Abstract
We report the molecular and cytogenetic characterization of a novel variant of acute promyelocytic leukemia (APL). The bone marrow showed 88% hypergranular promyelocytes, and the karyotype was 47,XY,+22 [5]/46,XY[30]. Fluorescence in situ hybridization (FISH) indicated disruption and deletion of the 5′-end of the RARA gene. Treatment with all-trans retinoic acid, idarubicin, and arsenic trioxide induced cytogenetic complete remission without morphologic evidence of residual leukemia. The diagnostic marrow was negative for PML-RARA transcripts by reverse transcription–polymerase chain reaction (RT-PCR), but an atypical product was observed. Sequencing showed partial homology to the PRKAR1A gene, encoding the regulatory subunit type I-α of cyclic adenosine monophosphate–dependent protein kinase. RT-PCR using specific primers for PRKAR1A and RARA amplified 2 transcript splice variants of a PRKAR1A-RARA fusion gene, and PRKAR1A and RARA FISH probes confirmed the fusion. This novel PRKAR1A-RARA gene rearrangement is the fifth variant APL in which the RARA partner gene has been identified and the second known rearrangement of PRKAR1A in a malignant disease. This trial was registered at www.actr.org.au with the Australian Clinical Trials Registry as number 12605000070639.
Introduction
The majority of acute promyelocytic leukemia (APL) cases are characterized by the PML-RARA fusion gene, usually as a consequence of the t(15;17)(q22;q21) translocation.1 As a result, retinoid sensitivity of the retinoic acid receptor α (RARα), which normally functions as a retinoid-inducible transcription factor, is reduced, causing a block in myeloid differentiation.2 Prolonged disease-free survival can be achieved by combining all-trans retinoic acid (ATRA) and chemotherapy.3 Arsenic trioxide (As2O3) is of proven value in relapsed disease4 and is also effective during initial induction or consolidation or both.5 In a small number of APL variants, RARA (at chromosome 17q21) is fused with an alternative partner gene; promyelocytic leukemia zinc finger (PLZF, 11q13),6 nucleophosmin (NPM, 5q35),7 nuclear mitotic apparatus (NUMA, 11q23),8 or signal transducer and activator of transcription 5b (STAT5B, 17q21).9 RARα fusions to PML, NPM, and NUMA are ATRA responsive, whereas PLZF-RARα is ATRA resistant.10,11 This report describes a new variant APL and identifies the RARA partner gene.
Study design
The patient concerned in this report was originally registered in a multicenter national trial for acute promyelocytic leukemia that had institutional ethics approval at the hospital concerned. Informed consent was obtained in accordance with the Declaration of Helsinki. The patient was subsequently deemed ineligible for that trial because he failed to meet the required genetic inclusion criterion. This report is concerned with an in vitro characterization of the patient's leukemic cells and did not involve any direct patient experimentation.
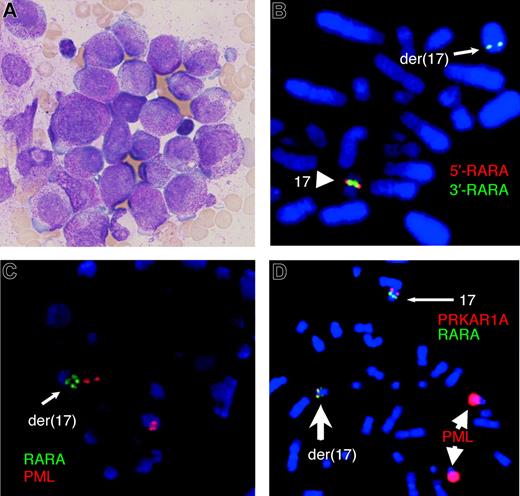
A 66-year-old man with a history of excessive alcohol consumption, clinical evidence of chronic liver disease, and a 14-month history of mild thrombocytopenia was investigated for lethargy, anorexia, and weight loss. A full blood count showed a hemoglobin level of 104 g/L, white cell count of 5.3 × 109/L, neutrophil count of 2.1 × 109/L, and platelet count of 93 × 109/L. The blood film was leukoerythroblastic with occasional hypergranular promyelocytes. Both creatinine (0.22 mmol/L) and γ-glutamyltransferase (5.17 μkat/L [310 U/L]) were mildly elevated. Coagulopathy was absent apart from mildly increased D-dimers (0.4 mg/L; normal, < 0.2 mg/L). A markedly hypercellular marrow contained 88% hypergranular promyelocytes, and most exhibited regular nuclear outlines (Figure 1A). Auer rods and faggot cells were absent. Flow cytometric analysis revealed strong intracellular myeloperoxidase expression; however, expression of CD13, CD33, and CD11b was weak. The cells were negative for CD2, CD19, CD34, CD56, CD117, and HLA-DR. The karyotype was 47,XY,+22[5]/46,XY[30], without the classic t(15;17) translocation. Fluorescence in situ hybridization (FISH) studies showed del(17)(q21)(5′RARA-,3′RARA+) (Figure 1B) plus a split RARA signal (Figure 1C) with RARA break-apart and PML-RARA dual fusion probes, respectively.
Morphology and metaphase FISH assays on bone marrow of the variant APL. (A) Bone marrow morphology showing hypergranular promyelocytes lacking Auer rods. Image of May-Grünwald-Giemsa–stained smear was acquired using an Olympus BX-40 microscope (Olympus, Tokyo, Japan) equipped with a 100×/1.3 NA oil objective and a mounted DP12 digital camera (Olympus). (B) The LSI RARA dual-color break-apart rearrangement probe (Vysis, Downers Grove, IL) resulted in a normal fusion signal on one chromosome 17 and a small green signal close to the q terminal region of der(17) with deletion of the proximal red signal. (C) FISH with the LSI PML-RARA dual-color dual-fusion translocation probe (Vysis) resulted in normal red PML signals, one normal green RARA signal (not shown), and one split RARA signal. (D) A PRKAR1A probe (BAC RP11–120M18), labeled with Spectrum Orange (Vysis), combined with LSI PML-RARA dual-fusion translocation probe (as in panel C) confirmed that the fusion gene expressed by the patient was the result of a complex chromosome rearrangement which disrupted both the RARA gene on 17q21 and PRKAR1A on 17q24; the normal chromosome 17 has a single green signal (RARA) and a distal red signal (PRKAR1A), whereas the der(17) has a split green signal (RARA) where the more distal green signal colocalizes with a weaker red signal (PRKAR1A), indicating deletion of part of the probe. Note that the strong red PML probe signals on the 2 chromosomes 15 are because of image enhancement that was necessary to visualize the weak signals on the der(17). All FISH was performed according to manufacturer's instructions, and chromosomes were counterstained with DAPI (Sigma, St Louis, MO). Analysis was undertaken using a Zeiss Axioskop fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a 100× /1.3 NA oil objective. Images captured using the Isis image analysis system (MetaSystems, Altlussheim, Germany) were processed with FISH imaging system version 5.2 software (MetaSystems).
Morphology and metaphase FISH assays on bone marrow of the variant APL. (A) Bone marrow morphology showing hypergranular promyelocytes lacking Auer rods. Image of May-Grünwald-Giemsa–stained smear was acquired using an Olympus BX-40 microscope (Olympus, Tokyo, Japan) equipped with a 100×/1.3 NA oil objective and a mounted DP12 digital camera (Olympus). (B) The LSI RARA dual-color break-apart rearrangement probe (Vysis, Downers Grove, IL) resulted in a normal fusion signal on one chromosome 17 and a small green signal close to the q terminal region of der(17) with deletion of the proximal red signal. (C) FISH with the LSI PML-RARA dual-color dual-fusion translocation probe (Vysis) resulted in normal red PML signals, one normal green RARA signal (not shown), and one split RARA signal. (D) A PRKAR1A probe (BAC RP11–120M18), labeled with Spectrum Orange (Vysis), combined with LSI PML-RARA dual-fusion translocation probe (as in panel C) confirmed that the fusion gene expressed by the patient was the result of a complex chromosome rearrangement which disrupted both the RARA gene on 17q21 and PRKAR1A on 17q24; the normal chromosome 17 has a single green signal (RARA) and a distal red signal (PRKAR1A), whereas the der(17) has a split green signal (RARA) where the more distal green signal colocalizes with a weaker red signal (PRKAR1A), indicating deletion of part of the probe. Note that the strong red PML probe signals on the 2 chromosomes 15 are because of image enhancement that was necessary to visualize the weak signals on the der(17). All FISH was performed according to manufacturer's instructions, and chromosomes were counterstained with DAPI (Sigma, St Louis, MO). Analysis was undertaken using a Zeiss Axioskop fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a 100× /1.3 NA oil objective. Images captured using the Isis image analysis system (MetaSystems, Altlussheim, Germany) were processed with FISH imaging system version 5.2 software (MetaSystems).
Treatment was commenced with ATRA (45 mg/m2/d), idarubicin (9 mg/m2/d on days 2, 4, 6, and 8), and As2O3 (0.15 mg/kg/d from day 9); however, arsenic was suspended on day 22 because of sepsis and cardiac toxicity. Complete remission was documented on day 35 by morphology, cytogenetics, and interphase FISH. Two cycles of consolidation (infusional cytarabine 100 mg/m2/d × 7 and amsacrine 100 mg/m2/d × 3) were administered, superimposed on continuous ATRA, and were followed by maintenance therapy (ATRA for 2 weeks every 3 months). Eleven months after diagnosis, a full blood count showed mild anemia (99 g/L), normal neutrophils, and marked thrombocytopenia (20 × 109/L), and the marrow showed essentially normal trilineage hemopoiesis. Cytogenetic analysis and interphase FISH were both normal (46,XY[40] and nuc ish(RARAx2)[72], respectively). Two years from diagnosis, the patient remains clinically free of leukemia with further hematologic improvement: normal hemoglobin level, normal neutrophil count, and moderate thrombocytopenia (platelets, 61 × 109/L).
Results and discussion
The combination of APL morphology and RARA disruption in the absence of PML involvement by FISH suggested the possibility of a novel translocation in the pathogenesis of this leukemia. A classic PML-RARA fusion was absent when PML- and RARA-specific primers were used (data not shown). However, an assay for bcr3 transcripts generated 2 products with high-quality sequence identity to exons 2 and 3 of the PRKAR1A gene, followed by the 5′ end of RARA exon 3 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). PRKAR1A encodes the regulatory subunit type I-α (RIα) of cyclic adenosine monophosphate (cAMP)–dependent protein kinase (PKA).12
To confirm the presence of the suggested PRKAR1A-RARA fusion (Figure S2), nested reverse transcription–polymerase chain reaction (RT-PCR) was performed with PRKAR1A-specific forward primers and reverse primers that would amplify substantial amounts of RARA (Figure 2A). Both first- and second-round PCR products of expected sizes were obtained (Figure 2B,C) and confirmed to be PRKAR1A-RARA fusion transcripts by DNA sequencing (Figure 2D). In contrast, RT-PCR for PRKAR1A-RARA was negative in the 11-month remission sample (Figure 2C).
Molecular analysis of PRKAR1A-RARA fusion transcripts. (A) The diagram shows the exon structure of the human PRKAR1A and RARA mRNAs (GenBank Accessions NM_002734 and NM_000964, respectively), with translated regions indicated by wide rectangles. Exploded views of exons 2 and 3 of PRKAR1A, and exons 3 and 4 of RARA are indicated with the positions of PCR primers used, PRKAR1A-F1 (5′-gaaccatggagtctggc-3′), PRKAR1A-F2 (5′-ggttggagaaggaggag-3′), RARA-R34o1 (5′-ggcggaagaagcccttgcag-3′), and RARA-R3 (5′-cagccctcacaggcgctgac-3′). (B) PRKAR1A-RARA RT-PCR analysis using primers PRKAR1A-F1 and RARA-R34o1 resulted in the detection of 2 transcripts (PCR products of 444 bp [base pairs] and 344 bp) in the diagnostic bone marrow (Dx) of the index patient but no products were amplified from either the 11-month remission sample (11 mo), from Meg01 cell line (Meg), used as a negative control, or in a “no template” control (NTC). DNA molecular weight markers (M) were 100 base pair ladder (GE Healthcare, Buckinghamshire, United Kingdom). (C) Reamplification of the reactions shown in panel B with nested PCR primers PRKAR1A-F2 and RARA-R3 (that exclusively amplify fusions between PRKAR1A exon 3 and RARA exon 3) resulted in production of a single 256-bp product in the diagnostic sample. The absence of a product in the remission sample confirmed PRKAR1A-RARA molecular remission. (D) Partial sequence trace of the predominant in-frame PRKAR1A-RARA fusion transcript product showing the junction of the 2 genes as the result of RNA splicing from a cryptic splice donor within PRKAR1A exon 3. (E) Diagram showing the exon structures (narrow rectangles), open reading frames (wide rectangles), and partial amino acid sequence at the fusion junctions of the 2 main PRKAR1A-RARA transcripts present at diagnosis. PRKAR1A-derived exons and sequences are indicated by gray shading. The in-frame fusion (GenBank Accession EF428110) would be capable of encoding a 495–amino acid protein. The shorter frame-shifted fusion transcript (GenBank Accession EF428111) would encode a truncated RIα protein with 11 carboxy terminal amino acids (in italics) derived from the frame-shifted RARA exon 3.
Molecular analysis of PRKAR1A-RARA fusion transcripts. (A) The diagram shows the exon structure of the human PRKAR1A and RARA mRNAs (GenBank Accessions NM_002734 and NM_000964, respectively), with translated regions indicated by wide rectangles. Exploded views of exons 2 and 3 of PRKAR1A, and exons 3 and 4 of RARA are indicated with the positions of PCR primers used, PRKAR1A-F1 (5′-gaaccatggagtctggc-3′), PRKAR1A-F2 (5′-ggttggagaaggaggag-3′), RARA-R34o1 (5′-ggcggaagaagcccttgcag-3′), and RARA-R3 (5′-cagccctcacaggcgctgac-3′). (B) PRKAR1A-RARA RT-PCR analysis using primers PRKAR1A-F1 and RARA-R34o1 resulted in the detection of 2 transcripts (PCR products of 444 bp [base pairs] and 344 bp) in the diagnostic bone marrow (Dx) of the index patient but no products were amplified from either the 11-month remission sample (11 mo), from Meg01 cell line (Meg), used as a negative control, or in a “no template” control (NTC). DNA molecular weight markers (M) were 100 base pair ladder (GE Healthcare, Buckinghamshire, United Kingdom). (C) Reamplification of the reactions shown in panel B with nested PCR primers PRKAR1A-F2 and RARA-R3 (that exclusively amplify fusions between PRKAR1A exon 3 and RARA exon 3) resulted in production of a single 256-bp product in the diagnostic sample. The absence of a product in the remission sample confirmed PRKAR1A-RARA molecular remission. (D) Partial sequence trace of the predominant in-frame PRKAR1A-RARA fusion transcript product showing the junction of the 2 genes as the result of RNA splicing from a cryptic splice donor within PRKAR1A exon 3. (E) Diagram showing the exon structures (narrow rectangles), open reading frames (wide rectangles), and partial amino acid sequence at the fusion junctions of the 2 main PRKAR1A-RARA transcripts present at diagnosis. PRKAR1A-derived exons and sequences are indicated by gray shading. The in-frame fusion (GenBank Accession EF428110) would be capable of encoding a 495–amino acid protein. The shorter frame-shifted fusion transcript (GenBank Accession EF428111) would encode a truncated RIα protein with 11 carboxy terminal amino acids (in italics) derived from the frame-shifted RARA exon 3.
FISH, using a RARA probe and a BAC probe (RP11–120M18) encompassing the PRKAR1A gene, confirmed the presence of a PRKAR1A-RARA fusion on der(17) (Figure 1D). The simplest explanation would be an insertion of RARA distal to PRKAR1A followed by a deletion removing 3′ PRKAR1A, 5′ RARA, and any intervening sequences.
Alternative splicing of PRKAR1A explains the presence of 2 PRKAR1A-RARA PCR products observed in the diagnostic sample. The longer transcript results from cryptic splicing of the first 100 bases of PRKAR1A exon 3 to RARA exon 3, producing an in-frame fusion transcript capable of encoding a 495–amino acid RIα-RARα fusion protein. Translation of the putative open reading frame would generate a protein containing the RIα protein dimerization domain fused to the same carboxy terminal end of the RARα protein shared by all RARA rearrangements in APL.11 Splicing of PRKAR1A exon 2 to RARA exon 3 generates a shorter out-of-frame fusion transcript possibly encoding a carboxy-truncated chimeric RIα protein.
PKA is a multimeric protein consisting of 2 catalytic subunits complexed with a regulatory subunit dimer.13 Binding of cAMP to the regulatory subunits produces a conformational change that causes dissociation and de-inhibition of the catalytic subunits.14 Type-I regulatory subunits are generally associated with cytosolic isoforms of PKA, whereas type-II regulatory subunits, capable of interacting with A kinase–anchoring proteins, are found in organelle-localized isoforms of PKA.13
PRKAR1A is involved in another gene rearrangement with the RET proto-oncogene in papillary thyroid carcinomas.15,16 Inherited null mutations of PRKAR1A have been reported in Carney complex17,18 PRKAR1A-inactivating mutations or down-regulation have also been found in some sporadic adrenocortical tumors.19 In our case, fusion of the RIα dimerization domain to RARα may be involved in dysregulated RARα homodimerization or heterodimerization with RXR and potentially deregulation of PKA through disruption of RIα.
Mouse models of APL have varying phenotypic features that depend on the RARA fusion partner.20 It is conceivable that dysregulation of PKA by disruption of RIα may have contributed to this patient's atypical APL presentation. The ATRA responsiveness in this case is unknown because multiple agents were used. However, the remarkable clinical response suggests that the fusion gene products were sensitive to either As2O3 or ATRA or to both. An in vitro model of this variant could help clarify this issue, but based on current knowledge of the mechanisms of As2O3 and ATRA in APL, we propose that both agents may be effective. First, because the RARα component of the RIα-RARα fusion protein is identical to other ATRA responsive RARα fusion proteins and the truncated RIα component lacks nuclear corepressor interacting domains, it is conceivable that the RIα-RARα protein is responsive to pharmacologic doses of ATRA. Second, there may be synergic effects because of the involvement of PRKAR1A in the fusion. Gene expression profiling of the APL-derived NB4 cell line identified PRKAR1A as one of the genes that is up-regulated in response to ATRA treatment.21 Thus, ATRA treatment could also up-regulate expression of the in-frame PRKAR1A-RARA fusion transcript or the shorter out-of-frame fusion transcript, both of which might encode defective RIα proteins with dominant-negative effects on normal RIα. The resulting constitutive activation of PKA activity could lead to increased responsiveness to As2O3, which is known to have synergic effects with cAMP in APL.22,23
Although this report describes a single case expressing PRKAR1A-RARA, the nature of the cryptic cytogenetic lesion raises the possibility that other cases of variant APL carrying the same lesion may have been missed, even with RARA-specific FISH probes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Arsenic trioxide was provided by Pharmalab for use in patients registered on the APML4 trial under the auspices of the Australasian Leukaemia and Lymphoma Group. The BAC probe (RP11–120M18) was kindly provided by Elizabeth Baker (King Edward Memorial Hospital, Perth, WA).
Authorship
Contribution: A.C. designed and performed research, analyzed and interpreted data, and wrote the paper. M.A.D. provided clinical care; collected, analyzed, and interpreted data, and wrote the paper. K.S. performed research and analyzed data. S.O. analyzed and interpreted data and supervised data collection. A.S. provided clinical care and collected data. L.J.C. designed research, analyzed and interpreted data, and wrote the paper. H.I. analyzed and interpreted data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alberto Catalano, Institute of Haematology, Royal Prince Alfred Hospital, Missenden Rd, Camperdown NSW 2050, Australia; e-mail: alberto.catalano@email.cs.nsw.gov.au.

![Figure 2. Molecular analysis of PRKAR1A-RARA fusion transcripts. (A) The diagram shows the exon structure of the human PRKAR1A and RARA mRNAs (GenBank Accessions NM_002734 and NM_000964, respectively), with translated regions indicated by wide rectangles. Exploded views of exons 2 and 3 of PRKAR1A, and exons 3 and 4 of RARA are indicated with the positions of PCR primers used, PRKAR1A-F1 (5′-gaaccatggagtctggc-3′), PRKAR1A-F2 (5′-ggttggagaaggaggag-3′), RARA-R34o1 (5′-ggcggaagaagcccttgcag-3′), and RARA-R3 (5′-cagccctcacaggcgctgac-3′). (B) PRKAR1A-RARA RT-PCR analysis using primers PRKAR1A-F1 and RARA-R34o1 resulted in the detection of 2 transcripts (PCR products of 444 bp [base pairs] and 344 bp) in the diagnostic bone marrow (Dx) of the index patient but no products were amplified from either the 11-month remission sample (11 mo), from Meg01 cell line (Meg), used as a negative control, or in a “no template” control (NTC). DNA molecular weight markers (M) were 100 base pair ladder (GE Healthcare, Buckinghamshire, United Kingdom). (C) Reamplification of the reactions shown in panel B with nested PCR primers PRKAR1A-F2 and RARA-R3 (that exclusively amplify fusions between PRKAR1A exon 3 and RARA exon 3) resulted in production of a single 256-bp product in the diagnostic sample. The absence of a product in the remission sample confirmed PRKAR1A-RARA molecular remission. (D) Partial sequence trace of the predominant in-frame PRKAR1A-RARA fusion transcript product showing the junction of the 2 genes as the result of RNA splicing from a cryptic splice donor within PRKAR1A exon 3. (E) Diagram showing the exon structures (narrow rectangles), open reading frames (wide rectangles), and partial amino acid sequence at the fusion junctions of the 2 main PRKAR1A-RARA transcripts present at diagnosis. PRKAR1A-derived exons and sequences are indicated by gray shading. The in-frame fusion (GenBank Accession EF428110) would be capable of encoding a 495–amino acid protein. The shorter frame-shifted fusion transcript (GenBank Accession EF428111) would encode a truncated RIα protein with 11 carboxy terminal amino acids (in italics) derived from the frame-shifted RARA exon 3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-06-095554/4/m_zh80240710050002.jpeg?Expires=1769462812&Signature=Ayua0lDHTtin0n67RrkbQKyyHGgFxBhQLxsoynLEg~KdWHgKQ2SmFw79GK~sivewLtr7~RjbdgI5KVVWHxeolCrCLr~dfPKhTENvuukOzeFrKi11CZsLQcpNSV5zjPkg5nUe369KEmv2siUKeEldmMgq5CSn48rDe2WVNpEIPewV0BmpB0k8E23tRGlVBIcHF8c2gqkh5eeYDSJlbxOJE~s0Thu3JFVofLaWihM5PhWDn9FEninG~GUoq3V4l94j-HEc-ZMeL0~YSae4N7SHUn9gBKhC-FQRClQhHh6xvr5YN44~84K4Csbhj1iTbw0oEEpLtU8qT-keQRdnwM~vNA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
