction. Sequence verification of tryptic fragments was performed by nano-electrospray tandem mass spectrometry on a hybrid quadrupole orthogonal acceleration time-of-flight mass spectrometer (QSTAR Pulsar i; Applied Biosystems/MDS Sciex, Foster City, CA) equipped with a nanoflow electrospray ionization source. Purified aliquots were loaded in a nano-electrospray needle (BioMedical Instruments, Zoellnitz, Germany), and tandem mass spectra were obtained by collision-induced decay of selected precursor ions. The instrument was calibrated externally. Database searches (NCBInr, nonredundant protein database) were performed using MASCOT software from Matrix Science (Boston, MA) with carboxymethylation of cysteine and methionine oxidations as variable modifications (P < .05).26
Western blotting
For protein extraction, cultured cells were homogenized on ice in lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% Na-desoxycholate, 5 mM EDTA, 1 mM NaF, 25 mM Na3VO4, and 0.1 mM PMSF. Lysates were left on ice for 10 minutes, and cellular debris was pelleted at 20 000g for 20 minutes at 4°C. The supernatant was frozen at −80°C. The protein concentration of the lysate was determined with the BCA Protein Assay Kit (Pierce, Rockford, IL). Proteins (20 μg) were separated by 12.5% or 15% SDS-PAGE and transferred onto nitrocellulose membranes with the Bio-Rad Transblot system. After blocking in PBS-Tween/5% wt/vol milk powder for 1 hour, membranes were incubated in primary antibody diluted in PBS-Tween/5% wt/vol milk powder. The following primary antisera were used: antivinculin and anti–Ran-binding protein (Upstate Biotechnology, Lake Placid, NY); anti–β-tubulin Ab-1 and anti–α-tubulin Ab-1 (Oncogene, Boston, MA); antinucleophosmin (Zymed, San Francisco, CA); anti–phospho-CrKL and anti–phospho-STAT5 (Cell Signaling, Danvers, MA); and antiphosphotyrosine (BD Biosciences, San Jose, CA). After washing, membranes were incubated for 1 hour with HRP-conjugated rabbit anti–goat immunoglobulin (1/10 000) or with rabbit anti–mouse immunoglobulin (1/10 000) (both from Amersham Pharmacia Biotech UK, Little Chalfont, Buckinghamshire, United Kingdom), diluted in PBS-Tween/5% wt/vol BSA. After washing, the enhanced chemiluminescence kit (Amersham Pharmacia Biotech UK) was used to visualize the secondary antibody.
RNA isolation and measurement of transcripts by semiquantitative RT-PCR
RNA was isolated from 5 × 106 K562, HL-60, BA/F3, BA/F3p210, BA/F3p210T315I, M351T, and E255K cells and from PBMCs from 2 healthy donors and 7 CML patients (4 in CP, 2 in AP, and 1 BC) using TRIzol (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol. cDNA was prepared by reverse transcription of 250 ng total RNA using oligo(dT)15 or random hexamer primer and Superscript II reverse transcriptase (Invitrogen, Karlsruhe, Germany). cDNA was amplified by 20 cycles of PCR using REDTaq polymerase (Sigma) and the following primers for eIF5A: sense, 5′-GCG GCC GCA CCA TGG CAG ATG ACT TGG ACT TCG AG-3′; antisense, 5′-CGC AAG CTT CTA TTT TGC CAT GGC CTT GAT TGC-3′, which gives a 476-bp product. In a comparable assay, the 18S ribosomal RNA gene was amplified by 15 cycles of PCR using Taq polymerase and the following primers: 5′-CCA TCC AAT CGG TAG TAG CG-3′ and 5′-GTA ACC CGT TGA ACC CCA TT-3′, which yields a product of 80 bp.
Lentiviral vectors, generation of pseudotyped viral particles, and infection experiments
The replication-incompetent and self-inactivating HIV-1–derived lentiviral vector was kindly provided by Dr Carol Stocking-Harbers (Heinrich-Pette-Institute, Hamburg, Germany) and contains an internal polymerase-III H1 RNA gene promoter for expression of either human eIF5A–specific (5′-GAGATCCTGATCACGGTGC-3′) or luciferase-specific (5′-CGTACGCGGAATACTTCGA-5′; negative control) siRNA. To generate lentiviral pseudotypes, 3 μg lentiviral vector plasmid DNA was contransfected with 1.5 μg pMDLg/pRRE,27 a vector expressing the HIV-1 gag and pol coding sequences, 0.75 μg Rev expression construct pRSV-Rev,27 and 0.75 μg VSV glycoprotein expression plasmid pCMV-VSV-G28 into 3 × 106 293T cells using Lipofectamine according to the manufacturer's protocol (Invitrogen). Forty-eight hours after transfection, viral supernatants were harvested and passed through 0.2-μm pore-size filters, and virus loads were determined (Innotest HIV p24 antigen mAb [Innogenetics NV, Gent, Belgium]). Subsequently, 3 × 105 K562 cells were infected with 100 ng of the respective virus by spin inoculation in 6-well plates at 1200g for 90 minutes at room temperature. After centrifugation, cells were incubated for another 2 hours at 37°C before viral supernatants were removed, and cells were washed twice with PBS and further incubated in RPMI 1640 medium for subsequent analyses.
MTT assay and combination index
K562, BA/F3p210, T315I, M351T, and CD34+ cells from CML patients were plated into 96-well flat-bottomed microtiter plates (Becton Dickinson, Heidelberg, Germany) at 1.5 × 104 cells/well in 150 μL of their respective media. Cells were preincubated for 24 hours before increasing concentrations of imatinib (0-3 μM), CPX (0-81 μM), or GC7 (0-300 μM) were added. All analyses were performed in triplicate. After 48 hours, the viable cells in each well were assayed as described previously.29 The dose-response effect for imatinib at the point of inhibitory concentration (IC50) (corresponding to the affected fraction, Fa, of 0.5) was analyzed by the median-effect method with CalcuSyn Software (Biosoft, Cambridge, United Kingdom).30,31 After an IC50 was obtained for each drug, the antileukemia activity of imatinib, in combination with CPX or GC7, was assayed in a single 96-well plate. Bcr-Abl–positive leukemia cells and CD34+/Bcr-Abl–positive primary cells were treated at 6 concentrations of each compounds.
Apoptosis
K562 and HL-60 cells (1.5 × 105 cells/mL) were cultured in 6-well tissue plates under the conditions described. After 24 hours of preincubation, cells were incubated in increasing concentrations of CPX (0-81 μM) or increasing concentrations of GC7 (0-300 μM) with or without 0.15 μM imatinib (the highest dose of a single-agent treatment that did not lead to a measurable effect). BA/F3, BA/F3p210, T315I, M351T, and E255K cells were incubated in 0.5 μM imatinib, 2.5 μM CPX, or 20 μM GC7 or in imatinib combined with one HI. Cells were sampled after 24 hours of incubation, and the fraction of apoptotic cells was measured by flow cytometry according to the method of Nicoletti et al.32
Short-term expansion of bulk CD34+ cells from healthy donors and CML patients
To determine the effect of hypusination inhibition on the short-term expansion of normal or leukemic hematopoietic progenitor cells, 103 mobilized CD34+ cells from healthy donors (n = 3) or CML patients (n = 2) were seeded in triplicate in 96-well plates (Nunc, Roskilde, Denmark) containing 100 μL serum-free medium (SFM) supplemented with human SCF (100 ng/mL), Flt-3 ligand (100 ng/mL), TPO (50 ng/mL), IL-3 and IL-6 (both 20 ng/mL) (all reagents were from CellSystems, St. Katharinen, Germany), and granulocyte–colony-stimulating factor (G-CSF; 20 ng/mL; Amgen, Munich, Germany).33 The same cytokine-supplemented SFM was used for all in vitro expansion experiments of primary cells reported here. CPX was added at increasing doses from 0 to 40 μM. After 5 days of culture, another 100 μL medium containing growth factor and CPX was added.
High-resolution cell-cycle analysis
Cell-cycle analysis was performed on CD34+-selected primary CML cells at baseline and again after 72 hours in culture with each of the treatment conditions. The flow cytometry method used combines staining of the nuclear activation antigen Ki-67 (Becton Dickinson, Oxford, United Kingdom) in FL1 with 7-amino-actinomycin D (7-AAD; Becton Dickinson) staining of DNA in FL3.34
High-resolution tracking of Bcr-Abl–positive CD34+ CML cell division
After recovery from cryopreservation, CD34+ cells from CML patients (n = 3) and healthy donors (n = 2) were stained with the fluorescent vital stain carboxy fluorescein succinimidyl ester (CFSE; Molecular Probes, Eugene, OR), and a CD34+ CFSE+ population was sorted using a FACSVantage (Becton Dickinson).35 Briefly, the CFSE-stained cells were labeled with anti–CD34-phycoerythrin (PE) (Becton Dickinson) and 1 μg/mL propidium iodide (PI; Sigma), and CFSEmax, CD34+ PI− cells were selected. CD34+ CFSE+ cells were then cultured in SFM for 2 cycles of 72 hours in the following conditions: no drug control; imatinib 0.5 μM; imatinib 5 μM; CPX 1 μM; CPX 9 μM; imatinib 0.5 μM and CPX 1 μM; imatinib 0.5 μM and CPX 9 μM; imatinib 5 μM and CPX 1 μM; imatinib 5 μM and CPX 9 μM; Colcemid (Biological Industries, Kibbutz Beit Haemek, Israel) control.
Results
Two-dimensional gel electrophoresis of extracts from untreated versus imatinib-treated K562 cells
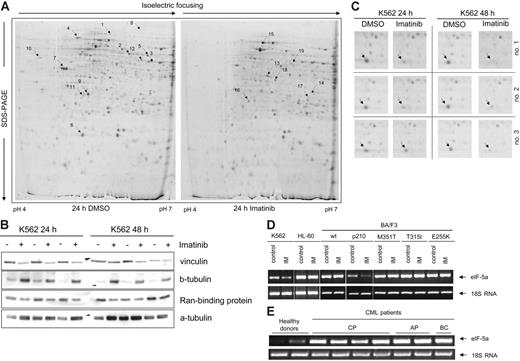
To detect proteins differentially expressed as a function of Bcr-Abl activity, K562 cells were incubated in the presence or absence of the Bcr-Abl inhibitor imatinib for 24 or 48 hours. Representative Coomassie-stained 2-D gels of K562 cell extracts are shown in Figure 1A. In 3 independent experiments making use of image analysis of the 2-D gels, we found 19 protein spots reproducibly displaying a significantly different density dependent on imatinib treatment at both time intervals (Student t test; P < .05). Twelve protein signals were down-regulated (highlighted in Figure 1A, left), and 7 were up-regulated (highlighted in Figure 1A, right). The 19 protein signals were investigated by peptide mass fingerprinting or peptide sequencing (Table 1). Expression patterns of selected differentially regulated candidate proteins were confirmed by Western blot analysis (Figure 1B; Table 2). Expression changes of α-vinculin and Ran-binding protein were consistent with 2-D electrophoretic analysis. One protein, eIF5A (Figure 1A [6 arrow], C [close-up of all 3 gels]), exhibited marked reduction with imatinib treatment. Down-regulation of eIF5A expression after 24-hour treatment with imatinib was confirmed by RT-PCR in Bcr-Abl–positive K562 and BA/F3p210 cells but was not seen in Bcr-Abl–negative HL60 or wt-BA/F3 cells or in BA/F3 cells carrying Bcr-Abl mutants M351T, T315I, or E255K (the latter confers resistance to imatinib; Figure 1D). Furthermore, we extended our analysis to RNAs extracted from healthy donor PBMCs and PBMCs from CML patients without cytogenetic response because they either were untreated or were treated with hydroxyurea at the time of analysis. Clearly, eIF5A mRNA was expressed at elevated levels in the PBMCs of CML patients compared with the PBMCs of healthy donors (Figure 1E).
Two-dimensional gel analysis of proteins from K562 cells. (A) Protein profiles of extracts from K562 cells incubated with DMSO (left) or with imatinib (right) for 24 hours. The marked protein spots were chosen for further characterization by MALDI-MS and ESI-MS/MS because they were expressed to elevated relative levels either in control cells (left) or in cells after imatinib treatment (right). (B) Immunoblots of some representative proteins to confirm the results of 2-D gels. Expression of vinculin and Ran-binding protein was down-regulated, and expression of β-tubulin was up-regulated in K562 cells after incubation with imatinib for 24 and 48 hours. Normalization was performed using α-tubulin. (C) Enlarged regions of Coomassie-stained 2-D gels. Arrows indicate the location of eIF5A. (D) RT-PCR for the detection of eIF5A expression in K562, HL-60, and BA/F3p210, including the mutants M351T, T315I, and E255K as well as wt-BA/F3 cells. Cells were incubated with imatinib for 24 hours and compared with untreated control. (E) RT-PCR for the detection of eIF5A mRNA levels in PBMCs from 2 healthy donors and 7 CML patients. Normalization was performed with human 18S rRNA.
Two-dimensional gel analysis of proteins from K562 cells. (A) Protein profiles of extracts from K562 cells incubated with DMSO (left) or with imatinib (right) for 24 hours. The marked protein spots were chosen for further characterization by MALDI-MS and ESI-MS/MS because they were expressed to elevated relative levels either in control cells (left) or in cells after imatinib treatment (right). (B) Immunoblots of some representative proteins to confirm the results of 2-D gels. Expression of vinculin and Ran-binding protein was down-regulated, and expression of β-tubulin was up-regulated in K562 cells after incubation with imatinib for 24 and 48 hours. Normalization was performed using α-tubulin. (C) Enlarged regions of Coomassie-stained 2-D gels. Arrows indicate the location of eIF5A. (D) RT-PCR for the detection of eIF5A expression in K562, HL-60, and BA/F3p210, including the mutants M351T, T315I, and E255K as well as wt-BA/F3 cells. Cells were incubated with imatinib for 24 hours and compared with untreated control. (E) RT-PCR for the detection of eIF5A mRNA levels in PBMCs from 2 healthy donors and 7 CML patients. Normalization was performed with human 18S rRNA.
HI induces antiproliferative effects in human leukemic cells
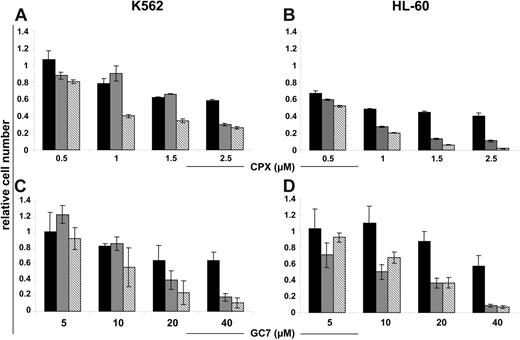
The activation of eIF5A is regulated by a 2-step posttranslational modification called hypusination. We investigated the in vitro effects of 2 HI, CPX, and GC7, which inhibit the first and second steps of this process, on the cellular viability and proliferation of K562 and HL-60 cells using trypan blue exclusion. Treatment with increasing doses of CPX or GC7, respectively, produced a dose-dependent reduction of cell growth after 24, 48, and 72 hours (Figure 2A-D; black, gray, and hatched bars) in Bcr-Abl–positive K562 and Bcr-Abl–negative HL60 cells.
HI inhibits proliferation of Bcr-Abl–positive and –negative human leukemic cells in a dose-dependent fashion. K562 and HL-60 cells were exposed to the indicated concentrations of CPX (A-B) and GC7 (C-D), for 24, 48, and 72 hours (black, gray, and hatched bars, respectively). CPX and GC7 impaired the proliferation of all tested cells independent of Bcr-Abl status. Data shown are mean ± SD of 3 independent experiments. The percentage of cell growth was normalized to the growth of cells in the absence of drug treatment.
HI inhibits proliferation of Bcr-Abl–positive and –negative human leukemic cells in a dose-dependent fashion. K562 and HL-60 cells were exposed to the indicated concentrations of CPX (A-B) and GC7 (C-D), for 24, 48, and 72 hours (black, gray, and hatched bars, respectively). CPX and GC7 impaired the proliferation of all tested cells independent of Bcr-Abl status. Data shown are mean ± SD of 3 independent experiments. The percentage of cell growth was normalized to the growth of cells in the absence of drug treatment.
Cotreatment with imatinib and HI synergized to induce cytotoxicity and apoptosis in K562 cells
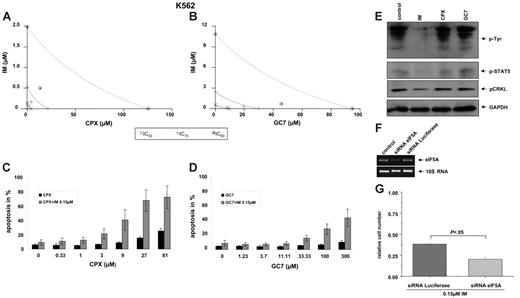
To test the pharmacologic synergism of HI plus imatinib by MTT assay, equitoxic ratios for testing the combination of imatinib plus CPX or GC7, respectively, were chosen on the basis of the single drug concentration that led to a 50% growth inhibition in the target cell line. Results for both combinations are shown in isobologram plots (Figure 3A-B). For 50% inhibition (ED50), 75% inhibition (ED75), and 90% inhibition (ED90), points that fall to the left of the predicted line of additive effect suggest a synergistic interaction of imatinib and CPX (Figure 3A) and of imatinib and GC7 (Figure 3B). When imatinib was combined with CPX, a significant (P < .05) synergistic effect became apparent at therapeutically relevant dose levels (P < .05 at IC50, IC75, and IC90; Table 3; combination index [CI] values less than 1). Similar results were obtained when imatinib was combined with GC7 (Table 3).
Synergistic effects of HI and imatinib on K562 cells. (A-B) MTT assay. Isobologram plots for the effects of imatinib and CPX (A) or GC7 (B) on cellular cytotoxicity. Cells were incubated with CPX or GC7 plus imatinib for 48 hours. Proliferation was assessed with an MTT-based assay. Results were analyzed with Calcusyn software. (C-D) Apoptosis. Cotreatment with subapoptotic concentrations of imatinib and CPX (C) or GC7 (D), respectively, increased apoptosis in K562 cells. Apoptotic cell fractions were measured by PI-staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs. (E) Phosphorylation. Compared with imatinib, no effects of HI on tyrosine, STAT5, or CRKL phosphorylation was observed in K562 cells. (F-G) Lentiviral delivery of siRNA directed against eIF5A in K562 cells. (F) RT-PCR analysis with total RNA from stable K562 cells was performed with a pair of primers amplifying an approximately 476-bp sequence of eIF5A. The mRNA level of eIF5A was normalized to the endogenous 18S rRNA level, as indicated. (G) eIF5A–specific siRNA sensitizes K562 cells to imatinib in the cell viability assay.
Synergistic effects of HI and imatinib on K562 cells. (A-B) MTT assay. Isobologram plots for the effects of imatinib and CPX (A) or GC7 (B) on cellular cytotoxicity. Cells were incubated with CPX or GC7 plus imatinib for 48 hours. Proliferation was assessed with an MTT-based assay. Results were analyzed with Calcusyn software. (C-D) Apoptosis. Cotreatment with subapoptotic concentrations of imatinib and CPX (C) or GC7 (D), respectively, increased apoptosis in K562 cells. Apoptotic cell fractions were measured by PI-staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs. (E) Phosphorylation. Compared with imatinib, no effects of HI on tyrosine, STAT5, or CRKL phosphorylation was observed in K562 cells. (F-G) Lentiviral delivery of siRNA directed against eIF5A in K562 cells. (F) RT-PCR analysis with total RNA from stable K562 cells was performed with a pair of primers amplifying an approximately 476-bp sequence of eIF5A. The mRNA level of eIF5A was normalized to the endogenous 18S rRNA level, as indicated. (G) eIF5A–specific siRNA sensitizes K562 cells to imatinib in the cell viability assay.
We next sought to determine whether imatinib could sensitize Bcr-Abl–dependent cells to HI-induced apoptosis. For this purpose, K562 cells were exposed for 24 hours to increasing concentrations of CPX or GC7 in the presence or absence of subapoptotic concentrations of imatinib (0.15 μM). CPX and GC7 by themselves induced apoptosis in a dose-dependent manner (Figure 3C-D, black bars). However, when either CPX (Figure 3C, gray bars) or GC7 (Figure 3D, gray bars) was combined with imatinib, remarkable increases in the proportion of apoptotic cells were observed. Thus, subapoptotic concentrations of imatinib dramatically increased HI-induced apoptosis in Bcr-Abl–positive K562 cells.
Treatment with HI does not affect the phosphorylation of Bcr-Abl downstream targets in K562 cells
To investigate the potential effects of HI treatment on specific phosphorylation of Bcr-Abl downstream targets, we studied global tyrosine phosphorylation and the phosphorylation of CRKL and STAT5 specifically under treatment with CPX and GC7, respectively. As expected, treatment of K562 cells with imatinib led to the inhibition of global tyrosine phosphorylation and to reduced levels of phosphorylated CRKL and STAT. In comparison, treatment with neither CPX nor GC7 significantly affected the phosphorylation of tyrosine residues in general or of CRKL or STAT5 specifically. These findings argue against a direct effect of HI on the kinase activity of Bcr-Abl.
Stable lentiviral expression of siRNA directed against eIF5A sensitizes K562 cells to imatinib with respect to proliferation
To further investigate the specificity of the antiproliferative effects of HI on Bcr-Abl–positive cells, we aimed to knock down eIF5A in K562 cells. For this purpose, we stably expressed siRNA against eIF5A and luciferase (negative control) in K562 cells by lentiviral gene transfer. First, we confirmed the effect of eIF5A siRNA on eIF5A expression in K562 cells at the mRNA level by RT-PCR (Figure 3F). For comparison, no significant down-regulation of eIF5A mRNA expression was detected in the cells after lentiviral transduction of luciferase siRNA. These results demonstrate that eIF5A–derived siRNA is highly specific for the suppression of eIF5A expression.
We then tested for synergism between eIF5A siRNA expression and imatinib treatment in K562 cells by comparing the antiproliferative effects of imatinib in K562 cells stably expressing siRNA against eIF5A or luciferase as a control. As shown in Figure 3G, treatment with 0.15 μM imatinib for 48 hours led to a significant decrease in proliferation of K562 cells expressing eIF5A siRNA compared with luciferase siRNA–expressing control K562 cells.
HI induces antiproliferative effects in imatinib-resistant BA/F3p210 cells expressing T315I, M351T, and E255K mutations
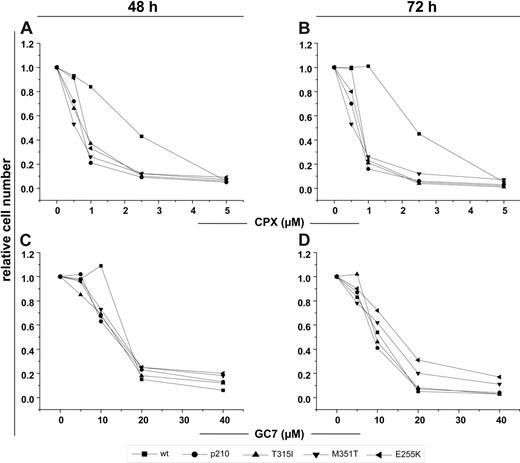
Next, we investigated the antiproliferative effects of the HI CPX and GC7 on wt-BA/F3 and BA/F3p210 cells, including their imatinib-resistant mutants T315I, M351T, and E255K. Analogous to the observations in K562 and HL-60 cells, treatment with increasing doses of CPX or GC7 alone produced a dose-dependent reduction of cell growth after 48 and 72 hours (Figure 4A-D) in BCR-ABL–expressing BA/F3 cells (including imatinib-resistant mutants) and wt-BA/F3 cells. Interestingly, however, particularly with CPX treatment, this antiproliferative effect was substantially more pronounced in Bcr-Abl–positive than in wt-Ba/F3 cells and seemed to be largely independent of Bcr-Abl mutational status (Figure 4A-B).
HI inhibits proliferation of BA/F3 and BA/F3p210 cells, including the mutants M351T, T315I, and E255K. Cells were exposed to the indicated concentrations of CPX (A-B) or GC7 (C-D) for 48 and 72 hours, respectively. CPX and GC7 impaired the proliferation of all tested cells independently of Bcr-Abl mutational status. CPX monotherapy rendered wt-Ba/F3 cells relatively unaffected compared with Ba/F3p210 cells at a dose range of 1 to 5 μm. Data shown are mean of 3 independent experiments. The percentage of cell growth was normalized to the growth of cells in the absence of drug treatment.
HI inhibits proliferation of BA/F3 and BA/F3p210 cells, including the mutants M351T, T315I, and E255K. Cells were exposed to the indicated concentrations of CPX (A-B) or GC7 (C-D) for 48 and 72 hours, respectively. CPX and GC7 impaired the proliferation of all tested cells independently of Bcr-Abl mutational status. CPX monotherapy rendered wt-Ba/F3 cells relatively unaffected compared with Ba/F3p210 cells at a dose range of 1 to 5 μm. Data shown are mean of 3 independent experiments. The percentage of cell growth was normalized to the growth of cells in the absence of drug treatment.
Cotreatment with imatinib and HI synergistically induces cytotoxicity and apoptosis in BA/F3 cells ectopically expressing Bcr-Abl (Ba/F3p210), but not in wt-BA/F3 cells.
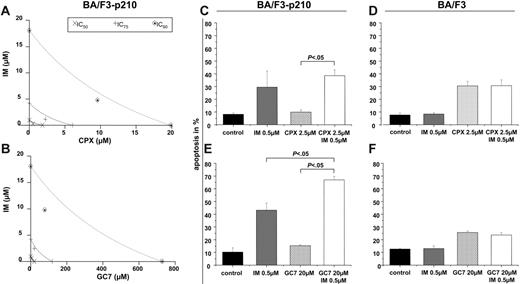
Next, we determined the effects of cotreatment of imatinib and HI on BA/F3p210 cells. Through the MTT assay, we demonstrated synergism between imatinib and both CPX (Figure 5A) and GC7 (Figure 5B), respectively. Compared with treatment with either agent alone, relatively low concentrations of imatinib (0.5 μM) plus CPX (2.5 μM) or GC7 (20 μM) for 24 hours led to an increased rate of apoptotic cells in BA/F3p210 cells (Figure 5C, E). In contrast, combined treatment of HI with imatinib resulted in no significant increase in the induction of apoptosis in BCR-ABL–negative HL-60 (not shown) or BA/F3 control cells (Figure 5D, F).
Synergistic effects of HI and imatinib on BA/F3p210 cells. (A-B) MTT assay. Isobolograms for the effects of imatinib and CPX (A) or GC7 (B), respectively, on cellular proliferation after 48 hours. (C-F) Apoptosis. Co-treatment with imatinib and CPX (C) or GC7 (D) increased apoptosis in BA/F3p210 cells. No synergistic effects in apoptosis induction were observed in wt-BA/F3 cells for the combination of imatinib with CPX (E) or GC7 (F). Apoptotic cell fractions were measured by PI staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs.
Synergistic effects of HI and imatinib on BA/F3p210 cells. (A-B) MTT assay. Isobolograms for the effects of imatinib and CPX (A) or GC7 (B), respectively, on cellular proliferation after 48 hours. (C-F) Apoptosis. Co-treatment with imatinib and CPX (C) or GC7 (D) increased apoptosis in BA/F3p210 cells. No synergistic effects in apoptosis induction were observed in wt-BA/F3 cells for the combination of imatinib with CPX (E) or GC7 (F). Apoptotic cell fractions were measured by PI staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs.
Synergistic effect of cotreatment of imatinib and HI is dependent on the type of resistance to imatinib
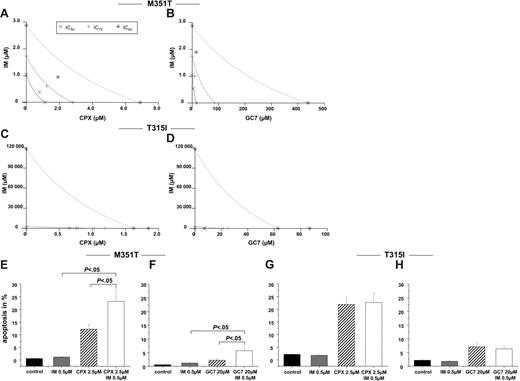
We then assessed the effect of combined treatment of HI and imatinib on BA/F3 cells harboring mutants conferring different degrees of resistance to imatinib. In MTT assays performed in BA/F3p210M351T (M351T) cells, isobolograms demonstrated a synergistic induction of cytotoxicity for the combination of imatinib and CPX or GC7, respectively (Figure 6A-B; Table 3). Calculated CI values were less than 1 at almost all dose effects for the BA/F3p210M351T cells, indicating that a combination of imatinib and HI had much more than an additive effect on inducing cytotoxicity of these leukemic cell lines with moderate resistance to imatinib. In addition, cotreatment with imatinib and CPX or GC7, respectively, significantly increased apoptosis in M351T cells than with either agent alone (Figure 6E-F; P < .05). In contrast, in BA/F3p210T315I (T315I) cells, which are resistant to imatinib as a single agent, the cotreatment of imatinib and HI did not lead to additive or synergistic effects (Figure 6C-D). Consistent with these data, no synergism between imatinib and HI over HI alone with respect to the induction of apoptosis could be observed (Figure 6G-H).
Combination of imatinib with HI acts synergistically only in BA/F3p210 cells with moderate resistance to imatinib. (A-D) MTT assay. Isobolograms for the effects of imatinib and CPX or GC7on BA/F3p210 M351T (A-B) and BA/F3p210 T315I (C-D) cells, respectively, after 48 hours. (E-H) Apoptosis. Cotreatment with imatinib and CPX (C) or GC7 (D) induced more apoptosis in BA/F3p210 M351T cells. Apoptotic cell fractions were measured by PI staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs.
Combination of imatinib with HI acts synergistically only in BA/F3p210 cells with moderate resistance to imatinib. (A-D) MTT assay. Isobolograms for the effects of imatinib and CPX or GC7on BA/F3p210 M351T (A-B) and BA/F3p210 T315I (C-D) cells, respectively, after 48 hours. (E-H) Apoptosis. Cotreatment with imatinib and CPX (C) or GC7 (D) induced more apoptosis in BA/F3p210 M351T cells. Apoptotic cell fractions were measured by PI staining. Values (mean ± SD of 3 experiments) are depicted as bar graphs.
Antiproliferative activity of CPX is enhanced in CD34+ cells from patients with CML compared with healthy donors
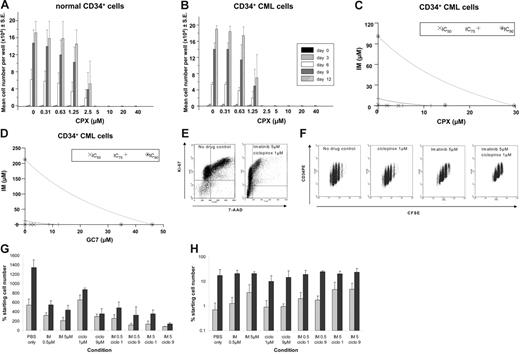
Mobilized CD34+ cells from healthy donors, seeded at 1 × 103 cells/well, were expanded for 12 days in SFM supplemented with human growth factors in the presence of CPX at concentrations up to 40 μM (Figure 7A). For doses up to 2.5 μM CPX, a continuous expansion of hematopoietic cells was observed. However, starting from a concentration of 2.5 μM CPX, significantly reduced expansion became evident compared with untreated cells (Figure 7A). In comparison, a significant suppression of expansion of CD34+ cells was already observed at a dose level of 1.25 μM when cells were derived from untreated patients with CML (Figure 7B), suggesting that dose ranges can be selected at which CPX acts synergistically with imatinib on Bcr-Abl–positive cells while normal cells remain largely unaffected.
Synergistic effects of CPX and imatinib on CD34+ cells from CML patients at diagnosis. Mobilized CD34+ cells from 2 CML patients (A) and 3 healthy donors (B) were expanded in SFM supplemented with growth factors starting with 1 × 103 cells/well with increasing doses of CPX (0-40 μM). The average cell number per well was determined every third day for a total of 12 days in culture. Mean ±SE of replicate experiments is shown. (C) Isobolograms for the effects of imatinib and CPX (C) or GC7 (D) on cellular proliferation. Cells were incubated with a combination of imatinib and CPX (C) or GC7 (D) for 48 hours, and cytotoxicity was assessed using an MTT-based assay. Results were analyzed using Calcusyn software. (E) High-resolution cell-cycle analysis and tracking of CML CD34+ cell division. Flow cytometry dot plot showing high-resolution cell-cycle analysis with Ki-67 (FL1) and 7-AAD (FL3) staining. (left) No drug control. (right) Imatinib 5 μM/CPX 1 μM combination. G1 arrest with combination therapy can be clearly seen in the right plot. (F) Representative flow cytometry dot plots of CFSE (FL1) versus CD34PE (FL2) for viable (PI-negative) primary CML cells remaining after treatment with no drug control, CPX 1 μM, imatinib 5 μM, or combination imatinib 5 μM/CPX 1 μM. Tracking of cell division demonstrates the antiproliferative effect of imatinib 5 μM, which is enhanced by the addition of CPX 1 μM. (G) Total cell numbers (expressed as percentage of no drug control ± SD) remaining at the end of treatment for each condition studied using CML (⊡) and normal (▪) cells. (H) Number of quiescent (CFSEmax) CML (⊡) and normal (▪) CD34+ cells remaining viable at the end of 72-hour culture in the presence of imatinib, CPX, and the combination.
Synergistic effects of CPX and imatinib on CD34+ cells from CML patients at diagnosis. Mobilized CD34+ cells from 2 CML patients (A) and 3 healthy donors (B) were expanded in SFM supplemented with growth factors starting with 1 × 103 cells/well with increasing doses of CPX (0-40 μM). The average cell number per well was determined every third day for a total of 12 days in culture. Mean ±SE of replicate experiments is shown. (C) Isobolograms for the effects of imatinib and CPX (C) or GC7 (D) on cellular proliferation. Cells were incubated with a combination of imatinib and CPX (C) or GC7 (D) for 48 hours, and cytotoxicity was assessed using an MTT-based assay. Results were analyzed using Calcusyn software. (E) High-resolution cell-cycle analysis and tracking of CML CD34+ cell division. Flow cytometry dot plot showing high-resolution cell-cycle analysis with Ki-67 (FL1) and 7-AAD (FL3) staining. (left) No drug control. (right) Imatinib 5 μM/CPX 1 μM combination. G1 arrest with combination therapy can be clearly seen in the right plot. (F) Representative flow cytometry dot plots of CFSE (FL1) versus CD34PE (FL2) for viable (PI-negative) primary CML cells remaining after treatment with no drug control, CPX 1 μM, imatinib 5 μM, or combination imatinib 5 μM/CPX 1 μM. Tracking of cell division demonstrates the antiproliferative effect of imatinib 5 μM, which is enhanced by the addition of CPX 1 μM. (G) Total cell numbers (expressed as percentage of no drug control ± SD) remaining at the end of treatment for each condition studied using CML (⊡) and normal (▪) cells. (H) Number of quiescent (CFSEmax) CML (⊡) and normal (▪) CD34+ cells remaining viable at the end of 72-hour culture in the presence of imatinib, CPX, and the combination.
Cotreatment of imatinib and HI synergistically induces cytotoxicity in CD34+ cells from CML patients at diagnosis
We next determined the synergistic antileukemic effects of imatinib and CPX or GC7 against primary CML cells isolated from patients at diagnosis. As shown in Figure 7C-D and Table 3, for most of the dose range studied, the combination of imatinib and CPX or GC7 was strongly synergistic (CI less than 1; P < .05) on CD34+ cells from CML patients at diagnosis.
Imatinib and CPX act synergistically to inhibit proliferation in primary CD34+ cells of CML patients
To further expand the analysis in primary patient cells, high-resolution cell-cycle analysis using Ki-67 and 7-AAD was performed after 72-hour culture of CD34+ cells with imatinib and CPX, alone and in combination. A relative accumulation of G0 cells compared with G1 and SG2M cells was detected for all conditions compared with the no drug control. The combination of imatinib and CPX led to profound G1 arrest compared with the no drug control (Figure 7E). CD34+ CFSEmax-sorted primary CML cells were cultured for 2 cycles of 72 hours in imatinib and CPX alone and in combination at different concentrations, as described. At the end of the first treatment cycle, CPX (1 μM) alone exerted a mild proliferative effect, whereas imatinib (5 μM) and the combination of imatinib and CPX (1 μM) demonstrated a marked antiproliferative effect (Figure 7F). Compared with the treatment of primary CML cells with imatinib alone at either 0.5 μM or 5 μM, there was an absolute reduction in total cell numbers when imatinib was combined with CPX at 1 μM or 9 μM (Figure 7G). This was accompanied by an increase in quiescent CD34+ CML stem cells (defined as CD34+ CFSEmax CML cells remaining after 72-hour culture), confirming the antiproliferative effect of the combination (Figure 7H). The effect was further enhanced at the end of cycle 2 (data not shown). Additional experiments were performed with normal CD34+ cells to determine the selectivity of the combination of imatinib and CPX for CML cells. After 72 hours, total cell numbers were higher in all treatment arms compared with CML (Figure 7G). However, we did see a reduction in total cell numbers in all treatment arms compared with the no drug control. Despite this, the combination of imatinib (5 μM) and CPX (1 μM) was significantly more toxic to CML than normal CD34+ cells (P = .007). The toxicity of imatinib to normal CD34+ cells is well described.22 Normal quiescent CD34+ cells were unaffected by imatinib or CPX alone or in combination (Figure 7H), providing further evidence for the selectivity of this combination in CML compared with normal cells.
Discussion
Delineation of intracellular signaling cascades activated by Bcr-Abl tyrosine kinase activity is essential to allow a better understanding of the biology of Ph+ leukemias. We used a proteomics approach to investigate the differential protein expression profile of BCR-ABL–positive K562 cells on treatment with the Bcr-Abl inhibitor imatinib. We detected 19 differentially expressed proteins, 7 of which were overexpressed and 12 of which were down-regulated by imatinib treatment. Consistent with recent findings by Unwin et al,13 in which Bcr-Abl–dependent alterations in the Rho pathway were described in a murine background, we found reduced expression of Rho-GDI under imatinib treatment. Furthermore, we detected imatinib-induced down-regulation of eIF5A, a protein highly conserved from yeast to mammalian cells.36,37 eIF5A precursor is the only cellular protein known to contain a specific lysine residue (Lys50), which is converted to the unique amino acid hypusine, thereby enabling activation of eIF5A. Hypusination is induced stepwise through 2 distinct mechanisms. In the first step, catalyzed by the enzyme deoxyhypusine synthase, deoxyhypusine intermediates are formed by the transfer of 4-aminobutyl to lysine residues of the eIF5A precursor. The second step involves hydroxylation of the side chain of the deoxyhypusine intermediates by a second enzyme, deoxyhypusine hydroxylase. Hypusination plays a key role in the regulation of eIF5A function. eIF5A precursors that do not contain hypusine are barely active.19,38 A series of observations suggests that eIF5A plays a key role in cell growth and differentiation.39,40 It has been proposed that eIF5A is an essential regulator of the nuclear export of some specific mRNAs.41,42 In addition, it has been suggested that a minor human isoform, eIF5A2, is an oncogene.43 Hypusinylated eIF5A contributes to the life cycle of human immunodeficiency virus by interacting with the retroviral REV protein, thereby participating in the nuclear export of unspliced and incompletely spliced viral mRNA.44,45 Recently, hypusination inhibition was shown to suppress the HIV-1 replication cycle. Accordingly, deoxyhypusine synthase could be used as a novel drug target in HIV treatment.46
Our observations of down-regulated eIF5A protein expression after imatinib treatment of Bcr-Abl–positive K562 cells and elevated eIF5A expression levels in the PBMCs of CML patients raise the intriguing possibility that eIF5A is critically involved in the Bcr-Abl–mediated malignant transformation of hematopoietic cells. One could speculate that imatinib might, at least in part, exert its antiproliferative effect through the inhibition of BCR-ABL–mediated eIF5A overexpression. To test this hypothesis, we investigated whether additive or even synergistic effects could be detected by treating BCR-ABL–positive leukemia cells jointly with HI and imatinib. Interestingly, in addition to the cytotoxic and proapoptotic effects of HI as single agents, the combination of imatinib and CPX was found to act synergistically on cellular cytotoxicity and the induction of apoptosis in BCR-ABL–positive K562 cells. These data were confirmed in CD34+ primary cells from 6 independent patients with newly diagnosed CML. By means of lentiviral overexpression of eIF5A siRNA, we could down-regulate eIF5A expression in K562 cells and thereby induce a significant increase in the antiproliferative effect of imatinib treatment compared with wild-type K562 cells. This observation mimics the effect of combined HI/imatinib treatment and strongly supports the specificity of HI treatment on eIF5A hypusination in the in vitro model used here. The effect of monotherapy with HI seemed largely independent of BCR-ABL mutational status, and HI did not seem to directly affect the phosphorylation of Bcr-Abl downstream targets. However, not unexpectedly, residual Bcr-Abl activity seemed to be a prerequisite for the synergistic effect between HI and imatinib. Although cells harboring the M351T mutation conferring relative resistance to imatinib did respond synergistically, no such effect was observed in T315I mutant-resistant cells. Taken together, these data suggest that treatment of BCR-ABL–positive cells with imatinib sensitizes the cells for HI and vice versa.
Recently, Li et al47 described a p53-dependent, proapoptotic function of eIF5A in nonleukemic cells. However, considering that K562 cells are p53 deficient and bearing in mind that loss of p53 is a typical feature of disease progression of CML from CP to AP/BC, these results are not in conflict with our observations. Interestingly, hypusination is also inhibited by IFN-α treatment in nonleukemic cells.48,49 In addition, combination treatment with cytarabine and HI leads to synergistic antiproliferative effects in human cancer cells.50 Given that both drugs are typically used for the treatment of CML patients and considering that both drugs act synergistically with imatinib on BCR-ABL–positive leukemias, these findings might provide the molecular background for the effects seen in vitro and in vivo.51,52 Thus, based on the results reported here, it is suggested that preclinical treatment strategies combining HI and imatinib should be evaluated to reduce the potential development of clinical resistance to imatinib monotherapy.
This study indicates that comparative global proteome analysis in BCR-ABL–positive cell lines represents a useful screening tool for potential new therapeutic targets in Ph+ leukemia. More sophisticated approaches, including comparative analysis of distinct cellular compartments and of subproteomes (eg, phosphoproteome),53 will allow more specific characterization of functional protein modifications such as those induced by small molecule kinase inhibitors.
Authorship
Contribution: S.B., W.K., T.L.H., A.N., and T.H.B. designed the research; S.B., A.G., P.Z., U.H., M.C., U.B., and I.H. performed the research; A.N., I.H., and J.H. contributed vital new reagents or analytical tools; W.K., M.P., G.S., and T.W. conducted data analysis and protein identification; and S.B., T.H.B., L.K., C.B., T.L.H., A.N., and J.H. wrote the paper.
Conflict-of-interest disclosure: These studies were, in part, supported by a grant from Novartis Pharma, Germany. S.B., U.H., W.K., A.N., and T.H.B. have filed a patent on the synergistic effect of HI and imatinib (US 60/531,563).
Correspondence: Tim H. Brümmendorf, Department of Oncology and Hematology, University Hospital Hamburg-Eppendorf, Martinistrasse 52, 20246 Hamburg, Germany; e-mail: t.bruemmendorf@uke.uni-hamburg.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
These studies were supported by a grant from Novartis Pharma (T.H.B., A.N.) and by grants from the Deutsche Forschungsgemeinschaft (DFG) (NO120/11-1) and the Fonds der Chemischen Industrie (A.N.). M.C. holds a Medical Research Council (MRC) personal research fellowship.