Abstract
Hematopoietic stem cells (HSCs) can both self renew and differentiate into precursors of all types of blood cells. HSCs are divided into an active pool and a quiescent reserve. Cells selected for the active pool contribute to hematopoiesis for many years. Mutations in HSCs can lead to neoplasms such as chronic myeloid leukemia, although the risk of neoplastic HSC disorders varies across mammals. We use allometric scaling relations combined with mutation-selection evolutionary dynamics to determine which mammalian species is most resistant to HSC disorders. We find that the advantage of large mammals at escaping the selective pressure of cancer cells is insufficient to overcome the increased risk of acquiring mutations. Hence, mutation dominates, which favors smaller stem-cell pools and, consequently, smaller mammals, since these minimize the development of mutations in the active stem-cell pool. Consequently, the smaller the active stem-cell pools, the better.
Introduction
The transformation of normal cells into cancer is a problem that emerged with the appearance of multicellular organisms. Cells acquire the tumor phenotype by the progressive accumulation of mutations in proto-oncogenes and/or tumor suppressor genes.1 One of the mechanisms that multicellular organisms have evolved to minimize the retention of mutated cells is the organization of tissues into 2 major types of cells: transient cells form the bulk of the tissue and replicate often but are retained for a relatively short period of time, and stem cells that maintain these tissues but replicate slowly. Initially identified in the bone marrow, there is increasing evidence that tissue-specific stem cells are present in every organ of the body.2 Tumors also appear to depend on cancer stem cells (CSCs) for their own maintenance.2 It is therefore no surprise that stem cells are the focus of intense research, also for curative therapy.3,4
Although tumors need not arise from mutations in tissue-specific stem cells, it is clear that hematopoietic stem cells (HSCs) can be transformed into CSCs and drive diseases such as chronic myeloid leukemia5 and polycythemia vera. Perhaps a single mutation in an HSC may be enough to explain the early development of these disorders.6-8 HSCs have the dual property of self renewal and production of cells that can differentiate into all different types of blood cells. Cells are at the highest risk of acquiring mutations during DNA replication in preparation for cell division. Hence, HSCs divide infrequently, reducing the accumulation of mutations.9 HSCs are operationally divided into an active pool of cells that may contribute to hematopoiesis for many years10 and a quiescent reserve. Scaling relations, which pervade at all levels of organization in living organisms,11 have been used to explain how the number of active HSCs (NSC) in adult mammals scales allometrically with their mass M as NSC= N0 M¾.9 This, together with other well-known allometric relations, has immediate consequences with respect to the size of mammalian species and the associated risk of neoplastic HSC disorders. In the following we use knowledge of (a) the scaling of the active HSC pool, (b) the rate of replication of these cells, and (c) the mammalian mutation rate, to determine the mammal at the lowest risk of neoplastic transformation of the HSCs.
Results and discussion
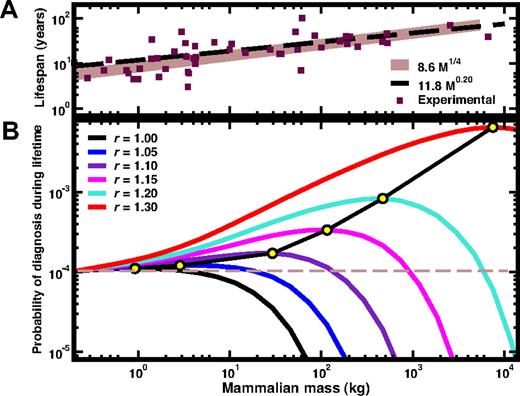
Details of the scaling and numerical methods used are provided in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). In Figure 1A, we show how the expected lifespan of terrestrial mammals qualitatively follows the M¼ scaling (using L0 ≈ 8.6 kg−¼ year, solid line). For comparison, we also plot the empirical fit11 (LE = 11.8M0.20 years, dashed line). Using these scaling relations and mutation-selection dynamics of the HSC pool (Document S1), we compute the probability that diagnosis of an HSC disorder, detectable by a 20% (or more12 ) contamination of the HSC pool, occurs during the lifespan of the mammal. Figure 1B shows the results as a function of M and relative fitness advantage r. For values of r marginally larger than 1, a large active HSC pool offers protection against acquired HSC disorders. However, for larger values of r, the opposite happens, due to the increased efficiency of CSC expansion within the active stem-cell pool during the mammal's lifespan. We have confirmed that the conclusions are robust with respect to (1) changes in the mutation probability μ by more than one order of magnitude in both directions; (2) changes in the fraction of CSC contamination necessary to make a diagnosis—in fact, the answer does not change even if we require 100% contamination; and (3) changes of the lifespan scaling, from the M¼ scaling to the M0.20 scaling in Figure 1A.
A. Relation between mammalian mass, lifespan and size of the active stem-cell pool, and the risk of HSC disorders. (A) Comparison between known values for the lifespan of several terrestrial mammals24 and the 2 scaling exponents considered in this work: the empirical scaling LE = 11.8M0.20 (- - -) and the ¼ scaling L = 8.6 M¼ (—). While both scaling relations account for the qualitative trend of the data, the conclusions drawn from the panel below do not depend on the specific scaling relation used to describe the lifespan of a given mammal. (B) The probability of diagnosis during the lifespan is plotted as a function of the mass of the mammal, for different values of the relative selective advantage r of CSC. For most usual tumors one expects r ≥ 1.7, which moves the maximum of probability (illustrated by the yellow circles) outside of the plotted (and terrestrial mammals) range. Consequently, for such tumors it is an advantage to be as smaller a mammal as possible. The curves plotted predict that, for r = 1.05, only mammals with M > 18 kg are more protected against mutations with this value of r than mice; for r = 1.10 the threshold is already 125 kg; for r = 1.15 we obtain 870 kg, whereas for r = 1.20 the threshold becomes 5800 kg. These values correspond to the ages at which each line crosses the dashed horizontal line.
A. Relation between mammalian mass, lifespan and size of the active stem-cell pool, and the risk of HSC disorders. (A) Comparison between known values for the lifespan of several terrestrial mammals24 and the 2 scaling exponents considered in this work: the empirical scaling LE = 11.8M0.20 (- - -) and the ¼ scaling L = 8.6 M¼ (—). While both scaling relations account for the qualitative trend of the data, the conclusions drawn from the panel below do not depend on the specific scaling relation used to describe the lifespan of a given mammal. (B) The probability of diagnosis during the lifespan is plotted as a function of the mass of the mammal, for different values of the relative selective advantage r of CSC. For most usual tumors one expects r ≥ 1.7, which moves the maximum of probability (illustrated by the yellow circles) outside of the plotted (and terrestrial mammals) range. Consequently, for such tumors it is an advantage to be as smaller a mammal as possible. The curves plotted predict that, for r = 1.05, only mammals with M > 18 kg are more protected against mutations with this value of r than mice; for r = 1.10 the threshold is already 125 kg; for r = 1.15 we obtain 870 kg, whereas for r = 1.20 the threshold becomes 5800 kg. These values correspond to the ages at which each line crosses the dashed horizontal line.
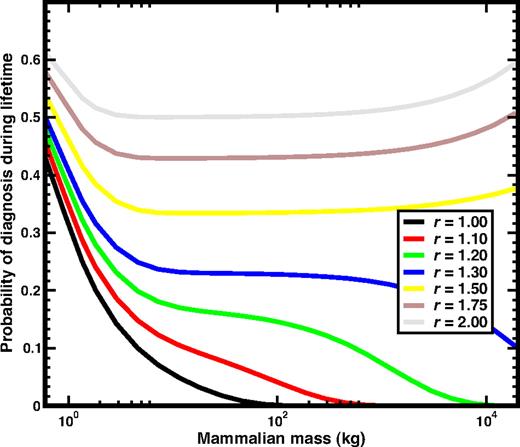
Since in most cases, selection dominates the dynamics once the first mutation occurs, it is interesting to ask what happens in mammals born with one CSC. We show the results in Figure 2. In the smallest mammals, the impact of a single mutation has severe effects, reflected in the high probability of diagnosis. Overall, one observes 2 regimes: For r < 1.4 the probability of diagnosis ultimately drops down to zero, favoring larger mammals, while for r > 1.4, the probability will ultimately converge to 1, although for masses that exceed those of existing terrestrial mammals.
The risk of neoplastic HSC disorders when a mutant (cancer) stem cell is present at birth. We plot the probability of diagnosis during the lifespan as a function of the mass of the mammal (using the same color codes as in Figure 1) for the case when the mammal is already born with a single CSC due to mutation. Consequently, in these curves selection plays a more important role than mutation during the life of the animal. Only for small values of r does the probability of diagnosis decrease monotonically, and already for moderate values of r the probability remains constant for over 2 orders of magnitude of mammalian mass, further increasing for large values of M.
The risk of neoplastic HSC disorders when a mutant (cancer) stem cell is present at birth. We plot the probability of diagnosis during the lifespan as a function of the mass of the mammal (using the same color codes as in Figure 1) for the case when the mammal is already born with a single CSC due to mutation. Consequently, in these curves selection plays a more important role than mutation during the life of the animal. Only for small values of r does the probability of diagnosis decrease monotonically, and already for moderate values of r the probability remains constant for over 2 orders of magnitude of mammalian mass, further increasing for large values of M.
The main source of genetic mutations in HSCs is DNA replication. In humans, the probability that a mutation occurs during replication has been estimated13 to be 10−7 ≤ μ ≤ 10−6. Assuming this probability remains unchanged across mammals,14 the expected number of mutations in the HSC pool during the lifetime of a mammal scales with M as nμ = μ · NSC · B · L ≈ n0 · M3/4, favoring smaller mammals. In the smaller mammals, a single mutation will immediately affect a large fraction of the active HSCs, whereas in larger mammals, the effect will not be readily perceived before expansion within the active HSC pool. For disorders of the HSCs, the relevant question to ask is whether a HSC mutation will occur and expand its lineage, leading to a diagnosis of cancer during the lifespan of the mammal. Conflicting problems come into play: While the risk of acquiring mutations and the average lifespan increase with animal mass, the probability to reach a given fraction of the HSC population, ignoring animal lifespan, decreases with increasing mass.
The results in Figure 1 show that for r > 1.20 no terrestrial mammal is better off compared with mice. Thus, a larger active HSC pool protects the mammal from the expansion of neutral mutations but not from mutations that give a fitness advantage to the cell.
Recently, it has been suggested12 that many cancer-inducing mutations require r ≥ 1.7. Assuming these values to extend across mammalian species, then smaller mammals have an advantage: The overall probability curve is a one humped function whose maximum location increases exponentially with r (r < 1.4, circles in Figure 1B). The combined message from Figures 1 and 2 is clear: Within the lifespan of each species, mutation is more important than selection.
The available literature supports (albeit indirectly) our model predictions. Spontaneous appearance of a chronic myeloproliferative disorder (CMPD) in mice has not been observed.15,16 The literature on CMPD in cats is essentially composed of small case reports,17 while in dogs, the literature implies that CMPDs are more common since the reported series include a larger number of subjects.18 Interestingly, in the latter paper, the author wonders why the incidence of CMPD is so low in dogs compared with humans when they are exposed to the same amount of radiation, the only known external cause of these disorders in both species. As discussed in the SI, humans start with a very small pool (< 40 cells) that expands linearly with mass to reach adult levels.19 The small number of active HSCs explains why CMPDs are so rare in children: fewer than 10% of cases of chronic myeloid leukemia occur in humans younger than 20 years of age.20
It has been predicted that the total number of HSCs is conserved across mammals and may be as low as 11 000 cells.21 Data from transplantation in rodents22 and subsequent extrapolation to larger mammals seem compatible with this proposition.23 Our model, based on allometric scaling, partitions the existing HSCs into 2 pools: an active HSC pool that scales with mass, reflecting the fact that different mammals have different requirements for bone marrow output, and a quiescent reserve.9 Our estimates for the active HSC pool are consistent with the limits predicted.21 Hence, there is no conflict between our estimates and those of Abkowitz et al since the latter applies to the total number of HSCs. In summary, our work shows that the smaller the mammal, the better. The advantage of a small active stem-cell pool may provide an evolutionary pressure toward the development of hierarchical and multicompartmental architectures, such as those found in hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Science and Technology Foundation Portugal (J.V.L., J.M.P.) and the Mayo Foundation (D.D.). The Program for Evolutionary Dynamics is supported by Jeffrey Epstein. We also thank the expert reviewers for their many insightful suggestions, which helped us improve our manuscript.
Authorship
Contribution: J.V.L. designed and implemented the computer program, ran the simulations, analyzed the data, and wrote the manuscript. J.M.P. developed the concept, reviewed the computer algorithm, analyzed the data, and wrote the manuscript. D.D. developed the concept, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Dingli, Mayo Clinic College of Medicine, 200 First St SW, Rochester, MN 55905; e-mail: dingli.david@mayo.edu.