To the editor:
Aberrant activation of the Wnt/β-catenin signaling pathway is a hallmark of cancers.1,2 A recent Blood report showed the down-regulation of multiple Wnt-signaling inhibitors including SFRPs, WIF1, DKK3, and DACT1 in acute lymphoblastic leukemia (ALL) cell lines and tumors due to promoter methylation, leading to the activation of Wnt signaling.3
WNT5A is another noncanonical and nontransforming Wnt protein.4 Although there are conflicting data regarding its expression and functions in tumorigenesis, WNT5A signals through the noncanonical Wnt/Ca++ pathway to block the Wnt-signaling cascade and inhibits B-cell proliferation.5 Its expression was down-regulated in most acute leukemia patients.5
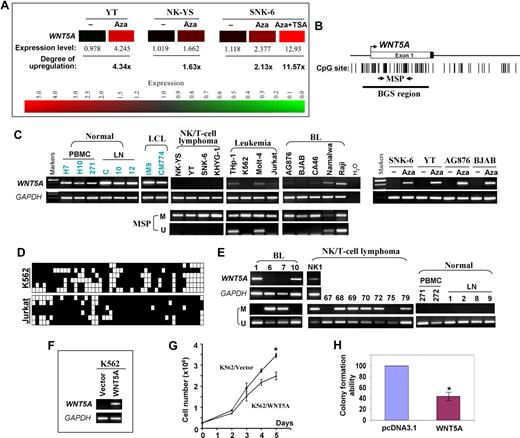
We identified WNT5A as an up-regulated gene in nasal NK/T-cell lymphoma (NL) after 5-aza-2′-deoxycitidine (Aza) global demethylation and microarray expression analysis (Figure 1A), indicating that WNT5A is a methylation-silenced gene. Semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) showed its down-regulation or silencing in multiple hematologic tumor cell lines including NL (4/4), leukemia (4/4), and Burkitt lymphoma (BL; 6/6) but not in normal lymph node (LN), peripheral blood mononuclear cell (PBMC), and lymphoblastoid cell lines (LCLs; Figure 1C). The WNT5A promoter is a typical CpG island (Figure 1B), thus subject to methylation silencing.6,7 We analyzed its methylation by methylation-specific PCR (MSP) and detected methylation in all silenced cell lines (Figure 1C). Detailed methylation analysis with bisulfite genomic sequencing (BGS) confirmed the MSP analysis and revealed a high density of methylated CpG sites in silenced cell lines (Figure 1D). After exposure to Aza, WNT5A expression was dramatically increased in silenced cell lines (Figure 1C), further elucidating that methylation directly contributes to WNT5A silencing in tumor cell lines. MSP analysis also showed that WNT5A was methylated in 50% (5/10) of BL, 73% (22/30) of NL, and 31% (11/36) of other types of non-Hodgkin lymphoma but not in 6 normal lymph nodes, 9 PBMC samples, and normal NK cells. Down-regulation of WNT5A was detected in methylated BL samples (Figure 1E). These results demonstrated that WNT5A methylation is common and tumor specific in lymphomas.
WNT5A is epigenetically silenced in hematologic malignancies and inhibits leukemic cell growth. (A) Expression levels of WNT5A mRNA in 5-aza-2′-deoxycytidine (Aza)–treated and untreated nasal NK/T-cell lymphoma (NK-YS and SNK-6) and NK leukemia (YT) cell lines. The data were obtained with microarray (Human Genome U133 Plus 2.0 Microarray, Affymetrix, Santa Clara, CA) expression analysis by GeneSpring version 7.3.1 software (Agilent, Palo Alto, CA). The degree of WNT5A up-regulation after Aza and TSA treatment is also shown. Red color represents high expression whereas green color represents low expression. (B) The CpG island (CGI) of the WNT5A promoter includes the core promoter, exon 1, and part of intron 1. The transcription start site is indicated by a curved arrow. The MSP and BGS regions analyzed in the CGI are indicated. (C) Semiquantitative RT-PCR and MSP analyses of WNT5A in normal cells and tissues and tumor cell lines. WNT5A expression in cell lines after treatment with 5 μM Aza7 is shown on the right. GAPDH was used as a control. The WNT5A expression levels among different PBMC samples could be variable. M indicates methylated; and U, unmethylated. (D) High-resolution methylation analysis of the WNT5A promoter in 2 leukemia cell lines by BGS, showing the methylation status of every CpG site in the studied region. Each row in the grid represents an individual allele of the WNT5A promoter. ■ and □ represent methylated or unmethylated CpG sites, respectively. (E) Representative MSP results of primary BL, nasal NK/T-cell lymphoma, normal PBMCs, and LN tissues. RT-PCR results of several primary BL and 1 nasal NK/T-cell lymphoma are also shown. (F) Expression levels of WNT5A in silenced K562 cells before and after transfection were determined by RT-PCR. (G) Growth curves of K562 cells after transfection with pcDNA3.1-WNT5A or control vector. At indicated time points after transfection, cell numbers were counted and plotted. Mean values plus or minus SD of triplicate experiments are shown (*P < .05). (H) Quantitative analysis of colony numbers after transfection with pcDNA3.1-WNT5A and G418 selection in K562. The number of G418-resistant colonies in the control vector–transfected cell line was set to 100%. Mean values plus or minus SD of 3 separate experiments are shown.
WNT5A is epigenetically silenced in hematologic malignancies and inhibits leukemic cell growth. (A) Expression levels of WNT5A mRNA in 5-aza-2′-deoxycytidine (Aza)–treated and untreated nasal NK/T-cell lymphoma (NK-YS and SNK-6) and NK leukemia (YT) cell lines. The data were obtained with microarray (Human Genome U133 Plus 2.0 Microarray, Affymetrix, Santa Clara, CA) expression analysis by GeneSpring version 7.3.1 software (Agilent, Palo Alto, CA). The degree of WNT5A up-regulation after Aza and TSA treatment is also shown. Red color represents high expression whereas green color represents low expression. (B) The CpG island (CGI) of the WNT5A promoter includes the core promoter, exon 1, and part of intron 1. The transcription start site is indicated by a curved arrow. The MSP and BGS regions analyzed in the CGI are indicated. (C) Semiquantitative RT-PCR and MSP analyses of WNT5A in normal cells and tissues and tumor cell lines. WNT5A expression in cell lines after treatment with 5 μM Aza7 is shown on the right. GAPDH was used as a control. The WNT5A expression levels among different PBMC samples could be variable. M indicates methylated; and U, unmethylated. (D) High-resolution methylation analysis of the WNT5A promoter in 2 leukemia cell lines by BGS, showing the methylation status of every CpG site in the studied region. Each row in the grid represents an individual allele of the WNT5A promoter. ■ and □ represent methylated or unmethylated CpG sites, respectively. (E) Representative MSP results of primary BL, nasal NK/T-cell lymphoma, normal PBMCs, and LN tissues. RT-PCR results of several primary BL and 1 nasal NK/T-cell lymphoma are also shown. (F) Expression levels of WNT5A in silenced K562 cells before and after transfection were determined by RT-PCR. (G) Growth curves of K562 cells after transfection with pcDNA3.1-WNT5A or control vector. At indicated time points after transfection, cell numbers were counted and plotted. Mean values plus or minus SD of triplicate experiments are shown (*P < .05). (H) Quantitative analysis of colony numbers after transfection with pcDNA3.1-WNT5A and G418 selection in K562. The number of G418-resistant colonies in the control vector–transfected cell line was set to 100%. Mean values plus or minus SD of 3 separate experiments are shown.
Further functional studies showed that ectopic expression of WNT5A resulted in inhibition of tumor cell growth and clonogenicity (Figure 1F-H), consistent with previous studies that WNT5A inhibited B-cell proliferation and WNT5A heterozygous mice developed myeloid leukemias and B-cell lymphomas,5 suggesting that WNT5A could serve as a tumor suppressor.
Thus, we showed that WNT5A is frequently silenced by methylation in hematologic malignancies in a tumor-specific manner. Mutations of APC and epigenetic silencing of other Wnt pathway components including APC, SFRPs, DKKs, and WIF1 lead to the activation of canonical Wnt/β-catenin signaling. The noncanonical Wnt proteins (such as WNT5A), by activating alternative signaling pathways (such as the orphan tyrosine kinase ROR2), inhibit β-catenin stabilization8,9 or induce Ca2+ flux to block canonical signaling downstream through inhibiting TCF-mediated transcription,5,10 also preventing inappropriate signaling of the canonical Wnt pathway. WNT5A silencing might act as an additional mechanism to activate the canonical Wnt-signaling pathway. As targeting the Wnt/β-catenin signaling pathway is an attractive therapeutic strategy, our findings also indicate a possibility that WNT5A might be used as a therapeutic target.
Authorship
Contribution: J.Y., G.S., and Q.T. designed and analyzed data; J.Y. and Q.T. wrote the paper; Z.G. contributed vital samples; and J.Y., H.L., and Y.-W.C. performed research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qian Tao, Cancer Epigenetics Laboratory, State Key Laboratory in Oncology in South China, Sir YK Pao Center for Cancer, Department of Clinical Oncology, Hong Kong Cancer Institute and Li Ka Shing Institute of Health Sciences, Chinese University of Hong Kong, Hong Kong; e-mail: qtao@clo.cuhk.edu.hk.