Abstract
The mechanisms underlying the human fetal-to-adult β-globin gene switch remain to be determined. While there is substantial experimental evidence to suggest that promoter DNA methylation is involved in this process, most data come from studies in nonhuman systems. We have evaluated human γ- and β-globin promoter methylation in primary human fetal liver (FL) and adult bone marrow (ABM) erythroid cells. Our results show that, in general, promoter methylation and gene expression are inversely related. However, CpGs at −162 of the γ promoter and −126 of the β promoter are hypomethylated in ABM and FL, respectively. We also studied γ-globin promoter methylation during in vitro differentiation of erythroid cells. The γ promoters are initially hypermethylated in CD34+ cells. The upstream γ promoter CpGs become hypomethylated during the preerythroid phase of differentiation and are then remethylated later, during erythropoiesis. The period of promoter hypomethylation correlates with transient γ-globin gene expression and may explain the previously observed fetal hemoglobin production that occurs during early adult erythropoiesis. These results provide the first comprehensive survey of developmental changes in human γ- and β-globin promoter methylation and support the hypothesis that promoter methylation plays a role in human β-globin locus gene switching.
Introduction
The genes of the human β-globin locus undergo 2 well-described developmental switches in expression. The first switch involves silencing of the embryonic ϵ-globin gene and up-regulation of the 2 fetal γ-globin genes. The second switch occurs in the perinatal period as erythropoiesis moves from the fetal liver (FL) to the bone marrow. At this developmental stage, the γ genes are gradually silenced, whereas transcription of the δ- and β-globin genes increases to adult levels. The later switch is partially recapitulated during adult erythroid development, where some early precursor cells express the γ-globin genes and produce fetal hemoglobin (Hb).1 There is a strong rationale for understanding the human γ- to β-globin gene switch, as reversing it can provide therapeutic benefits to people with β hemoglobinopathies.2,3
Epigenetic mechanisms, including DNA methylation, have been associated with gene silencing and activation in a wide variety of experimental contexts. Such changes in chromatin structure are key steps in the binding of repressive or activating complexes to genetic regulatory elements.4 There is substantial data, primarily from model systems, implicating changes in globin locus DNA methylation with globin gene switching.5 Recent studies also show a correlation between gene expression and globin promoter methylation in baboons, which have a fetal-to-adult globin switch that is similar to the one seen in humans.6 Targeted deletion of the methyl-CpG binding protein MBD2 gene in transgenic mice carrying a human β-globin yeast artificial chromosome (YAC) delays γ-globin developmental silencing.7 Evidence that DNA methylation is involved in the human fetal-to-adult β-globin gene switch comes largely from clinical studies in which the DNA methyltransferase (DNMT) inhibitors 5-azacytidine (5-Aza) and 5-aza-2′-deoxycytidine (decitabine) were administered to β-hemoglobinopathy patients.8–11 These trials demonstrated activation of fetal globin gene expression, production of fetal Hb, and therapeutic benefit. This gene activation has been shown to correlate with hypomethylation of a specific CpG site located 53-bp upstream of the γ-globin transcriptional start site.12 Later work, using extracts from immortalized erythroid cell lines, demonstrated methylation-dependent binding of specific transcription factors at this site, including the stage-selector protein (SSP) complex.13,14 Further evidence for the role of DNA methylation in globin gene switching was provided by Perrine et al,15 who showed that the establishment of an adult methylation pattern across the locus was delayed in infants of diabetic mothers in whom the gene switch was also delayed. While these studies suggest that promoter CpG methylation plays an important role in the γ- to β-globin gene switch and in the reactivation of γ-globin expression by DNMT inhibitors, methylation of the human gene promoters has not been comprehensively studied, particularly in relation to developmental and differentiation-related erythropoiesis. In the early 1980s, 2 groups used methylation-sensitive restriction endonucleases (MSREs) to evaluate β-globin locus DNA methylation in primary human erythroid cells.16,17 While these studies showed developmental changes in locus methylation patterns, only 1 γ-promoter CpG and no β-promoter sites were evaluated because most CpGs are not associated with MSRE sites.
We have several reasons for studying γ- and β-globin promoter methylation. The first is to test the hypothesis that methylation of the promoter CpGs is inversely related to globin gene expression. While data from chicken and baboon systems suggest that this is likely, it has yet to be demonstrated in primary human cells. This information will be important in helping define the mechanisms by which DNMT inhibitors activate globin gene expression, as different models propose that this effect is the result of either demethylation of promoter DNA or of changes in the kinetics of erythroid differentiation.18 A third reason to investigate changes in globin promoter methylation during differentiation and development is that methylation at specific CpG residues may be differentially regulated. Such a finding would imply targeted regulation of methylation at specific sites, in contrast to a general regulation of methylation across larger domains. Similarly, it would be of interest to know whether the methylation status of specific CpGs correlates to gene expression, as is the case with the one at −53 of the γ-globin promoter. If this is found, then these other CpGs may also be sites of differential transcription factor binding. A fourth reason is that most studies of promoter-related DNA methylation have examined promoters that contain CpG islands. These islands have been defined as regions of DNA of at least 200 bp with at least 50% C+G content and an actual/predicted CpG content ratio of greater than 0.6.19 Neither the γ- or β-globin promoters come close to fitting this definition and may therefore have alternative methyl-CpG–dependent mechanisms of gene regulation. Finally, while significant advances are being made in understanding the regulation of globin gene expression in model systems, it is likely that before these advances can be translated into therapies for people with hemoglobinopathies, we will need to verify findings and therapeutic approaches in primary human cells. This study provides a baseline for understanding the role of DNA methylation in globin gene switching.
To investigate the role of DNA methylation in the human fetal-to-adult globin switch in humans, we have used the bisulfite conversion method to determine the methylation status of specific CpGs in the regions of the γ- and β-globin gene promoters in purified fetal liver and adult bone marrow (ABM) erythroid cells and during in vitro differentiation of adult erythroid cells. We have also studied the methylation of the promoters in fetal hepatocytes and human peripheral blood CD34+ cells, the expression of DNMT genes during differentiation, and the patterns of globin promoter histone acetylation. These studies provide the first comprehensive description of the developmental and differentiation-specific patterns of DNA methylation in human γ- and β-globin gene promoters in primary human cells and provide insights into the regulation of these clinically important genes.
Materials and methods
Tissue samples
Fetal liver samples and adult CD34+ peripheral blood progenitor cells were obtained using a protocol approved by the University of Washington Medical School Institutional Review Board (IRB). Human bone marrow samples were obtained using a protocol approved by the Dartmouth Medical School IRB. All donors gave informed consent in accordance with the Declaration of Helsinki. ABM samples were obtained from healthy donors at Dartmouth-Hitchcock Medical Center. FL samples ranging in age from 80 days to 110 days were provided by the Birth Defects Research Laboratory of the University of Washington Medical School. Peripheral blood CD34+ cells were obtained from the National Heart, Lung, and Blood Institute (NHLBI) Programs of Excellence for Gene Therapy Hematopoietic Cell Processing Core at the University of Washington.
Purification of primary erythroid cells
Fetal livers were shipped by overnight express mail in DMEM + 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mM l-glutamine supplemented with erythropoietin at 5 units per mL. Single-cell suspensions were made by gently compressing the tissue between 2 frosted glass slides in cold PBS. BM samples were obtained from healthy donors. Heparin was present in the aspiration syringe and the sample was immediately put into a heparinized blood collection tube at room temperature. ABM samples were processed within 1 hour of collection. Cell suspensions of FL or BM were diluted 2-fold in room-temperature PBS. Fifteen milliliters of suspension was laid over 25 mL room-temperature Ficoll-Paque PLUS (Sigma, St Louis, MO) in 50-mL conical tubes. Cells were then centrifuged at 1800 rpm for 45 minutes at 18°C. Enucleated erythrocytes and fetal hepatocytes were pelleted by this procedure. For analysis of hepatocyte DNA methylation, erythrocytes were lysed in hypotonic saline and hepatocytes were then washed and their DNA isolated. The interphase layers, containing nucleated hematopoietic cells, were then pooled and the samples were washed twice in 50 mL PBS/EDTA. Cells were then resuspended in PBS/EDTA at 80 μL per 1 × 107 cells, and glycophorin-A human microbeads (Miltenyi Biotec, Auburn, CA) were added at 20 μL per 1 × 107 cells. Cells were incubated at 6°C for 15 minutes, then washed twice in PBS/EDTA, and brought to a final volume of 500 μL PBS/EDTA/BSA per 1 × 108 cells. A Miltenyi LS Column and magnet were then used to isolate glycophorin-A-positive erythroid cells per the manufacturer's protocol. Purity of the isolated cells was analyzed by flow cytometry using anti-glycophorin-A antibody and by Wright-Giemsa staining of cytospin preparations. Microscopy was performed using an Olympus model CHT light microscope (Olympus Optical, Tokyo, Japan). Magnification of Figure 1A was 100× (10×/10×/0.25 NA) and Figure 6A was 400× (10×/40×/0.65 NA). Images were acquired with a FujiFilm FinePix A345 digital camera and visualized with Fujifilm FinePix Viewer 4.3 (Fujifilm USA, Valhalla, NY). Images were processed using iPhoto version 6.0.6 (Apple Computer, Cupertino, CA).
Quantitative RT-PCR and Hb HPLC
Total RNA was isolated from purified erythroid cells using an RNeasy mini kit (Qiagen, Valencia, CA) with on-column DNase digestion. Reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using a GeneAmp RNA PCR kit (Roche, Branchburg, NJ). Two microliters RT-PCR product was used in each real-time reaction using iQ Sybr Green Super mix (Bio-Rad, Hercules, CA). Levels of globin mRNA were calculated by the method of Larionov et al20 using β-actin and G3PD as normalizing genes. Levels of DNMT mRNA were normalized to those found on day 3 of in vitro differentiation. The primers used for RT-PCR analysis are shown in Table 1. High-performance liquid chromatography (HPLC) Hb analysis was performed on total cell lysates as described by Ou and Rognerud21 using a PolyCAT A cation exchange column (The Nest Group, Southboro, MA).
Bisulfite sequencing
Bisulfite modification was performed on genomic DNA using the CpGenome Fast DNA Modification Kit (Chemicon, Temecula, CA). The γ-globin and β-globin promoter regions were amplified by PCR using seminested primers to amplify both regions. For the γ-globin promoter, the initial round of PCR was performed using primers Gf (5′-GGTATATGTTTTAATTTTAAATTATAG-3′) and Gr (5′-CAAATTACCAAAACTATCAAAAAACC-3′). The second round used primers Gf and Gr2 (5′-CAAAACTATCAAAAAACCTCTAAA-3′). These primer sets are complimentary to both the γG and γA promoter sequences. Because there are differences in the sequences between the primers, the DNA from each sequencing reaction could be assigned to the appropriate promoter. For the β-globin promoter, the initial round of PCR was performed using primers Bf (5′-ATTAGAAGGTTTTAATTTAAATAAGGA-3′) and Br (5′-ACCCAATTTCTATTAATCTCCTTAAAC- 3′). The second round used primers Bf and Br2 (5′-ACCTTAATACCAACCTACCCAAAAC-3′). PCR products were cloned into the Topo TA pCR2.1 vector (Invitrogen, Carlsbad, CA). Sequence analysis of 6 to 20 clones from each sample was performed using Prism 300 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Acetylated histone H3 ChIP
For chromatin immunoprecipitation (ChIP) of primary erythroid cells, 1 × 106 to 3 × 106 cells per IP were suspended in 90 mL cold PBS plus Complete Mini Protease Inhibitors (Roche). One milliliter 37% formaldehyde was added and the suspension shaken gently at room temperature for 15 minutes. The cross-linking reaction was stopped with the addition of 5 mL of 2.5 M glycine for 5 minutes. Cells were then washed 2 times at 4°C with PBS plus protease inhibitors. The remainder of the ChIP was then performed using the Upstate ChIP Assay Kit and protocol (Lake Placid, NY). Anti-acetyl-histone H3 antibody (Upstate) and, as a negative control, AffiniPure rabbit antirat IgG (Jackson ImmunoResearch, West Grove, PA) were used in immunoprecipitation reactions. Immunoprecipitation of γ- and β-globin promoter regions was analyzed using quantitative real-time PCR. For each set of primers and for each experiment, an input DNA standard curve was constructed. The amount of target DNA in each immunoprecipitate was quantified using the standard curve. For each tissue sample, 2 to 3 precipitations were performed and each precipitate was then analyzed in triplicate PCR reactions. The positive control H3 hyperacetylation was the globin LCR HS3 core. The negative control for H3 hyperacetylation was the human necdin gene promoter (NDN). Primer sequences are as follows: HS3-F (5′-ATAGACCATGAGTAGAGGGCAGAC-3′), HS3-R (5′ TGATCCTGAAAACATAGGAGTCAA-3′), NDN-F (5′GTGTTATGTGCGTGCAAACC-3′), NDN-R (5′-CTCTTCCCGGGTTTCTTCTC-3′), GammaF (5′-GGCCTCACTGGAGCTACAGACAAG-3′), GammaR (5′-GAAATGACCCATGGGGTCTG-3′), BetaF (5′-TGGTATGGGGCCAAGAGATA-3′), and BetaR (5′-TGCTCCTGGGAGTAGATTGG-3′).
In vitro erythroid differentiation
Our in vitro erythroid differentiation system was derived from the previously reported methods of Fibach and Rachmilewitz22 and Wojda et al.23 Erythroid cells were generated by culturing purified CD34+ peripheral blood stem cells in IMDM supplemented with 100 ng/mL rhSCF, 100 ng/mL rhFlt-3 ligand, and 50 ng/mL rhIL-3 (cytokines from R&D Systems, Minneapolis, MN) for 7 days, followed by growth in DMEM supplemented with 1% BSA, 30% FBS (Gibco, Carlsbad, CA), 1 μM dexamethasone, 10 μM BME, 0.3% wt/vol transferrin, and 4 U/mL Epo (Ortho Biotech, Bridgewater, NJ) for 12 to 14 days.
Results
Characterization of primary erythroid cells
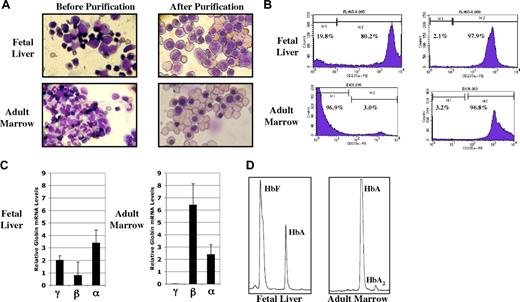
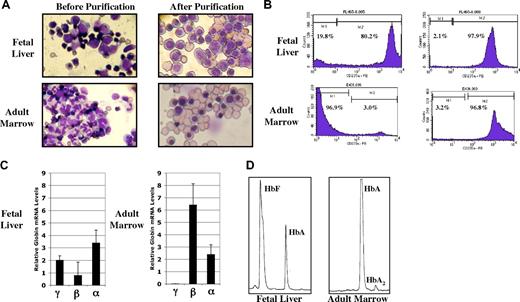
Our initial experiments were designed to determine the methylation status of the human γ- and β-globin gene promoters in purified populations of fetal and adult primary erythroid cells. FL and ABM samples were obtained using IRB-approved protocols and informed consent documents. Single-cell suspensions of each tissue were first processed by centrifugation over ficol-hypaque to remove mature red blood cells and fetal hepatocytes. Anti-glycophorin-A (CD235a)–conjugated magnetic beads were then used to purify erythroid cells. Results from a typical purification experiment are shown in Figure 1 and Table 2. Photomicrographs of fetal liver and adult marrow cells before and after this purification procedure are shown in Figure 1A. Counting by light microscopy showed that the purified nucleated cells were 95% to 100% erythroid. While enucleated erythroid cells are present in the final populations, these cells do not contribute to, or interfere with, the analysis of DNA methylation patterns in the nucleated erythroid cells. This level of purity was confirmed by flow cytometry using anti-CD235a antibody, as shown in Figure 1B. To further characterize the populations of purified erythroid cells, we performed quantitative RT-PCR analysis to determine the relative steady-state expression levels of the γ-, β-, and α-globin genes. These results are presented in Figure 1C. While α-globin gene expression is similar in erythroid cells from both tissues, β-globin gene expression is nearly 10-fold higher in the adult marrow cells. γ-Globin expression is more than 3 times that seen for β-globin in fetal liver cells but is only detectable in adult marrow cells at levels below those shown in Figure 1C. Finally, as shown in Figure 1D and Table 2, we performed HPLC analysis to characterize the production of Hb in our 2 cell sources. Both HbF and HbA were detected at significant proportions in lysates from fetal liver erythroid cells. As expected, HbA was the predominant Hb in adult cells. While β-globin gene expression and HbA are generally thought to be associated with adult erythropoiesis, the low-level production of β-globin mRNA and HbA we found in fetal liver cells is in agreement with previous reports.24,25 These results demonstrate that we have produced purified populations of fetal and adult erythroid cells that exhibit expected patterns of globin gene expression and Hb production.
Characterization of purified fetal and adult nucleated erythroid cells. (A) Wright-Giemsa–stained fetal liver and adult marrow cells before and after purification by ficol density gradient and anti-glycophorin-A–conjugated magnetic beads. (B) Percentage of glycophorin-A (CD235a)–positive cells before and after purification. (C) Relative levels of globin gene mRNA in purified cells from fetal and adult erythroid cells determined by quantitative RT-PCR. Results are normalized to the geometric mean of β-actin and G3PD mRNAs as described in “Materials and methods; Quantitative RT-PCR and Hb HPLC.” Values are plus or minus 1 standard deviation. (D) Hb HPLC patterns from purified fetal liver and adult erythroid cells.
Characterization of purified fetal and adult nucleated erythroid cells. (A) Wright-Giemsa–stained fetal liver and adult marrow cells before and after purification by ficol density gradient and anti-glycophorin-A–conjugated magnetic beads. (B) Percentage of glycophorin-A (CD235a)–positive cells before and after purification. (C) Relative levels of globin gene mRNA in purified cells from fetal and adult erythroid cells determined by quantitative RT-PCR. Results are normalized to the geometric mean of β-actin and G3PD mRNAs as described in “Materials and methods; Quantitative RT-PCR and Hb HPLC.” Values are plus or minus 1 standard deviation. (D) Hb HPLC patterns from purified fetal liver and adult erythroid cells.
γ- and β-globin promoter methylation in fetal and adult erythroid cells
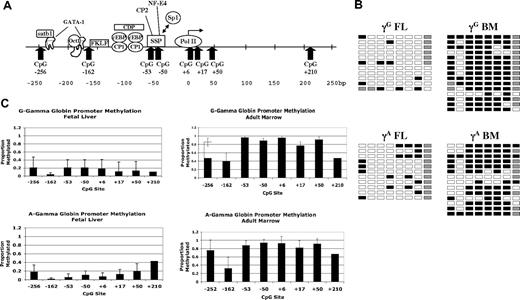
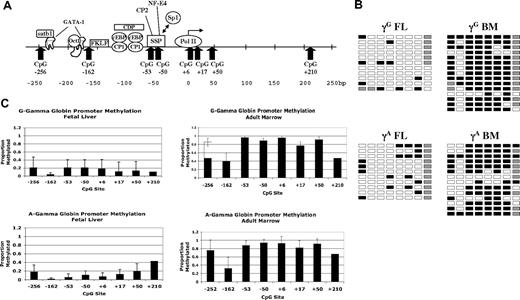
We next investigated the methylation status of CpGs near the γ- and β-globin promoters using the bisulfite conversion method. In contrast to previous studies of β-globin locus methylation patterns in primary human cells that used methylation-sensitive restriction endonucleases, this method allows analysis of all potential methylation sites within a given region. We used tissue samples from at least 5 individual donors for each promoter analysis. Results of DNA methylation studies for the 2 γ-globin promoters are presented in Figure 2. Figure 2A shows the location of the 8 CpGs that were evaluated. These sites are contained in an approximately 500-bp region centered on the transcriptional start site. Also shown are the binding locations of several transcription factors known to bind the γ-globin gene promoter. Figure 2B shows representative primary data for the γ-globin genes in fetal liver and adult marrow erythroid cells; the columns of boxes correspond to the CpG sites shown in Figure 2A, and the rows of boxes represent sequences of individual clones following bisulfite treatment. Figure 2C shows the average proportions of methylated CpGs at each site for all tissue samples analyzed. Five to 7 individual tissue samples were used for each promoter analysis. Seven to 20 individual sequences were analyzed for each promoter in each tissue sample. The CpG at +210 of the γ promoters was not readable in many of our sequencing runs because of its distance from the sequencing primer. We therefore pooled the data from all samples for this site for Figure 2C. Our results indicate that the 2 γ-globin promoters are generally hypomethylated in fetal erythroid cells and are hypermethylated in adult erythroid cells. The one exception to this is the CpG at −162, which is significantly less methylated in adult cells (P = .002) compared with the other sites. This CpG is adjacent to the −158 (C/T) hereditary persistence of fetal hemoglobin (HPFH) mutation, raising the possibility that this differential methylation pattern is involved in gene regulation. In agreement with earlier studies, methylation of the −53 CpG is inversely correlated with γ-globin expression.16,17
Methylation of γ-globin promoter sequences. (A) Diagram of the human γ-globin promoter regions showing the locations of binding sites for transcription factors and CpG dinucleotides. Adapted from Stamatoyannopoulos and Grosveld.26 (B) Representative methylation data for the 2 γ-globin promoters in fetal and adult erythroid cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing. (C) Summary methylation data from multiple independent tissue samples. Error bars indicate plus or minus 1 standard deviation.
Methylation of γ-globin promoter sequences. (A) Diagram of the human γ-globin promoter regions showing the locations of binding sites for transcription factors and CpG dinucleotides. Adapted from Stamatoyannopoulos and Grosveld.26 (B) Representative methylation data for the 2 γ-globin promoters in fetal and adult erythroid cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing. (C) Summary methylation data from multiple independent tissue samples. Error bars indicate plus or minus 1 standard deviation.
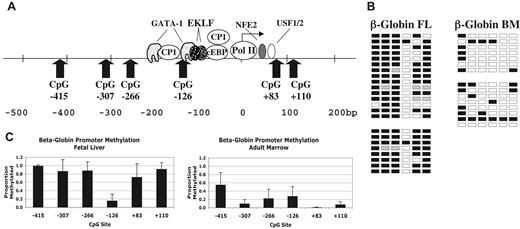
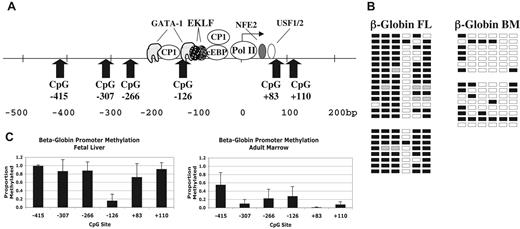
Results for the β-globin promoter are shown in Figure 3. Figure 3A shows the locations of the 6 CpG sites evaluated between −415 and +110 of the β-globin gene. Figure 3B shows representative primary data from 2 individual fetal liver and 2 adult marrow samples. Figure 3C shows the average methylation for 5 FL and 5 ABM samples. Here too, overall promoter methylation correlates inversely with gene expression. However, in FL erythroid cells, the CpG at −126 is significantly hypomethylated (P < .001) compared with surrounding CpGs. This CpG is adjacent to a GATA-1 binding site at −120 and is near sites for CP1 and EKLF (Figure 3A).27,28 In adult marrow, the CpG at −415 is relatively hypermethylated compared with the other sites in this setting.
Methylation of β-globin promoter sequences. (A) Diagram of the human β-globin promoter region showing the locations of binding sites for transcription factors and CpG dinucleotides. Adapted from Stamatoyannopoulos and Grosveld.26 (B) Representative methylation data for the β-globin promoter in fetal and adult erythroid cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing. (C) Summary methylation data from multiple independent tissue samples. Error bars indicate plus or minus 1 standard deviation.
Methylation of β-globin promoter sequences. (A) Diagram of the human β-globin promoter region showing the locations of binding sites for transcription factors and CpG dinucleotides. Adapted from Stamatoyannopoulos and Grosveld.26 (B) Representative methylation data for the β-globin promoter in fetal and adult erythroid cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing. (C) Summary methylation data from multiple independent tissue samples. Error bars indicate plus or minus 1 standard deviation.
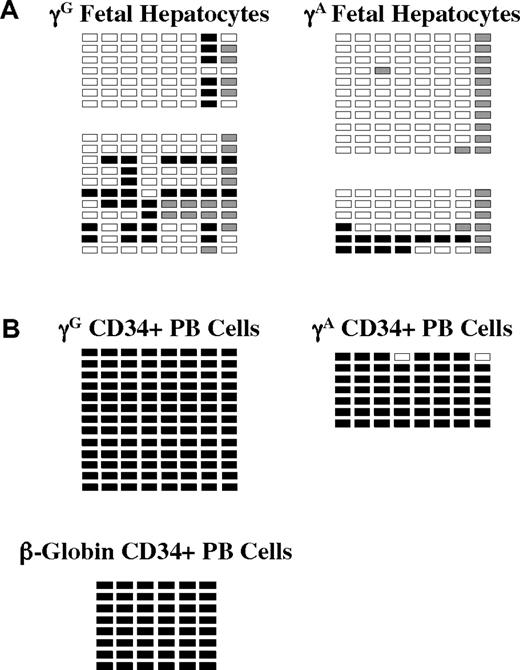
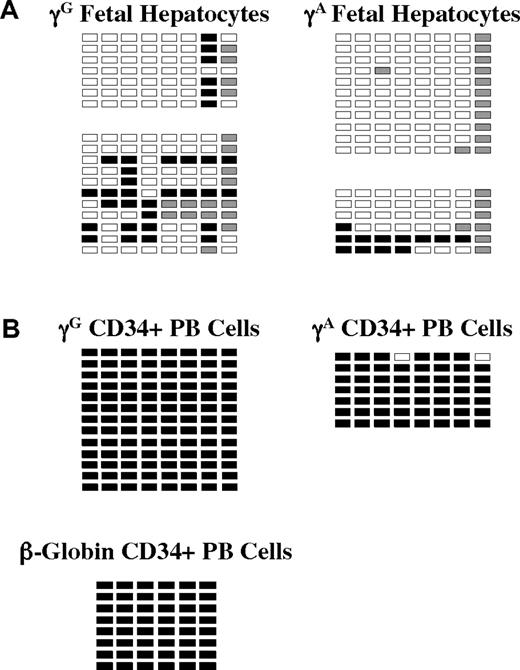
We also studied globin promoter methylation in purified fetal hepatocytes from 2 specimens and in G-CSF–mobilized peripheral blood CD34+ cells from one healthy donor. These results are shown in Figure 4. Despite the fact that the globin genes are not expressed in either of these nonerythroid cell types, they exhibit dramatically different patterns of promoter methylation. In fetal hepatocytes, all γ-globin promoter CpGs were hypomethylated (< 20% Me-CpG), whereas in CD34+ cells, γ- and β-promoter CpGs were greater than 99% methylated. These results may imply different mechanisms of globin gene suppression in the 2 cell types.
Methylation of γ- and β-globin promoter sequences in fetal hepatocytes and adult CD34+ peripheral blood hematopoietic cells. (A) Methylation of the 2 γ-globin promoters and the β-globin promoter in purified fetal hepatocytes. Data from 2 independent samples are shown. (B) Methylation of the γ- and β-globin promoters in CD34+ peripheral blood cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing.
Methylation of γ- and β-globin promoter sequences in fetal hepatocytes and adult CD34+ peripheral blood hematopoietic cells. (A) Methylation of the 2 γ-globin promoters and the β-globin promoter in purified fetal hepatocytes. Data from 2 independent samples are shown. (B) Methylation of the γ- and β-globin promoters in CD34+ peripheral blood cells. □ indicates unmethylated; ■, methylated; and ▩, indeterminate sequencing.
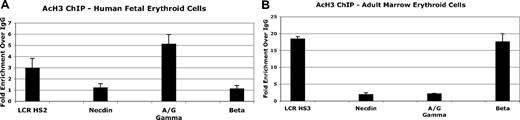
Because low-level β-globin mRNA is detectable in FL cells despite promoter hypermethylation, we studied the general histone H3 acetylation status at the γ- and β-globin promoters in FL and ABM by ChIP assay. These results are shown in Figure 5. Using LCR HSs as internal positive controls and the necdin gene promoter as a negative control, we found hyperacetylation of γ-promoter histones in the FL and β-promoter-associated histones in the adult erythroid cells. Hyperacetylation of the β-globin promoter was not seen in fetal cells despite detectible gene expression. These results are consistent with those recently reported for baboon FL and ABM by Lavelle et al.6
Histone H3 acetylation at the γ- and β-globin promoters in purified fetal and adult erythroid cells determined by ChIP assay. (A) Relative promoter H3 acetylation in nucleated fetal erythroid cells. (B) Relative promoter H3 acetylation in nucleated adult erythroid cells. Results are expressed as fold enrichment using anti-AcH3 antibody compared with nonspecific IgG. Error bars indicate plus or minus 1 standard deviation. β-Globin LCR HS2 and HS3 core regions serve as positive controls. The necdin gene promoter, which is not expressed in erythroid cells, serves a negative control.
Histone H3 acetylation at the γ- and β-globin promoters in purified fetal and adult erythroid cells determined by ChIP assay. (A) Relative promoter H3 acetylation in nucleated fetal erythroid cells. (B) Relative promoter H3 acetylation in nucleated adult erythroid cells. Results are expressed as fold enrichment using anti-AcH3 antibody compared with nonspecific IgG. Error bars indicate plus or minus 1 standard deviation. β-Globin LCR HS2 and HS3 core regions serve as positive controls. The necdin gene promoter, which is not expressed in erythroid cells, serves a negative control.
The γ-globin promoters undergo transient demethylation during adult erythroid differentiation
It has been previously observed that the human γ-globin genes are expressed early in erythroid differentiation and are then silenced later in differentiation when β-globin gene expression predominates.1 We have shown that the γ-globin gene promoters are highly methylated in multi-potential CD34+ cells and in adult primary erythroid cells. This led us to hypothesize that the γ-globin expression seen during early differentiation is, at least in part, the result of a transient demethylation of the γ promoters. The alternative to this is that the observed γ-globin expression occurs despite promoter hypermethylation. We are also interested in this process because early fetal gene expression is an important component of the 2 major models of how DNMT inhibitors activate fetal globin gene expression in hemoglobinopathy patients.18 In the differentiation kinetics model, 5-Aza and other cytotoxic agents are thought to speed the differentiation process, allowing for increased γ-globin gene expression.29,30 In the demethylation model, it is proposed that γ-globin expression during differentiation is enhanced by a nonspecific demethylation of genomic DNA, including the γ-globin promoters.
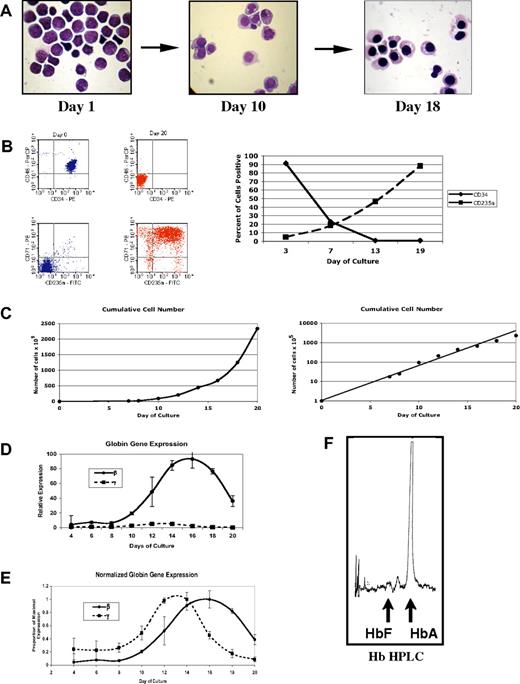
To investigate how γ-globin promoter methylation changes during erythroid differentiation, we have used an in vitro differentiation system modified from the previously reported methods of Fibach and Rachmilewitz22 and Wojda et al.23 CD34+ peripheral blood cells were cultured with stem cell factor (SCF), Flt3 ligand, and IL-3 for 7 days. These cytokines were then removed and erythropoietin (Epo) was added for an additional 12 to 14 days of culture. As shown in Figure 6, this results in a pure population of hemoglobinized erythroid cells with condensed nuclei (Figure 6A), loss of the CD34 marker and expression of glycophorin A (Figure 6B), and an exponential expansion of erythroid cells (Figure 6C). When globin gene expression was examined in the differentiating cells using quantitative RT-PCR, the expected early expression of γ-globin was seen. γ-Globin mRNA levels were found to be much lower than for β-globin (Figure 6D) and peaked on approximately day 13 compared with β-globin mRNA, which peaked at day 16 (Figure 6E). Thus, this system recreated the low-level, early γ-globin gene expression seen in vivo. By the end of the culture period, globin gene expression had decreased to baseline levels. Finally, we performed HPLC on lysates from cells at the end of the culture period to determine the relative levels of HbA and HbF. As shown in Figure 6F, nearly all of the Hb produced in this culture system is HbA, again reproducing what is seen in vivo.
In vitro erythroid differentiation from purified human CD34+ peripheral blood hematopoietic cells. (A) Wright-Giemsa–stained cytospin samples during in vitro differentiation. (B) Flow cytometric characterization of cells during differentiation. Scatter plots show CD45/CD34 staining and CD71 (transferrin)/CD235 (glycophorin A) at start and completion of differentiation. Graph shows the changing patterns of CD34 and CD235a cell surface expression during in vitro differentiation. (C) Expansion of cells during in vitro differentiation. (D) Relative levels of γ- and β-globin mRNA during in vitro differentiation as determined by quantitative RT-PCR. Values are normalized to the geometric mean of β-actin and G3PD mRNAs as described in “Materials and methods; Quantitative RT-PCR and Hb HPLC.” (E) Patterns of steady-state γ- and β-globin mRNA during differentiation. Values are expressed as the proportion of maximal expression to highlight the time course of expression. (F) Hb HPLC performed on cellular extracts at the end of the culture period.
In vitro erythroid differentiation from purified human CD34+ peripheral blood hematopoietic cells. (A) Wright-Giemsa–stained cytospin samples during in vitro differentiation. (B) Flow cytometric characterization of cells during differentiation. Scatter plots show CD45/CD34 staining and CD71 (transferrin)/CD235 (glycophorin A) at start and completion of differentiation. Graph shows the changing patterns of CD34 and CD235a cell surface expression during in vitro differentiation. (C) Expansion of cells during in vitro differentiation. (D) Relative levels of γ- and β-globin mRNA during in vitro differentiation as determined by quantitative RT-PCR. Values are normalized to the geometric mean of β-actin and G3PD mRNAs as described in “Materials and methods; Quantitative RT-PCR and Hb HPLC.” (E) Patterns of steady-state γ- and β-globin mRNA during differentiation. Values are expressed as the proportion of maximal expression to highlight the time course of expression. (F) Hb HPLC performed on cellular extracts at the end of the culture period.
Using this in vitro differentiation system, we next evaluated γ-globin promoter methylation during erythroid differentiation. Because our previous results showed that the γG and γA promoters had equivalent patterns of methylation, we combined data from the 2 promoters for this experiment. We also focused on the 7 CpGs between −256 and +50 for this experiment. As shown in Figure 7A, all of the γ-globin promoter CpGs are highly methylated on day 0 of differentiation. By day +3, there is a decrease in methylation at most of the sites, but this is more pronounced at the CpGs located upstream of the transcriptional start site. This upstream hypomethylation persists through day +10 of differentiation but is then lost by day +14. The −53 CpG exhibits the most pronounced hypomethylation, falling to approximately 20%. Because of this differential methylation, we compared methylation at the upstream and downstream sites during differentiation (Figure 7B). The upstream promoters show significantly less methylation at days +3 and +10 but then return to the highly methylated state seen in adult nucleated erythroid cells by day +14.
Changing patterns of γ-globin promoter methylation during in vitro differentiation. (A) Percentage methylation of specific CpGs in the γ-globin promoters during in vitro differentiation. (B) Comparison of percentage methylation of the 3 CpGs upstream of the transcriptional start site to that of the 3 CpGs downstream of the start site. (C) Relative mRNA levels of DNMT genes during in vitro differentiation. Error bars indicate plus or minus 1 standard deviation.
Changing patterns of γ-globin promoter methylation during in vitro differentiation. (A) Percentage methylation of specific CpGs in the γ-globin promoters during in vitro differentiation. (B) Comparison of percentage methylation of the 3 CpGs upstream of the transcriptional start site to that of the 3 CpGs downstream of the start site. (C) Relative mRNA levels of DNMT genes during in vitro differentiation. Error bars indicate plus or minus 1 standard deviation.
To investigate the mechanism underlying these changing patterns of γ-globin promoter CpG methylation during differentiation, we performed RT-PCR to characterize expression of the DNMT genes during development. Amounts of mRNA for each gene were normalized to the day +3 values. As shown in Figure 7C, there is a pronounced increase in expression of the maintenance methyltransferase DNMT1 at day +5 of differentiation. DNMT3a, DNMT3b, and actin genes are increased to a lesser extent at this point. There is also a second increase in the expression of these genes later in differentiation. The initial increase occurs before the induction of erythroid differentiation by Epo addition on day +7, whereas the second peak is seen during erythroid differentiation.
Discussion
A major goal of the hemoglobinopathy field is to understand the mechanisms underlying γ- to β-globin developmental gene switching so that clinical strategies to reverse γ gene silencing can be developed. Despite progress in model systems, relatively little is known about the molecular events underlying this process in primary human erythroid cells. Investigation of switching in primary human cells is important because these are the cells that will be manipulated in the clinical setting and because there are significant differences in globin gene switching between humans and model systems. For example, neither chickens nor mice exhibit the fetal-to-adult β-like globin gene switch. Even baboons, which do have this switch, may have subtle differences in fetal globin gene regulation, as they exhibit a more robust activation of fetal globin gene expression following the induction of anemia31 than is typically seen in humans. Our goal is to investigate the chromatin-based mechanisms underlying human globin gene switching in their natural context. In the current work, we have focused on characterizing the changes in promoter methylation that are associated with differential γ- and β-globin gene expression.
Patterns of γ and β-globin promoter DNA methylation during development
We have shown that the overall methylation patterns of the γ- and β-globin promoter regions are inversely related to gene expression. These data are consistent with the observation that promoter methylation is generally associated with gene silencing and the hypothesis that promoter methylation plays a role in the regulation of these genes. The methylation status of the −53 CpG of the γ promoter has been previously determined in fetal and adult erythroid cells. The presence or absence of methylation at this site was later shown to mediate differential transcription factor binding.13,14 Our findings that the methylation states of several other γ- and β-globin promoter CpGs also correlate with gene expression and are near known transcription factor binding sites makes these potential targets for additional differential transcription factor binding.
While an inverse correlation between DNA methylation and expression of the γ- and β-globin genes exists, it is not a simple on/off relationship. For example, while the γ genes are not expressed in CD34+, adult erythroid, or fetal liver cells, the methylation patterns are strikingly different between the hematopoietic and liver cells. Also, while the β-globin promoter is generally hypermethylated (and the histones relatively hyperacetylated) in fetal erythroid cells, there is a relatively high level of gene expression, as roughly 30% of β-like globin mRNA is from the β-globin gene. Of note is that the −126 CpG of the β promoter is hypomethylated in both FL and ABM. It may be that positive-acting factors are able to bind this site in both cell types but that FL-specific hypermethylation of the other promoter CpGs results in the loss of additional positive-acting factors or the recruitment of repressive factors. The result of these changes is that β-globin gene expression is only about 10% that seen in ABM erythroid cells, thereby generating the high-γ/low-β expression pattern of FL.
Our finding that the γ-globin promoters are hypomethylated in fetal hepatocytes but are hypermethylated in adult erythroid cells implies that different mechanisms produce the lack of expression of these genes in the 2 different cell types. One model is that a simple absence of positive transcription factors is sufficient to produce a lack of γ-globin gene expression in nonerythroid cells such as the fetal hepatocytes. Whereas, in adult erythroid cells, an environment that contains a large complement of erythroid and hematopoietic transcription factors and requires the expression of other similarly regulated genes, mechanisms for specifically targeting DNA methylation to the fetal globin gene promoters may have evolved. If this model plays a role in γ-globin developmental silencing, then determining how developmentally specific patterns of methylation are achieved will be key to understanding the switching process. In support of this model, we have recently shown that ectopic expression of GATA-1 protein in the nonerythroid Hela cell line is sufficient to mediate the expression of several erythroid genes, including those for γ- and β-globin.32
Our results also raise questions about how domains of differential CpG methylation are established around the globin gene promoters at different developmental stages, how far these domains extend beyond the regions we have studied, and how, despite the overall pattern of promoter DNA methylation, there are 2 CpGs that show patterns substantially different from adjacent sites. These issues are not unique to the globin locus and are being investigated in other experimental contexts.33
Promoter methylation and γ-globin gene expression during adult erythropoiesis
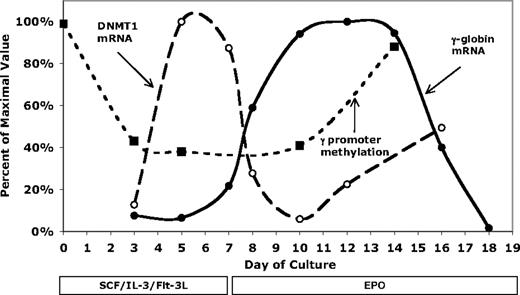
Our investigation of how γ-globin promoter methylation changes during differentiation showed that in pluripotent CD34+ hematopoietic precursor cells, the γ-globin promoter regions are highly methylated. We were interested in determining whether this high level of methylation would decrease during differentiation, providing a potential explanation for the observation of γ-globin gene expression early in adult erythropoiesis. Consistent with this hypothesis, we observed a transient decrease in upstream γ-globin promoter methylation (Figure 7). To get a better picture of the temporal changes involved in this process, we have combined γ-globin and DNMT1 gene expression and upstream γ-globin promoter DNA methylation data in Figure 8. As shown in this figure, γ-globin promoter methylation is dramatically decreased by day +3 of culture, well before the erythroid phase of differentiation. By day 7, when the cytokines are changed to Epo, γ-globin mRNA and glycophorin-A protein (Figure 6) are beginning to rise. Levels of γ-globin mRNA then increase during a period when the promoter is hypomethylated (days 7-9). Promoter methylation begins to increase after day 10, a period associated with leveling off and then falling γ-globin mRNA levels. This time course suggests that demethylation of the promoter early in differentiation is necessary for γ-globin gene expression and that remethylation of the promoter late in differentiation is associated with loss of expression. At least 2 observations suggest that a specific process transiently protects the upstream promoter from DNMT1 (the primary maintenance methyltransferase) mediated methylation during differentiation. The first is that downstream CpG sites remain highly methylated throughout differentiation (Figure 7). The second is that the upstream promoter remains hypomethylated during a time of peak DNMT1 expression (Figure 8). If there is a specific mechanism responsible for this effect, it is interesting to speculate that low-level γ-globin expression during early adult erythropoiesis may serve a physiologic purpose and is not just an artifact of evolution.
Comparison of γ-globin mRNA, DNMT1 mRNA, and upstream γ-globin promoter methylation levels during in vitro differentiation. The maximum levels of γ-globin and DNMT1 mRNA were set to 100%. DNA methylation is the percentage methylation of the upstream (−162, −53, and −50) CpGs and is not normalized.
Comparison of γ-globin mRNA, DNMT1 mRNA, and upstream γ-globin promoter methylation levels during in vitro differentiation. The maximum levels of γ-globin and DNMT1 mRNA were set to 100%. DNA methylation is the percentage methylation of the upstream (−162, −53, and −50) CpGs and is not normalized.
The role of promoter methylation in globin gene switching and γ-globin reactivation
Our experiments have provided baseline data on the changes in human γ- and β-globin promoter methylation that occur during erythroid development and differentiation. These results are consistent with a mechanistic connection between promoter DNA methylation and differential expression of the human γ- and β-globin genes during differentiation. The transient modification of γ-globin promoter methylation we have observed is likely to be relevant to understanding how inducers of fetal Hb work. In fact, it may be that instead of “inducing” or “reactivating” fetal globin gene expression, these agents work by prolonging γ-globin gene expression into the later phases of differentiation. For example, DNMT inhibitors may prevent the remethylation and subsequent silencing of the γ promoter. Similarly, HDAC inhibitors may act to prevent deacetylation of the γ promoter at the time of silencing. While these hypotheses will need to be tested, they offer a new way of thinking about the mechanisms of fetal Hb inducers. However, this still does not provide an explanation for how other cytotoxic agents such as HU and cytosine arabinoside are able to induce fetal Hb production. It is possible that these cytotoxic agents work by different mechanisms and that promoter demethylation is only one facet of induction. Understanding the molecular mechanisms by which the currently known fetal Hb-inducing agents function is likely to aid in the development of future targeted strategies for the therapeutic induction of fetal Hb.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Birth Defects Research Laboratory of the University of Washington Medical School for providing fetal liver samples, the NHLBI Programs of Excellence for Gene Therapy Hematopoietic Cell Processing Core at the University of Washington for providing peripheral blood CD34+ cells, and Dr Lionel Lewis and Bernie Beaulieu for assistance with Hb HPLC.
This work was supported by National Institutes of Health (NIH) grants HL52243 and HL73442 (C.H.L.) and HL73431 (S.F.) and from funding provided by the Knights of the York Cross of Honour (C.H.L.).
National Institutes of Health
Authorship
Contribution: R.M., C.A.R., K.J., M.H., and S.F. performed and/or helped design experiments. C.H.L. designed the project and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
R.M. and C.A.R. contributed equally to this project.
Correspondence: Christopher H. Lowrey, Section of Hematology/Oncology, Dartmouth-Hitchcock Medical Center, One Medical Center Drive, Lebanon, NH 03756; e-mail: c.lowrey@dartmouth.edu.