Abstract
The C-Myb transcription factor is essential for hematopoiesis, including in the T-cell lineage. The C-Myb locus is a common site of retroviral insertional mutagenesis, however no recurrent genomic involvement has been reported in human malignancies. Here, we identified 2 types of genomic alterations involving the C-MYB locus at 6q23 in human T-cell acute leukemia (T-ALL). First, we found a reciprocal translocation, t(6;7)(q23;q34), that juxtaposed the TCRB and C-MYB loci (n = 6 cases). Second, a genome-wide copy-number analysis by array-based comparative genomic hybridization (array-CGH) identified short somatic duplications that include C-MYB (MYBdup, n = 13 cases of 84 T-ALL, 15%). Expression analysis, including allele-specific approaches, showed stronger C-MYB expression in the MYB-rearranged cases compared with other T-ALLs, and a dramatically skewed C-MYB allele expression in the TCRB-MYB cases, which suggests that a translocation-driven deregulated expression may overcome a cellular attempt to down-regulate C-MYB. Strikingly, profiling of the T-ALLs by clinical, genomic, and large-scale gene expression analyses shows that the TCRB-MYB translocation defines a new T-ALL subtype associated with a very young age for T-cell leukemia (median, 2.2 years) and with a proliferation/mitosis expression signature. By contrast, the MYBdup alteration was associated with the previously defined T-ALL subtypes.
Introduction
C-Myb is a leucine zipper transcription factor, the expression of which is associated with immature and proliferative cellular stages and turned off during the maturation of the hematopoietic lineage.1,2 Silencing strategies in mice have shown that c-Myb plays a major role in hematopoiesis, including lineage commitment, proliferation, and differentiation.3–6 In the T-cell lineage, c-Myb is involved at several key steps throughout the maturation process.7–11 The c-Myb gene was first identified as the cellular homologue of the transforming v-Myb gene of 2 avian retroviruses that induce leukemia, AMV and E26.12–15 Transgenic expression of v-Myb, which is a truncated and mutated form of mammalian c-Myb, induces lymphoid or myeloid tumors in mice.16,17 The murine c-Myb locus is also a common site of retroviral insertion in lymphoid and myeloid leukemia.18–23 In humans, the C-MYB gene is located at chromosomal band 6q23.3, and interestingly the chromosome 6q is frequently involved in chromosomal abnormalities in human cancer, including hematologic malignancies.24 In addition, C-MYB is frequently expressed in human cancer.14,25–27 However, despite intensive studies in a large range of human neoplasias including leukemia, no clear recurrent involvement of the C-MYB locus in genomic abnormalities has been reported to date.23,28–31
T-cell acute lymphoblastic leukemias (T-ALLs) are highly malignant tumors that derive from T-cell progenitors.32–34 Immunophenotypic and gene expression analyses of leukemic cells have revealed heterogeneity that partially reflects distinct stages of T-cell maturation arrest.35–38 Correlations between oncogene expression, immunophenotype, and large-scale expression profiles have allowed the definition of distinct oncogenic T-ALL subtypes.37,38 Genetic studies have pointed out an increasing number of oncogenes in T-ALLs, further demonstrating the complexity of T-ALL oncogenesis and the requirement for several cooperative oncogenic events.33,34,38 T-ALL oncogenes include aberrantly activated transcription factor, namely bHLH (TAL1, TAL2, LYL1, and BHLHB1), LIM-only genes (LMO1 and LMO2), homeobox genes (HOXA, TLX1/HOX11, TLX3/HOX11L2, NKX2-5), and the CALM-AF10 and MLL fusion genes.33,34,38–42 In addition, the tumor suppressor locus CDKN2A/p16/ARF is inactivated in most T-ALL cases,43,44 and NOTCH1 is activated by mutations in half the cases.45,46 The Cyclin D2 gene and the NUP214-ABL1 fusion can also be involved in multistep oncogenesis.47,48 Notably, most T-ALL oncogenes were identified initially by their involvement in recurrent genomic abnormalities. It is expected that the identification of genetic lesions underlying each subtype of T-ALL will help to adapt therapy, including the development of specific agents against oncogenic pathways—so-called targeted therapy.49
Using combined cytogenetic, genomic, and molecular tools, we demonstrate for the first time a clear recurrent genomic involvement of the C-MYB gene locus at 6q23.3 in a human cancer, namely T-ALL, by 2 distinct types of somatic alteration: TCRB-related translocation and cryptic duplication of a short genome region. Expression analysis suggested that a deregulated C-MYB expression may be oncogenic in T-ALL. Moreover, the t(6;7) TCRB-MYB translocation defines a new T-ALL subtype that is associated with a very young age and a proliferation/mitosis expression signature.
Patients, materials, and methods
Patient samples and molecular annotations
A series of 92 T-ALL patients (56 children, 36 adults) were diagnosed and treated at Saint-Louis Hospital, Paris, France. The patients' age at diagnosis ranged from 13 months to 66 years (median age, 15 years). Informed consent was obtained from the patients or relatives in accordance with the Declaration of Helsinki. The study was approved by the Hopital Saint-Louis and Institut Universitaire d'Hematologie Institutional Review Board. Four T-cell lines, CCRF-CEM, HSB-2, Jurkat, and MOLT-4, were included in the analysis. This series was previously characterized for oncogene expression and classified for oncogenic groups according to combined immunophenotypic data, oncogene expression, and global gene expression analysis using Affymetrix (Santa Clara, CA) data.38 Three additional T-ALL cases were added to this series, one from Saint-Louis Hospital (TL93) and 2 that were analyzed by W.A.D. and A.W.L. (T142), and B.M. and B.N. (UPN5846). Large-scale expression data were obtained for case TL93 and were used after normalization using previously described methods38 for a new global analysis. The array-based comparative genomic hybridization (array-CGH) study was performed on 84 samples with available material of the 92 initial cases (80 T-ALL cases and the 4 T-cell lines), as part of the CIT National French program. Constitutional genomic DNA was obtained from follow-up samples of 6 patients in complete remission; these DNA samples were used as paired controls in array-CGH experiments to rule out copy-number polymorphisms. Additional T-ALL oncogenic annotations were obtained as follows: CDKN2A/p16/ARF genomic data using array-CGH analysis and/or locus-specific fluorescent in situ hybridization (FISH), and NOTCH1 mutations by sequencing polymerase chain reaction (PCR)–amplified genomic fragments of exons 26, 27, and 34 (encoding the heterodimerization domain [HD]), and the PEST domain.45
Cytogenetic and molecular analyses
Interphasic fluorescence in situ hybridization (FISH) analyses were performed on cryopreserved leukemic cells. All FISH probes were prepared from bacterial artificial chromosome (BAC) or P1-derived artificial chromosome (PAC) clones obtained from the BAC/PAC Resource Center at Children's Hospital Oakland Research Institute Oakland, CA (http://bacpac.chori.org). Whole chromosome painting FISH was performed using WCP6 and WCP7 from Abbott Laboratories (http://www.vysis.com). The TCRB-flanking FISH probes have been described previously.38 The C-MYB locus FISH probes were RP11–845K5, RP11–184J4, RP11–104D9, RP11–141K5, RP11–55H4, and RP11–166A21. For molecular combing analysis (fiber-FISH), DNA was combed using the Molecular Combing System (Genomic Vision, Paris, France) as described.50 All FISH micrograph images were performed with an upright epifluorescence microscope (Leica DM6000 B LT, Rueil-Malmaison, France) equipped with appropriate filter blocks for fluorescence analysis (fluoroscein isothiocyanate [FITC], Texas Red, and 4′,6-diamidino-2-phenylindole [DAPI]). The images were captured through a PlanApo 63×/1.32 oil immersion objective (0.132 mm/pixel; Leica) with a CCD camera (3 × ½″) JAI M300 (http://www.jai.com) and Isis FISH analysis software (Metasystems, Altlussheim, Germany). Images shown in the figures are at original magnification, ×630. Southern blot analysis for the C-MYB locus was performed using EcoRI, HindIII, and XbaI digestions and a panel of PCR-amplified probes from BAC RP3–388E23 DNA. Molecular cloning of the TCRB translocation breakpoint sequences was performed using ligation-mediated PCR methods as previously reported.38
Genome wide array-CGH analysis
A 4K BAC-PAC array-CGH (Curie Institute–French Ligue Contre le Cancer, Paris, France) was used on 84 T-ALL samples. This array contained 3922 BAC and PAC DNAs, as previously described.51 This array was designed to regularly cover the genome and also included additional clones for putative oncogenes such as C-MYB, or for regions known to be commonly rearranged in cancer. Hybridization, scanning, normalization, and data analysis were performed using standard procedures.51
High-density 244K oligonucleotide arrays (Agilent Technologies, http://www.agilent.com) were used to map the C-MYB duplication precisely, and to confirm somatic gain in paired tumor and constitutional genomic DNA from the same patients in cases TL29 and TL59.
Genomic databases and bioinformatics tools
The following databases and tools were used: the UCSC Genome Browser (http://genome.ucsc.edu), Ensembl (http://www.ensembl.org/index.html), the Immunogenetics (IMGT) repertoire for immunoglobulins and T-cell receptors (http://imgt.cines.fr/textes/IMGTrepertoire), the NCBI blast tools (http://ncbi.nlm.nih.gov/blast), the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman), the Database of Genomic Variants (http://projects.tcag.ca/variation), the Mouse Retrovirus Tagged Cancer Gene Database (http://rtcgd.ncifcrf.gov), the Entrez databases (http://www.ncbi.nlm.nih.gov/), and the Gene Ontology project database (http://www.geneontology.org).
Array-CGH data were analyzed using the Curie Institute VAMP tools52 (http://www.curie.fr/recherche/themes/detail_equipe.cfm/lang/_gb/id_equipe/303.htm) and the CGH Analytics 3.2 software (Agilent Technologies, http://www.agilent.com).
Large-scale gene expression data were normalized, and the analysis was performed using dChip (http://www.dchip.org), as previously described.38
C-MYB expression and sequence analyses
RNA samples were reverse transcribed and cDNAs were analyzed for C-MYB transcripts by real-time quantitative PCR (RQ-PCR) using Taqman methods (Applied Biosystems, http://www.appliedbiosystems.com/). Results were normalized on expression of the housekeeping gene TBP, according to the formula: level of C-MYB = 2[CtTBP−CtMYB]. RQ-PCR system for analysis of MYB expression was as follows: MYB-F: 5′-CTATTACCACATTTCTGAAGCACAAAA-3′, MYB-R: 5′-GCTGAGGGACATTGACTATATTTACATG-3′, and MYB probe: 5′-6-FAM-CTCCAGTCATGTTCCATACCCTGTAGCGTT-TAMRA-3′; TBP housekeeping gene: TBP-F: 5′-CACGAACCACGGCACTGATT-3′, TBP-R: 5′-TTTTCTTGCTGCCAGTCTGGAC-3′, and TBP probe: 5′-6-FAM-TGTGCACAGGAGCCAAGAGTGAAGA-TAMRA-3′. Additional PCR systems for the analysis of alternative transcripts and differential use of promoters are shown in the Figure S4 (available on the Blood website; see the Supplemental Materials link at the top of the online article). In selected cases, the entire C-MYB open reading frame was sequenced on both strands from leukemic cDNA after PCR of cDNA from leukemic cells; primers sequences are available in Figure S4. A polymorphic poly-T (T8/9) microsatellite found in the 3′-UTR region was used for analysis of C-MYB allelic expression. Fragment size analysis of a fluorescent-labeled PCR fragment using Genescan methods (Applied Biosystems) allowed discrimination of the T8/T9 alleles. PCR primers were as follows: S217F-5′-6-FAM-AGGTAATGAATTGTAGCCAG-3′ and S218R-5′-AAATACTGATCTGTTGGATCC-3′. Leukemic genomic DNA samples were first analyzed to determine the heterozygous cases, and then leukemic cDNAs from informative cases were analyzed to determine C-MYB allele expression.
Results
Identification of a recurrent translocation t(6;7)(q23;q34) in T-ALL involving the TCRB and C-MYB loci
In the course of conventional cytogenetic analysis at diagnosis of acute leukemia patients in Saint-Louis Hospital, we identified a chromosomal translocation t(6;7)(q23;q34) in 2 pediatric T-ALL cases (TL34 and TL92, Figure 1A,B). Dual-color FISH using TCRB flanking probes demonstrated involvement of the TCRB locus at 7q34 in both cases (Figure 1B). By using inverse PCR from the TCRB sequence, we amplified and characterized the breakpoints' derivative sequences in case TL34 (Figure 1C). The breakpoint on chromosome 6q23.3 was mapped in the vicinity of the C-MYB oncogene. Probes were derived from the C-MYB locus and demonstrated in FISH and Southern blot experiments that the same locus was involved in case TL92 (Figure 1D). The t(6;7)(q23;q34) involves telomeric regions in both chromosomes 6 and 7, hence we assumed that this translocation could have been missed in systematic T-ALL karyotype series. We therefore used C-MYB flanking FISH probes that allowed us to identify 2 additional cases with translocations involving the C-MYB and TCRB loci (TL33 and TL93, Figure 1E) from a series of 84 T-ALL (80 T-ALL patients and 4 T-cell lines). Finally, 2 isolated cases with a TCRB-MYB translocation (UPN5846 and T142) were detected from independent T-ALL series by systematic molecular analysis of oncogenic TCRB genes rearrangements, as performed by B.M. and B.N., and W.A.D. and A.W.L. These cases were added to the present study, giving a total of 6 TCRB-MYB cases, including 3 cases fully characterized at the molecular level on both derivative chromosomes (Figure 1C; Figure S1). Sequence analysis of the TCRB derivative sequences was suggestive of a specific mechanism of V(D)J-mediated translocation typically seen at the TCRB locus.53,54 These translocations were reciprocal and balanced, and led to the juxtaposition of the C-MYB proto-oncogene near to the TCRB regulatory sequence (Figure 1F), which suggested deregulated expression.
A recurrent translocation t(6;7)(q23;q34) in T-ALL involves the TCRB and C-MYB loci. (A) Partial R-banded karyotype with translocation t(6;7)(q23;q34). (B, left panel) FISH whole chromosome painting of chromosomes 6 (green) and 7 (red) on metaphases from leukemic cells confirmed the reciprocal translocation t(6;7). (B, right panel) Dual-color FISH analysis of the TCRB locus using TCRB-flanking probes (centromeric, CTD-3092H9 labeled in red; telomeric, RP11–168I15, labeled in green). Dissociation of the probes in leukemic cells with t(6;7) demonstrated the involvement of the TCRB locus as partner of the translocation. (C) Derivative sequences of t(6;7)(q23;q34) breakpoints in case TL34 and corresponding germ-line sequences. Recognition sequence signal (RSS) heptamer and putative heptamer-like sequence are indicated according to consensus53 ; untemplated nucleotides (N-diversity) are typed in lowercase. GL indicates germ line; der, derivative chromosomes. Breakpoint sequences for cases T142 and UPN5846 are shown in Figure S1. (D) Southern blot mapping of the translocation breakpoint in case TL92 using a 6q23.3 probe derived from BAC RP11–388E23. Germ-line (GL) and TL34 (TCRB-MYB) DNAs are shown as negative and positive controls, respectively. EcoRI rearranged bands are shown by arrows. (E) Interphasic FISH screening of T-ALL using flanking C-MYB locus probes RP11–845K5 (green) and RP11–184J4 (red) identified additional MYB-translocated cases TL33 and TL93, further shown to juxtapose the TCRB and C-MYB loci using combinations of FISH probes. (F) Schematic representation of the der(6) genomic region of t(6;7), according to data from the IMGT database and the UCSC Genome Bioinformatics site, and to the DNA derivative sequence of the breakpoint region in case T142. An arrow indicates the breakpoint (BP). See “Patients, materials, and methods; Cytogenetic and molecular analyses” for details about FISH image acquisition and manipulation.
A recurrent translocation t(6;7)(q23;q34) in T-ALL involves the TCRB and C-MYB loci. (A) Partial R-banded karyotype with translocation t(6;7)(q23;q34). (B, left panel) FISH whole chromosome painting of chromosomes 6 (green) and 7 (red) on metaphases from leukemic cells confirmed the reciprocal translocation t(6;7). (B, right panel) Dual-color FISH analysis of the TCRB locus using TCRB-flanking probes (centromeric, CTD-3092H9 labeled in red; telomeric, RP11–168I15, labeled in green). Dissociation of the probes in leukemic cells with t(6;7) demonstrated the involvement of the TCRB locus as partner of the translocation. (C) Derivative sequences of t(6;7)(q23;q34) breakpoints in case TL34 and corresponding germ-line sequences. Recognition sequence signal (RSS) heptamer and putative heptamer-like sequence are indicated according to consensus53 ; untemplated nucleotides (N-diversity) are typed in lowercase. GL indicates germ line; der, derivative chromosomes. Breakpoint sequences for cases T142 and UPN5846 are shown in Figure S1. (D) Southern blot mapping of the translocation breakpoint in case TL92 using a 6q23.3 probe derived from BAC RP11–388E23. Germ-line (GL) and TL34 (TCRB-MYB) DNAs are shown as negative and positive controls, respectively. EcoRI rearranged bands are shown by arrows. (E) Interphasic FISH screening of T-ALL using flanking C-MYB locus probes RP11–845K5 (green) and RP11–184J4 (red) identified additional MYB-translocated cases TL33 and TL93, further shown to juxtapose the TCRB and C-MYB loci using combinations of FISH probes. (F) Schematic representation of the der(6) genomic region of t(6;7), according to data from the IMGT database and the UCSC Genome Bioinformatics site, and to the DNA derivative sequence of the breakpoint region in case T142. An arrow indicates the breakpoint (BP). See “Patients, materials, and methods; Cytogenetic and molecular analyses” for details about FISH image acquisition and manipulation.
A genome-wide array-CGH screen in T-ALL identified recurrent somatic genomic duplications at the C-MYB locus
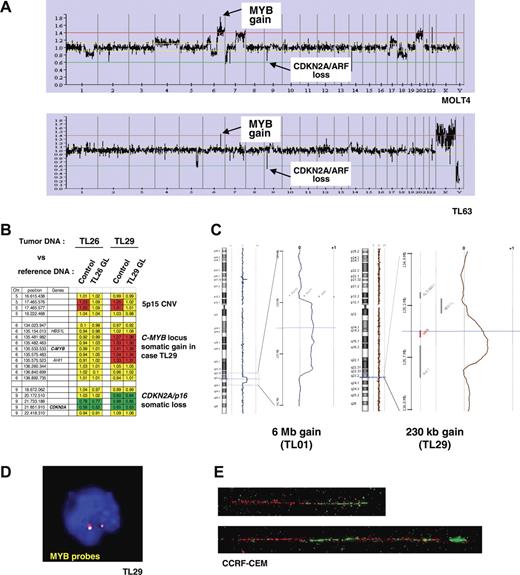
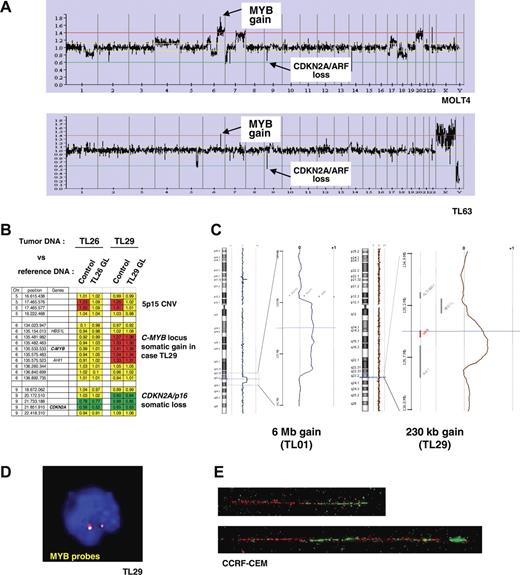
A genome-wide analysis of genomic copy number was performed in 84 of the 92 T-ALL cases (80 primary samples and 4 T-cell lines), in order to detect genomic imbalances in T-ALL. Leukemic DNA samples and a normal DNA XY control (GYPT) were cohybridized on a 4K BAC/PAC array (“Patients, materials, and methods”). Remarkably, examination of the data for chromosome 6 revealed a DNA copy gain of a 6q23.3 region in 13 cases of 84 T-ALL (11 T-ALL patients and 2 T-cell lines, Figure 2A). In 2 cases, the DNA copy gain consisted of a large chromosomal gain (6.0 Mb in TL01, and 22.4 Mb in TL76), while in the other cases the amplified region was restricted to a region less than 2 Mb including the C-MYB locus. The MOLT4 cell line showed a more complex profile with a gain of a large region of chromosome 6q in a pseudotetraploid karyotype reinforced by higher level of amplification in a region less than 2 Mb at the C-MYB locus (Figure 2A top panel).
Identification of recurrent somatic genomic duplications at the C-MYB locus in T-ALL. (A) Global representation of large-scale analysis of genomic copy number using a 4K array-CGH in MOLT4 cell line (upper panel) and TL63 primary T-ALL case (lower panel). DNA copy gain at the C-MYB locus at 6q23 is indicated, as well as the DNA copy loss at the CDKN2A/p16/ARF locus at 9p21 as an example. (B) Compared analysis of array hybridization using as normal DNA either a healthy subject DNA (control) or germ-line DNA (GL) of the same patient as DNA reference. This analysis allowed us to distinguish copy-number variations (CNVs) and somatic genomic imbalances. As an example, a CNV at 5p15 was resolved in the TL26 and TL29 cases (a gain was observed when the leukemic DNA was cohybridized with an unrelated control DNA, but it disappeared using paired leukemic and GL controls). A somatic loss of CDKN2A/p16/ARF was evidenced in both the TL26 and TL29 cases, because the imbalance persisted after cohybridization with paired control; similarly the somatic gain at the 6q23/C-MYB locus was confirmed in case TL29. For each BAC/PAC of the array, gains were represented in red; losses, in green; and balanced signals, in yellow. (C) Copy-number analysis using a very high-density oligonucleotide array (Agilent), focused on the C-MYB region, enabled the minimal region of genomic gain to be mapped to approximately 230 kb. (C, right panel) A 6-Mb–sized copy gain region was found in case TL01; (left panel) a short minimal genomic gain including the C-MYB gene was evidenced in case TL29 (array-CGH performed with paired leukemic and GL DNAs). The genomic region of gain of chromosome 6 is magnified, with the horizontal cursor (blue) pointing out the C-MYB gene (shown in red in the right panel). The so-called “moving average” ratio between leukemic and GL DNA appears as a blue line in case TL01, and a brown line in case TL29. Two copies (alleles) of the locus appear as a moving average close to 0, whereas a DNA copy gain of an allele shifts the line close to ratio + 0.5 along the corresponding genomic region. (D) Interphasic C-MYB FISH using the RP1–32B1 (green) and RP3–388E23 (red) probes in the MYBdup cases showed no extra signal (except in the complex MOLT4 pseudotetraploid cell line, not shown), which suggests local duplication. (E) Molecular combing analysis using C-MYB locus probes RP11–55H4 (red) and RP11–166A21 (green) demonstrated a local duplication (bottom); the normal allele is shown on the top. See “Patients, materials, and methods; Cytogenetic and molecular analyses” for details about FISH image acquisition and manipulation.
Identification of recurrent somatic genomic duplications at the C-MYB locus in T-ALL. (A) Global representation of large-scale analysis of genomic copy number using a 4K array-CGH in MOLT4 cell line (upper panel) and TL63 primary T-ALL case (lower panel). DNA copy gain at the C-MYB locus at 6q23 is indicated, as well as the DNA copy loss at the CDKN2A/p16/ARF locus at 9p21 as an example. (B) Compared analysis of array hybridization using as normal DNA either a healthy subject DNA (control) or germ-line DNA (GL) of the same patient as DNA reference. This analysis allowed us to distinguish copy-number variations (CNVs) and somatic genomic imbalances. As an example, a CNV at 5p15 was resolved in the TL26 and TL29 cases (a gain was observed when the leukemic DNA was cohybridized with an unrelated control DNA, but it disappeared using paired leukemic and GL controls). A somatic loss of CDKN2A/p16/ARF was evidenced in both the TL26 and TL29 cases, because the imbalance persisted after cohybridization with paired control; similarly the somatic gain at the 6q23/C-MYB locus was confirmed in case TL29. For each BAC/PAC of the array, gains were represented in red; losses, in green; and balanced signals, in yellow. (C) Copy-number analysis using a very high-density oligonucleotide array (Agilent), focused on the C-MYB region, enabled the minimal region of genomic gain to be mapped to approximately 230 kb. (C, right panel) A 6-Mb–sized copy gain region was found in case TL01; (left panel) a short minimal genomic gain including the C-MYB gene was evidenced in case TL29 (array-CGH performed with paired leukemic and GL DNAs). The genomic region of gain of chromosome 6 is magnified, with the horizontal cursor (blue) pointing out the C-MYB gene (shown in red in the right panel). The so-called “moving average” ratio between leukemic and GL DNA appears as a blue line in case TL01, and a brown line in case TL29. Two copies (alleles) of the locus appear as a moving average close to 0, whereas a DNA copy gain of an allele shifts the line close to ratio + 0.5 along the corresponding genomic region. (D) Interphasic C-MYB FISH using the RP1–32B1 (green) and RP3–388E23 (red) probes in the MYBdup cases showed no extra signal (except in the complex MOLT4 pseudotetraploid cell line, not shown), which suggests local duplication. (E) Molecular combing analysis using C-MYB locus probes RP11–55H4 (red) and RP11–166A21 (green) demonstrated a local duplication (bottom); the normal allele is shown on the top. See “Patients, materials, and methods; Cytogenetic and molecular analyses” for details about FISH image acquisition and manipulation.
We then investigated whether the short DNA copy gain of the C-MYB locus was a somatic rearrangement that originated in leukemic cells, or whether it could be a constitutional large-size copy-number variation (CNV) as was recently reported in the human genome.55–57 Notably, no CNV was reported in this region in the Database of Genomic Variants (Figure S2). Then we compared our data to those from 2 independent array-CGH studies on breast cancer samples that were performed using the same array and the same control DNA (GYPT, a unique XY healthy subject whose DNA was used in several studies); no recurrent genomic imbalance in the C-MYB locus was observed in these other studies, which suggested that there was no frequent CNV in this region and that the C-MYB gain was associated with T-ALL (data not shown). Finally, we were able to demonstrate definitively the somatic origin of the C-MYB gain by cohybridizing on the same array paired leukemic and constitutional DNAs from the same patient in 2 T-ALL cases with available constitutional material (Figure 2B).
Thereafter, we defined a minimal region of DNA copy gain using high-density 244K oligonucleotide array-CGHs (Agilent) to a consistent region of length approximately 230 kb including the C-MYB gene (Figure 2C). A panel of probes covering the C-MYB locus detected no extra signal in metaphases or interphase nuclei in cases with C-MYB copy gain (Figure 2D), which suggests a local duplication. That was demonstrated by molecular combing analysis showing a direct tandem duplication of the C-MYB locus (Figure 2E). Cases with C-MYB locus duplication are further referred to in the study reported herein as MYBdup cases.
Genomic location of the C-MYB locus rearrangements in human and mouse T-cell leukemias
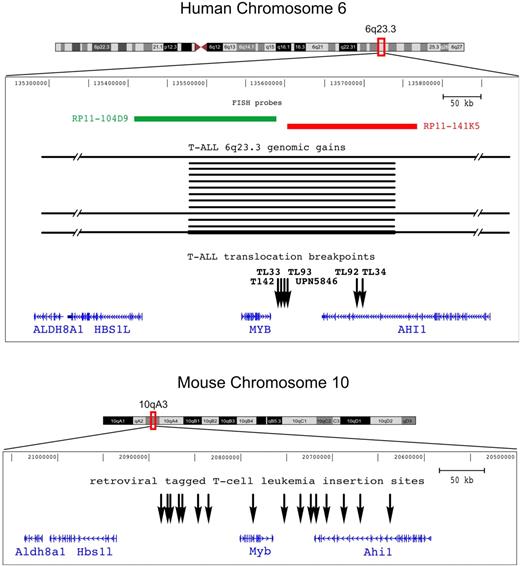
In the present study 2 types of recurrent somatic rearrangement were identified in T-ALL, namely reciprocal chromosomal translocation TCRB-MYB, and short genomic duplications MYBdup, which target a unique 6q23.3 region that includes the C-MYB proto-oncogene. Molecular mapping of the 6 t(6;7)(q23;q34) chromosomal breakpoints, which included complete breakpoint sequencing in 3 cases (TL34, T142, and UPN5846), showed 2 discrete breakpoint clusters at 6q23.3 (Figure 3 top panel): one located 5 kb telomeric, 3′ of the C-MYB gene (4 cases: TL33, TL93, T142, and UPN5846), and the other 50 kb more telomeric (2 cases: TL34 and TL92). In all cases, the translocation placed the C-MYB proto-oncogene in the vicinity of the TCRB regulatory sequence, which suggests that abnormal regulation of C-MYB expression could confer oncogenic properties. The 6q23 genomic duplication was mapped to a short region of approximately 230 kb that encompassed the entire C-MYB gene in all duplicated cases (Figure 3 top panel), which reinforced the view of a targeting of this gene by oncogenic somatic events in T-ALL. Another gene, known as AHI1, was located in the vicinity of the t(6;7) breakpoints and was disrupted in 2 of 6 cases with the t(6;7), but not in the other 4 cases. It was also partially included in the minimal region of duplication.
Genomic rearrangements target the C-MYB locus in both human and mouse T-cell leukemias. Representation of the C-MYB locus in both the human and mouse genomes according to the UCSC database; note that the orientation of the mouse locus was inverted in this figure in order to maintain the orientation of the human locus and facilitate comparison. The ALDH8A1, HSB1L, C-MYB, and AHI1 genes are shown according to annotations in UCSC. The genomic rearrangements we described in this work in human T-ALL are indicated: breakpoints (vertical black arrows) of the 6 TCRB-MYB cases, and duplicated genomic regions (horizontal black bars) in the 13 T-ALL cases (TL01, TL29, TL38, TL40, TL49, TL59, TL61, TL63, TL66, TL76, TL77, CCRF-CEM, and MOLT4). The darker horizontal bottom line refers to a MYBdup in addition to a larger 6q gain in MOLT4, leading to extra copies of the locus. Two FISH probes that were used for breakpoint mapping are shown. On the mouse genome (bottom panel), all the reported retroviral insertion sites of T-cell leukemias are indicated (vertical black arrows), based on data from the Retrovirus Tagged Cancer Gene Database.
Genomic rearrangements target the C-MYB locus in both human and mouse T-cell leukemias. Representation of the C-MYB locus in both the human and mouse genomes according to the UCSC database; note that the orientation of the mouse locus was inverted in this figure in order to maintain the orientation of the human locus and facilitate comparison. The ALDH8A1, HSB1L, C-MYB, and AHI1 genes are shown according to annotations in UCSC. The genomic rearrangements we described in this work in human T-ALL are indicated: breakpoints (vertical black arrows) of the 6 TCRB-MYB cases, and duplicated genomic regions (horizontal black bars) in the 13 T-ALL cases (TL01, TL29, TL38, TL40, TL49, TL59, TL61, TL63, TL66, TL76, TL77, CCRF-CEM, and MOLT4). The darker horizontal bottom line refers to a MYBdup in addition to a larger 6q gain in MOLT4, leading to extra copies of the locus. Two FISH probes that were used for breakpoint mapping are shown. On the mouse genome (bottom panel), all the reported retroviral insertion sites of T-cell leukemias are indicated (vertical black arrows), based on data from the Retrovirus Tagged Cancer Gene Database.
Reminiscent of the 6q23 rearrangements that we identified in human T-ALL, the C-Myb locus is known to be a frequent insertion site in retrovirally induced leukemias. The lower panel of Figure 3 shows the insertion sites in murine T-cell leukemia as reported in the Retrovirus Tagged Cancer Gene Database.19,21–23 These sites are distributed around the c-Myb gene, within a region that extends from approximately 100 kb upstream to 150 kb downstream of the gene. Importantly, insertion sites flank both sides of c-Myb, which suggests that this gene is an oncogenic target in this chromosomal region.
Therefore the combined genomic data from human (present study) and murine T-cell leukemias (from previous reports) demonstrate that this locus is recurrently targeted in T-cell oncogenesis and point to the C-MYB gene as a strong candidate oncogene in these T-ALL cases.
C-MYB expression analysis in human T-ALLs
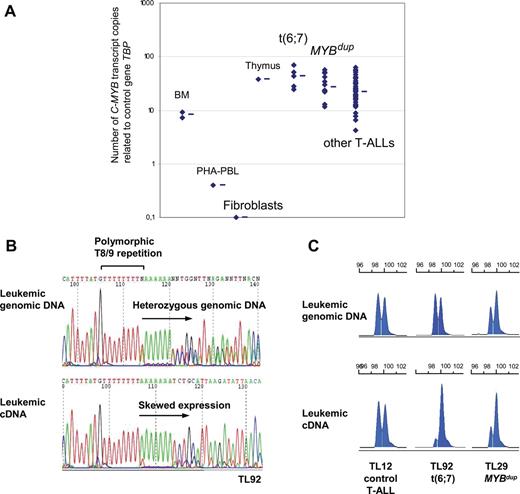
The murine c-Myb gene has been shown to be strongly expressed in normal thymus, and it is necessary for progression through several stages of T-cell differentiation.7–11 To gain insights in the putative oncogenic role of C-MYB rearrangements in human T-ALL, we analyzed C-MYB expression using real-time quantitative PCR (RQ-PCR) in the MYB-rearranged T-ALL cases in comparison with other T-ALL cases and control cells. We found strong C-MYB expression in the T-ALL cases (ie, not only in the rearranged cases), compared with normal PHA-stimulated peripheral blood leukocytes (PBLs), normal bone marrow, and growing fibroblasts (Figure 4A). In this high range, the median expression of C-MYB transcripts was stronger in the t(6;7) and MYBdup cases than in other T-ALLs (Figure 4A). Consistent results were obtained by analyzing C-MYB expression from large-scale expression data, whereas no difference was found for the other genes of the region among the T-ALL groups (Figure S3). An analysis was performed using RQ-PCR systems that were designed to evaluate several alternative transcripts (Figure S4). Systems included the main C-MYB transcripts, long variant transcripts that include the so-called alternative exon 9A, the use of distinct promoters (an alternative transcription site has been reported just upstream of exon 2 in humans and mice), and the use of the 3′UTR region in exon 15. Also, standard RT-PCR was performed to search for additional small exons. This analysis did not reveal changes in the differential pattern of alternative transcripts in the MYB-rearranged cases compared with other T-ALL cases and healthy controls, nor did it reveal additional exons (Figure S4). C-MYB immunoblot analysis of a number of T-ALL primary cases and cell lines did not reveal aberration of C-MYB levels or size (Figure S4). Then we sequenced the C-MYB open reading frame from leukemic cDNA in 2 t(6;7) cases (TL34, TL92) and in the 4 T-cell lines including the 2 MYBdup cell lines (CCRF-CEM and MOLT4), and no mutation was found. At this point, we investigated C-MYB allelic expression in the T-ALL leukemic cells, taking advantage of a T8/T9 polymorphic repetition in the 3′UTR of the C-MYB gene (Figure 4B). Forty-one cases of 84 T-ALLs showed a heterozygous T8/T9 polymorphism in leukemic genomic DNA and were therefore informative, including 3 TCRB-MYB cases (TL33, TL34, and TL92) and 7 MYBdup cases (TL01, TL29, TL38, TL49, TL61, TL76, and MOLT4). Strikingly, analysis at the RNA level demonstrated a dramatically imbalanced C-MYB expression in the 3 TCRB-MYB cases, suggesting a skewed TCRB-driven expression and, importantly, a low expression of the other allele (Figure 4B-C). In the 7 informative MYBdup cases, allelic expression of C-MYB was imbalanced in the same way as genomic DNA copy number, or even more in some cases (Figure 4C). By contrast, a balanced C-MYB expression was found in all nonrearranged T-ALL cases (n = 27 informative cases, Figure 4C). In all, our data suggest that the expression of C-MYB transcripts can be sustained inappropriately at strong levels due to the translocation in the TCRB-MYB cases, and reinforced due to copy-number increase in the MYBdup cases.
Expression analysis of the C-MYB gene in T-ALL. (A) C-MYB transcript levels were quantified by RQ-PCR in a series of samples: normal bone marrow (BM, n = 2), PHA-stimulated PBL (PHA-PBL, n = 1), growing fibroblasts (n = 1), thymus (n = 1), t(6;7) TCRB-MYB cases (n = 5, one case not available), MYBdup (n = 13), other T-ALL (n = 54) as indicated. Median value is indicated by a horizontal bar for each group. (B) Sequencing of C-MYB in TL92 tumor genomic DNA showed heterozygosity for a polymorphic T8/9 repetition in the 3′UTR region; sequencing of the TL92 leukemic cDNA in this TCRB-MYB case demonstrated a skewed allelic expression. (C) Analysis of the T8/9 polymorphic microsatellite using fragment size analysis. Heterozygous cases without C-MYB rearrangement (n = 27) showed balanced expression; heterozygous cases with t(6;7) translocation (n = 3; TL33, TL34, and TL92) showed a skewed expression of one C-MYB allele; and heterozygous cases with MYBdup (n = 7) showed an imbalanced profile of both leukemic genomic DNA and leukemic cDNA.
Expression analysis of the C-MYB gene in T-ALL. (A) C-MYB transcript levels were quantified by RQ-PCR in a series of samples: normal bone marrow (BM, n = 2), PHA-stimulated PBL (PHA-PBL, n = 1), growing fibroblasts (n = 1), thymus (n = 1), t(6;7) TCRB-MYB cases (n = 5, one case not available), MYBdup (n = 13), other T-ALL (n = 54) as indicated. Median value is indicated by a horizontal bar for each group. (B) Sequencing of C-MYB in TL92 tumor genomic DNA showed heterozygosity for a polymorphic T8/9 repetition in the 3′UTR region; sequencing of the TL92 leukemic cDNA in this TCRB-MYB case demonstrated a skewed allelic expression. (C) Analysis of the T8/9 polymorphic microsatellite using fragment size analysis. Heterozygous cases without C-MYB rearrangement (n = 27) showed balanced expression; heterozygous cases with t(6;7) translocation (n = 3; TL33, TL34, and TL92) showed a skewed expression of one C-MYB allele; and heterozygous cases with MYBdup (n = 7) showed an imbalanced profile of both leukemic genomic DNA and leukemic cDNA.
The TCRB-MYB translocation defines a new distinct T-ALL oncogenic subtype, associated with very young age and a proliferation/mitosis signature by microarray large-scale expression analysis, whereas the MYBdup is found in other T-ALL subtypes
Clinical and biologic data were collected for patients with C-MYB rearrangements (Table 1). Strikingly, 5 of the 6 cases with the translocation t(6;7) were diagnosed in very young children (1.1, 1.3, 1.8, 2.5, and 2.9 years old [Table 1]; median age of the TCB-MYB patients, 2.2 years). This clustered age range is very unusual in T-ALL where patients are generally older than in precursor B-ALL. For comparison, the median age in pediatric T-ALL was 9.4 years (range, 1 to 19.5 years) in the French randomized multicentric pediatric trials FRALLE 93 and FRALLE 2000 (n = 355 pediatric T-ALL patients), and only 26 (7.3%) of 355 children were younger than 3 years (P < .001). Such a unifying feature associated with the TCRB-MYB translocation suggests strongly that it defines a new distinct entity in T-ALL. Consistently, the common oncogenic abnormalities defining other T-ALL subtypes were not found in the TCRB-MYB patients (ie, SIL-TAL, TLX3/HOX11L2, TLX1/HOX11, HOXA, MLL, and CALM-AF10; Table 1). As in other oncogenic subtypes, the TCRB-MYB cases frequently displayed the common additional genomic events, CDNK2A/p16/ARF deletion and NOTCH1 mutations; this demonstrates genetic multievent oncogenesis in this new T-ALL subtype (Table 1).
The situation was clearly different in T-ALL patients with genomic C-MYB duplication. Patients had various ages (median, 17 years; range, 5 to 52 years; Table 1). The MYBdup leukemic cells frequently expressed various other oncogenic transcripts: SIL-TAL (cases TL29 and CCRF-CEM), TLX1 (TL49, TL59), TLX3 (TL61, TL63, TL66), MLL-ENL (TL38), and CALM-AF10 (TL40), and 2 cases were classified as immature (TL76 and TL77). Additional genomic events were also found in the MYBdup cases: CDNK2A/p16/ARF deletion, NOTCH1 mutations, and NUP214-ABL1 amplification (Table 1).
In order to characterize further the MYB-rearranged T-ALL, we analyzed large-scale gene expression data. Three cases with the TCRB-MYB rearrangement (TL33, TL34, and TL92) and all MYBdup cases have been previously included in a series of large-scale gene expression analysis experiments.38 A fourth case (TL93) was also tested and included in a new global analysis. The MYB-rearranged cases, as other T-ALLs, displayed high expression of cyclin D358 and of genes belonging to a global T-ALL signature59 (data not shown). An unsupervised classification was performed demonstrating that the 4 TCRB-MYB cases clustered closely (Figure S5). These cases were then compared by t test analysis to the 3 major T-ALL subtypes as defined by the molecular abnormalities SIL-TAL1, TLX1, and TLX3. A large number of differentially expressed probe sets was obtained in each comparison (median P value, .001; median, FDR < 10%). A merged list of genes was built (Table S1), and hierarchical clustering analysis was performed. The 4 TCRB-MYB cases stably coclustered with 3 cell lines, namely Jurkat, CCRF-CEM, and MOLT4, the thymus sample, and the SIL-TAL case TL29 (Figure 5A). This cluster was included in a large branch that contains all cases with a SIL-TAL1 rearrangement. The cluster was characterized by high expression of 19 genes (Figure 5B), 18 of them linked by Gene Ontology (GO) terms to cell cycle, mitosis, and cell proliferation (Table S2). Interestingly, no expression of the ectopic genes previously associated with the SIL-TAL1, TLX1, or TLX3 subtypes was found in any of the 4 cases. By contrast, the MYBdup cases were distributed in the clusters according to oncogene expression (SIL-TAL1, TLX1, TLX3, CALM-AF10, MLL, immature), and they did not cluster together (Figure S5).
Profiling of the TCRB-MYB cases by large-scale gene expression analysis. (A) Heat map representation of the hierarchical clustering of genes and samples using expression values for genes expressed differentially between the 4 TCRB-MYB cases and the SIL-TAL1, TLX1, and TLX3 cases (gene list in Table S1). One thymus sample and 4 T-cell lines are also included in the analysis. Cytogenetic/genomic annotations are as follows: Y indicates TCRB-MYB; L, TLX3; E, TLX1; and S, SIL-TAL1. Oncogenic groups: Y indicates TCRB-MYB; H, HOX-related; T, TAL-related; and c, cell lines.38 Oncogene expression levels are shown as follows: TAL1 positive indicates range from 1 to 6; −, not analyzed; others are negative. LMO1 and LMO2: P indicates positive; −, not analyzed; others are negative. Data were normalized as described38 and redundant probe sets were masked. The 4 TCRB-MYB cases (boxed in pink) are included in a cluster of cases indicated in blue, together with 3 cell lines (CCRF-CEM, Jurkat, and HSB2), the normal thymus sample, and a SIL-TAL case (TL29). Representative genes from the 3 major clusters are shown, and ectopic genes are indicated in red. (B) Average expression (± SD) of the genes that characterize the TCRB-MYB and cell line cluster. This cluster is specifically enriched in genes linked to cell cycle, cell proliferation, and mitosis, according to Gene Ontology (GO) terms (Table S2).
Profiling of the TCRB-MYB cases by large-scale gene expression analysis. (A) Heat map representation of the hierarchical clustering of genes and samples using expression values for genes expressed differentially between the 4 TCRB-MYB cases and the SIL-TAL1, TLX1, and TLX3 cases (gene list in Table S1). One thymus sample and 4 T-cell lines are also included in the analysis. Cytogenetic/genomic annotations are as follows: Y indicates TCRB-MYB; L, TLX3; E, TLX1; and S, SIL-TAL1. Oncogenic groups: Y indicates TCRB-MYB; H, HOX-related; T, TAL-related; and c, cell lines.38 Oncogene expression levels are shown as follows: TAL1 positive indicates range from 1 to 6; −, not analyzed; others are negative. LMO1 and LMO2: P indicates positive; −, not analyzed; others are negative. Data were normalized as described38 and redundant probe sets were masked. The 4 TCRB-MYB cases (boxed in pink) are included in a cluster of cases indicated in blue, together with 3 cell lines (CCRF-CEM, Jurkat, and HSB2), the normal thymus sample, and a SIL-TAL case (TL29). Representative genes from the 3 major clusters are shown, and ectopic genes are indicated in red. (B) Average expression (± SD) of the genes that characterize the TCRB-MYB and cell line cluster. This cluster is specifically enriched in genes linked to cell cycle, cell proliferation, and mitosis, according to Gene Ontology (GO) terms (Table S2).
These data show that the TCRB-MYB translocation defines a new T-ALL subtype, associated with very young age, no association to the other common oncogenic transcripts, and a proliferation/mitosis signature, whereas the MYBdup abnormality can be found in association with other T-ALL subtypes.
Discussion
The C-MYB gene has been a candidate oncogene in humans for years, based on its homology with the viral transforming oncogene v-Myb,12–17 frequent targeting by retroviral insertions in mice,18–23 and its major role in hematopoiesis.3–11 However, there has been a striking lack of genomic involvement in human cancer, making the C-MYB oncogenic role questionable in humans, and a general view has been that it could be expressed as a reflection of the immature and proliferative stages of neoplastic cells rather than to be causal in cancer.23,28,30 Here we report for the first time recurrent abnormalities of the C-MYB locus by 2 distinct types of genomic abnormalities in human T-cell lymphoblastic leukemia, with a deregulated expression with respect to other T-ALLs. These results point again to the C-MYB gene as a strong candidate oncogene in human neoplasia.
First, we detected and characterized fully a recurrent t(6;7)(q23;q34) reciprocal translocation. A few t(6;7) cases had been reported in karyotype studies of acute leukemias, but partner genes were largely unresolved (Mitelman Database of Chromosome Aberrations in Cancer).60–62 Here, we showed that this translocation juxtaposed the TCRB and the C-MYB loci, and localized precisely the breakpoints at the molecular level. It should be noted that the t(6;7) translocation can be missed due to the subtelomeric location of the breakpoints and frequent poor morphology of T-ALL karyotypes. Indeed, we identified t(6;7) cases that were undiscovered previously, using locus-specific FISH and molecular approaches. This translocation was found in very young children compared with the median age for T-ALL; hence, it may be a common abnormality in these patients. Systematic searches in young children with T-ALLs will allow evaluation of the occurrence of C-MYB abnormalities in this group of patients.
Using systematic genome-wide copy-number analysis, we identified a second recurrent genomic abnormality involving C-MYB in T-ALL, as a short genomic tandem duplication. This cryptic abnormality was undetectable using conventional cytogenetics and locus-specific FISH, which may explain why it was not identified in previous studies. We mapped precisely the MYBdup to a 230-kb region that contained the C-MYB gene using high-density oligonucleotide array-CGH. Local tandem duplication was demonstrated using molecular combing. These data highlight the strength of high-density array-CGH to identify cryptic copy-number abnormalities in leukemia, as was shown recently for the short LMO2 activating deletions in T-ALL.59 Importantly, we demonstrated that the duplication was somatic and not due to one of the constitutional copy-number variations (CNVs) that was recently discovered to be relatively common in the human genome.55–57 For this purpose, the availability of samples from the same patients in remission, which allowed paired constitutional and tumor analysis, was invaluable (Figure 2B). Interestingly, the MYBdup abnormality was frequently found in association with the known classifying oncogene transcripts (SIL-TAL, CALM-AF10, MLL-ENL, TLX1, TLX3),37,38 which suggests that MYBdup could be an additional oncogenic event.
In contrast with the MYBdup abnormality, clinical and biologic data suggest that the TCRB-MYB translocation defines a new T-ALL subtype. Strikingly, patients with this translocation were very young (no adult; median age, 2.2 years) compared with the age range of T-ALL patients. Moreover, the ectopic transcripts that have been associated previously with the principal T-ALL subtypes37,38 were not found in these cases, which supports further the view that this rearrangement is associated with distinct oncogenic pathways. Coclustering of these cases with T-cell lines and normal thymus sample in a subgroup that is characterized by a high expression of cell cycle, cell proliferation, and mitosis genes suggests a specific related biology (Figure 5, Figure S5, and Table S2).
The c-Myb locus is known as a frequent insertion site in retrovirus-mediated oncogenesis in animals.19,21–23 A detailed map of the mouse insertion sites based on the RTCG database and comparison with human data shows that the c-Myb insertions mimic the genomic abnormalities here described in human leukemia (Figure 3), and provides new validation of the power of these experimental approaches in animals to pinpoint human oncogenes. In all, genomic data in mice and humans involve a genomic region encompassing the c-Myb gene, with retroviral sites at both sides of the c-Myb gene in mice, which suggests that this gene is a major oncogenic target in this chromosomal region. Three other genes, namely AHI1 (Abelson Helper Integration), ALDH8A1 (aldehyde dehydrogenase 8 family, member A1), and HBS1L (HBS1-like, S cerevisiae), are located by this genomic region. The AHI1 gene was disrupted in 2 of 6 t(6;7) T-ALL cases. AHI1 is a WD40 and SH3-containing protein that has a complex pattern of isoforms.63 It was found to be mutated in a congenital brain malformation syndrome known as Joubert syndrome,64 and was investigated as a potential oncogene at retroviral insertions Ahi1 in Abl+ mice, and in human leukemic cell lines and Ph+ samples.63,65 Although we found no deregulation of AHI-1 gene expression, or of the other genes of the region, in the t(6;7) or in the MYBdup T-ALL cases (Figure S3), the possibility remains that AHI-1 gene could play a role as an oncogenic cofactor, in a “1 hit, 2 targets” genomic event. Comprehensive c-Myb and Ahi1 deregulation in cellular and animal models should be useful to investigate this issue.
The molecular characterization of the 6 TCRB-MYB cases that was performed in the present work showed that this translocation juxtaposed the Cβ enhancer in the vicinity of C-MYB and suggested transcriptional deregulation. In MYBdup cases, copy-number gain could be associated with reinforced C-MYB expression. Notably, the transcriptional consequences of the C-MYB genomic abnormalities appear unusual considering classical models of clearly ectopic oncogene expression, as seen in TLX1 and TLX3 cases for instance. Considering that C-MYB is expressed at high levels in normal thymus, it is likely that it is a strong and sustained, rather than ectopic, expression that may be oncogenic. It has been previously reported in mice that c-Myb levels are tightly regulated throughout hematopoietic differentiation, including during T-cell differentiation, with brief up and down level changes that regulate transition between differentiation stages and a final down-regulation.4,6–11 Abrogation of subtle levels of C-MYB into a stable massive expression due to TCRB regulatory sequences is likely to disturb progression through T-cell differentiation. This view is strongly supported by our finding that C-MYB expression was skewed massively toward single allele expression in the TCRB-MYB translocated cases, which suggests a cellular attempt to down-regulate C-MYB in these cases (Figure 4C). In the MYBdup cases, an increase of C-MYB expression related to copy-number gain may also reinforce oncogenic pathways. For both types of abnormality, in vitro and in vivo models should be useful to investigate the role of c-Myb in T-cell oncogenesis.
A number of published reports have shown that C-MYB knockdown by antisense inhibit the in vitro growth of leukemic cells, including T-cell lines.66–68 Although these experiments have to be confirmed in primary T-ALL samples using new silencing strategies, they suggest that C-MYB expression is necessary for the growth of leukemic T cells. Interestingly, C-MYB expression was recently involved in an oncogenic MLL → HOXA → C-MYB transcriptional pathway, which suggests that indirect biallelic stimulation of C-MYB could also be involved in oncogenic pathways.69
In conclusion, we report for the first time a recurrent somatic involvement of the C-MYB locus in human leukemia due to 2 distinct genomic events, TCRB-associated translocation and cryptic duplication of a short genome region, and leading to deregulated C-MYB expression. Moreover, a new T-ALL clinicobiologic entity has been here identified that associates a very young age, the t(6;7) TCRB-MYB translocation, no association with other common oncogenic transcripts, and a proliferation/mitosis expression signature. These results suggest that C-MYB could play an oncogenic role in T-ALL, and point to this gene as a potential target for therapeutic intervention in human malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
J.S. and F.S. were supported by INSERM and Paris 7 University, the Cancéropôle d'Ile-de-France, and a grant from the Ligue Nationale Contre le Cancer (Programme Cartes d'Identité des Tumeurs, CIT). The construction of the 4K array-CGH at the Curie Institute by A.A. and O.D. was supported by grants from the CIT (Ligue Nationale Contre le Cancer). W.C. was supported by a grant Médaille d'or de l'Internat CHU de Reims. W.A.D. and A.W.L. were supported by a grant from the Dutch Cancer Society (EMC 2002-2707). A.K. is supported by a grant from the NIH to A.M.G. (RO1 CA101859). A.M.G. is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation (DDCF).
We thank Lucie Hernandez, Claire Pichereau, Nathalie Rodriguez, Xavier Fund, Laurence Grollet, Yannick Fourne, Charles Decraenes, Marie-Françoise Auclerc, Jean-Pierre Kerckaert, Vahid Asnafi, Elizabeth Macintyre, and Hélène Cavé for helpful contributions. We are grateful to Anne Janin for support. We also thank Jean-Christophe Bories and Didier Auboeuf for critical reading of the paper and helpful comments.
Authorship
Contribution: E.C. performed experiments, analyzed the data, and wrote the paper; W.C. performed the array-CGH experiments; A.K. and A.C. performed experiments; J.-M.C. contributed to the molecular annotations of T-ALL cases; W.A.D., A.W.L., B.M., and B.N. characterized additional t(6;7) T-ALL cases; P.W. performed molecular combing experiments; O.D. and A.A. developed the 4K array; H.D., T.L., and A.B. managed the patients and conducted the French national trials GRAALL and FRALLE; A.M.G. contributed to data analysis; F.S. led the T-ALL project at Saint-Louis Hospital and performed the gene expression profiling analysis; J.S. designed the study, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean Soulier, Genome Rearrangements and Cancer Group, INSERM U728, Institut Universitaire d'Hématologie, Hôpital Saint-Louis, 1 Avenue Claude Vellefaux 75010 Paris, France; e-mail: jean.soulier@sls.aphp.fr.