Abstract
Suppressor of cytokine signaling 3 (SOCS3) is a negative regulator of granulocyte-colony stimulating factor (G-CSF) signaling in vivo. SOCS proteins regulate cytokine signaling by binding, via their SH2 domains, to activated cytokine receptors or their associated Janus kinases. In addition, they bind to the elongin B/C ubiquitin ligase complex via the SOCS box. To ascertain the contribution of the SOCS box of SOCS3 to in vivo regulation of G-CSF signaling, we generated mice expressing a truncated SOCS3 protein lacking the C-terminal SOCS box (SOCS3ΔSB/ΔSB). SOCS3ΔSB/ΔSB mice were viable, had normal steady-state hematopoiesis, and did not develop inflammatory disease. Despite the mild phenotype, STAT3 activation in response to G-CSF signaling was prolonged in SOCS3ΔSB/ΔSB bone marrow. SOCS3ΔSB/ΔSB bone marrow contained increased numbers of colony-forming cells responsive to G-CSF and IL-6. Treatment of the mice with pharmacologic doses of G-CSF, which mimics emergency granulopoiesis and therapeutic use of G-CSF, revealed that SOCS3ΔSB/ΔSB mice were hyperresponsive to G-CSF. Compared with wild-type mice, SOCS3ΔSB/ΔSB mice developed a more florid arthritis when tested using an acute disease model. Overall, the results establish a role for the SOCS box of SOCS3 in the in vivo regulation of G-CSF signaling and the response to inflammatory stimuli.
Introduction
Granulocyte colony-stimulating factor (G-CSF) is part of a network of hematopoietic growth factors and cytokines involved in regulating blood cell production. G-CSF exerts its activity via the G-CSF receptor, a member of the hematopoietin receptor superfamily. The ligand-receptor complex recruits cytoplasmic tyrosine kinases including the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) proteins. Other signaling molecules, such as growth receptor–bound protein 2 (GRB2) and Src homology and collagen protein (SHC), members of the Ras/mitogen-activated protein kinase (MAPK) pathway, and the Src homology 2–containing protein tyrosine phosphatase-2 (SHP-2), are also recruited. G-CSF has been demonstrated to influence the proliferation, survival, maturation, and functional activation of all cells of the neutrophil lineage and is a potent mobilizer of hematopoietic stem cells from bone marrow into peripheral blood. Clinically, it is widely used to mobilize stem cells for blood stem cell transplantation and to reduce the duration of neutropenia in patients receiving intensive chemotherapy or undergoing a bone marrow transplantation.1
SOCS proteins are critical regulators of cytokine signaling.2-4 They are cytokine inducible and act in a classical negative feedback loop to inhibit signal transduction via the JAK/STAT pathway (reviewed in Alexander5 and Kubo et al6 ). In bone marrow cells STAT3 is required for induction of SOCS3 in response to G-CSF.7 SOCS3 is recruited to phosphorylated Tyr729 (728 in the mouse) of the G-CSF receptor.8,9 Recently, we have shown that SOCS3 regulates the response of myeloid cells to G-CSF.10 Studies of SOCS3 function were initially hampered by the death of SOCS3-null embryos at midgestation due to placental defects resulting from exaggerated leukemia inhibitory factor (LIF) signaling.11-14 Subsequently, conditional deletion of SOCS3 in bone marrow cells demonstrated that SOCS3 is a key physiologic regulator of G-CSF signaling.10,15,16 When stimulated with G-CSF in vitro, SOCS3-deficient bone marrow cells exhibited prolonged STAT3 phosphorylation and enhanced cellular responses to G-CSF, including increased frequency of granulocytic progenitors, survival, and proliferative capacity.10,15 Mice with a hematopoietic-specific deletion of SOCS3 developed neutrophilia as they aged and showed hyperresponsiveness to G-CSF administration with enhanced bone marrow myelopoiesis, neutrophilia, splenomegaly, and the development of pathologic extramedullary neutrophilic infiltrates at multiple sites including the intrathecal space.10,15
The 8 SOCS proteins (CIS and SOCS1–7) contain an N-terminal region of variable length and limited homology, a central SH2 domain, and a highly conserved C-terminal region termed the SOCS box.17 The SH2 domains are responsible for recognizing phosphotyrosine motifs of substrates such as JAK18-20 or activated cytokine receptors.8,21-24 The SOCS box has been identified in several families of protein adaptors including the ankyrin repeat–containing proteins (ASB1–18), the WD-40 repeat–containing family (WSB1, 2, and Tubby-like protein), the SPRY domain–containing proteins (SSB1–4), as well as 4 RAR-like proteins.17 It is an approximately 40–amino acid motif containing a BC box and a Cul5 box that binds Elongin B/C, Cullin 5, and RING finger protein Rbx2. Binding specificity is dictated by the conserved amino acid sequence in the C-terminal portion of the SOCS box, known as the Cul2 or Cul5 box.25 The resulting complex, termed the ECS (Elongin B/C–Cullin2/5–SOCS-box protein) complex, acts as an E3 ubiquitin ligase. Together with a ubiquitin-activating enzyme (E1) and a ubiquitin-conjugating enzyme (E2), the ECS complex recognizes and recruits target proteins for polyubiquitylation and degradation via the 26S proteasome.26 SOCS3 interacts with Elongin B, Elongin C, Cul5, and Rbx2.25,27,28 SOCS1, the SOCS protein most closely related to SOCS3, has been shown to target JAK2, TEL-JAK2 fusion protein, VAV, FAK, IRS1, and IRS2 for ubiquitylation and proteasomal degradation in a SOCS box–dependent manner.29-33 To date, the endogenous proteins shown to be polyubiquitylated and degraded via SOCS3 are FAK, IRS-1, and IRS-2.33,34 The significance of the interaction between SOCS proteins and the other components of the ECS complex is unclear, as some studies propose that the association targets SOCS box–containing proteins for degradation31,35 whereas other data suggest it may stabilize SOCS proteins.18,28,36,37 The stability of the SOCS3 protein is also regulated by nonproteasomal degradation via a PEST motif, by phosphorylation of tyrosine residues within the SOCS box, and by ubiquitylation of lysine 6, as well as other unidentified residues.37-39 Functional studies of the SOCS1 and SOCS3 proteins lacking the SOCS box have largely failed to demonstrate an impact of SOCS box deletion on inhibition of cytokine signal transduction in vitro18,39,40 ; however, a physiologic role for the SOCS1 SOCS box was revealed by creation of mice in which DNA encoding the SOCS box was deleted.27 Whereas mice lacking SOCS1 die in the neonatal period from a disease characterized by hyperresponsiveness to interferon-γ,41 these mice survived for several months before succumbing to inflammatory disease, demonstrating that, in vivo, the SOCS1 ECS complex is essential for appropriate regulation of cytokine signaling.
To elucidate the physiologic role of the SOCS box of SOCS3 in hematopoiesis, we examined mice with a deletion of the DNA sequence encoding this domain and examined the response of these mice to G-CSF stimulation and inflammatory stimuli.
Materials and methods
Generation of SOCS3ΔSB/ΔSB mice
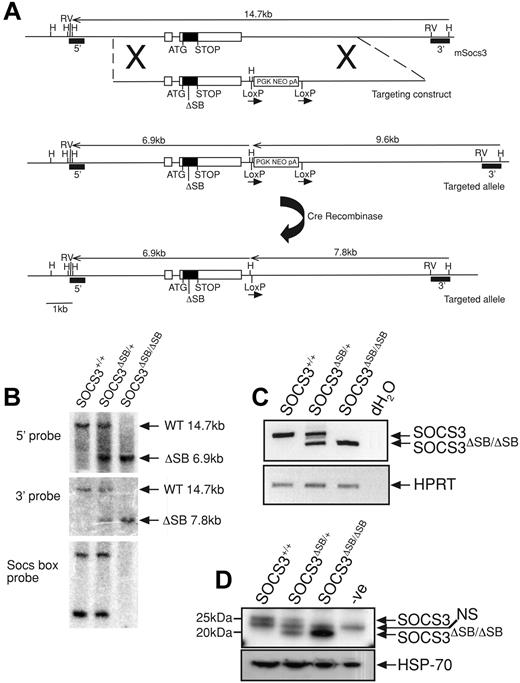
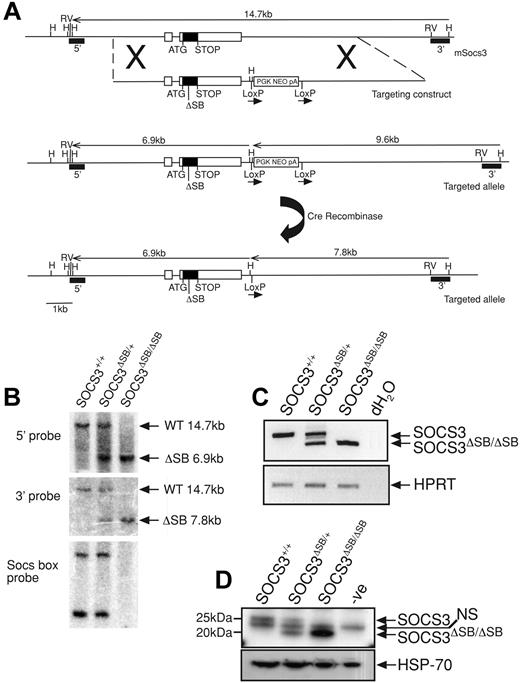
Using a nested polymerase chain reaction (PCR) approach, an 830-bp product lacking the 120 nucleotides that encode the 40 c-terminal amino acids (SOCS box) was amplified from the SOCS3 coding region. The product was digested with NotI and EcoRV and ligated into the pLoxpNeo vector containing the 3′ arm. The 5′ arm was amplified by PCR, digested with NotI, and ligated into the above plasmid at the NotI site (Figure 1A). The vector was linearized with AscI and electroporated into C57BL/6 embryonic stem (ES) cells (Bruce 4 line). G418-resistant clones were screened by Southern blotting of HindIII-digested DNA using the 3′ probe. Correctly targeted clones were injected into C57BL/6 blastocysts. Mice heterozygous for the mutation were crossed to CMV-Cre recombinase mice42 to remove the neomycin cassette and the offspring were interbred to produce wild-type (WT; SOCS3+/+), heterozygous (SOCS3ΔSB/+), and homozygous (SOCS3ΔSB/ΔSB) SOCS3 SOCS box–deleted mutants. Mice were genotyped by PCR with tail DNA using primers spanning the SOCS3 SOCS box (F-aagattccgctggtactgagcc, R-agctctactctgcgggagtgga) and primers to confirm the deletion of the neomycin cassette (F-agatggattgcacgcaggttctcc, R-agaaaagcggccattttccaccatg). For confirmation at the RNA/protein level, WT, SOCS3ΔSB/+, and SOCS3ΔSB/ΔSB mice were injected intraperitoneally with 5 μg IL-6 and killed 60 minutes later, and the liver was removed. RNA was isolated using an RNeasy Mini extraction kit (Qiagen, Valencia, CA). cDNA was transcribed using Superscript III (Invitrogen, Carlsbad, CA) and subjected to reverse transcriptase–PCR (RT-PCR) with primers spanning the SOCS box (F-cccgcgggcacctttcttatcc, R-ccccctctgacccttttgctcctt) and HPRT as a loading control (F-tccctggttaagcagtacagc, R-gatggccacaggactagaaca). Remaining liver tissue was homogenized in KALB buffer (1% [vol/vol] Triton X-100; 50 mM Tris [Tris(hydroxymethyl) aminomethane]-Hcl, pH 7.4; 150 mM NaCl; and 1 mM EDTA) supplemented with 1 mM sodium vanadate, 10 mM sodium fluoride, and complete protease inhibitor (Roche, Indianapolis, IN). Protein content of lysates was determined with a BCA protein quantitation assay (Pierce Biosciences, Rockford, IL), and 10 mg total protein was immunoprecipitated with a monoclonal antibody raised against SOCS3. Antibody/protein complexes were captured using protein A Sepharose, eluted in 2 × SDS sample buffer, run through Bis/Tris gels, transferred onto PVDF membranes, and probed with a polyclonal antibody raised against SOCS3. Membranes were reprobed with an antibody to HSP-70 (Santa Cruz Biotechnology, Santa Cruz, CA) as a loading control. Protein bands were detected with Millipore Western Chemiluminescent HRP substrate on x-ray film (Amersham, Arlington Heights, IL) using a Kodak Xomat 3000-RA developer (Rochester, NY). All animal experimental procedures were approved by the Melbourne Health Research Directorate Animal Ethics Committee.
Generation and confirmation of SOCS3ΔSB/ΔSB mice. (A) The SOCS3 locus is shown at the top with the targeting construct and targeted allele below. The structure of the knock-in allele after removal of the neomycin selection cassette (PGKNEOpA) by cre-mediated excision is also shown. Exons are indicated as boxes with the coding region shaded. The 120-bp deletion in exon 2 of nucleotides 573 to 692 of the SOCS3 transcript that encodes the SOCS box is indicated by ΔSB. 5′ and 3′ probes used for Southern analysis are indicated. H indicates HindIII; RV, EcoRV. (B) Correct homologous recombination and deletion of the selection cassette was confirmed by Southern blot analyses of tail DNA digested with HindIII using 5′ and 3′ probes. DNA digested with PvuII was probed with a 130-bp SOCS box probe, confirming the absence of this sequence in SOCS3ΔSB/ΔSB samples. (C) RT-PCR analysis of cDNA prepared from RNA from livers from WT, SOCS3ΔSB/+, and SOCS3ΔSB/ΔSB mice injected with IL-6 (5 μg, intraperitoneally 60 minutes prior to the time that they were killed). Primers spanning the SOCS3 SOCS box were used. In SOCS3ΔSB/ΔSB samples, only the 200-bp transcript lacking the SOCS box was amplified. HPRT was amplified as a loading control. (D) Immunoblot of liver lysates prepared from mice injected with IL-6 (5 μg, intraperitoneally 60 minutes prior to the time that they were killed). SOCS3 was immunoprecipitated with a monoclonal antibody to SOCS3 and immunoblotted using a polyclonal SOCS3 antibody. In SOCS3ΔSB/ΔSB lysates, only the smaller (20 kDa) SOCS3 protein was detectable. NS indicates nonspecific band.
Generation and confirmation of SOCS3ΔSB/ΔSB mice. (A) The SOCS3 locus is shown at the top with the targeting construct and targeted allele below. The structure of the knock-in allele after removal of the neomycin selection cassette (PGKNEOpA) by cre-mediated excision is also shown. Exons are indicated as boxes with the coding region shaded. The 120-bp deletion in exon 2 of nucleotides 573 to 692 of the SOCS3 transcript that encodes the SOCS box is indicated by ΔSB. 5′ and 3′ probes used for Southern analysis are indicated. H indicates HindIII; RV, EcoRV. (B) Correct homologous recombination and deletion of the selection cassette was confirmed by Southern blot analyses of tail DNA digested with HindIII using 5′ and 3′ probes. DNA digested with PvuII was probed with a 130-bp SOCS box probe, confirming the absence of this sequence in SOCS3ΔSB/ΔSB samples. (C) RT-PCR analysis of cDNA prepared from RNA from livers from WT, SOCS3ΔSB/+, and SOCS3ΔSB/ΔSB mice injected with IL-6 (5 μg, intraperitoneally 60 minutes prior to the time that they were killed). Primers spanning the SOCS3 SOCS box were used. In SOCS3ΔSB/ΔSB samples, only the 200-bp transcript lacking the SOCS box was amplified. HPRT was amplified as a loading control. (D) Immunoblot of liver lysates prepared from mice injected with IL-6 (5 μg, intraperitoneally 60 minutes prior to the time that they were killed). SOCS3 was immunoprecipitated with a monoclonal antibody to SOCS3 and immunoblotted using a polyclonal SOCS3 antibody. In SOCS3ΔSB/ΔSB lysates, only the smaller (20 kDa) SOCS3 protein was detectable. NS indicates nonspecific band.
Immunoblotting
Bone marrow was flushed from femora and tibiae into Dulbecco modified Eagle medium (DME) supplemented with 0.5% (vol/vol) fetal calf serum (FCS; Sigma Aldrich, St Louis, MO), and red cells were lysed in red cell removal buffer (RCRB). White blood cell pellets were resuspended in DME supplemented with 0.5% FCS and allowed to rest at 37°C for 30 minutes before stimulation. For detection of SOCS3, cells were stimulated with 10 ng/mL recombinant human G-CSF (Amgen, Thousand Oaks, CA) in the presence of 10 μM of the proteosome inhibitor MG-132 (Calbiochem, San Diego, CA) at 37°C. For analysis of JAK/STAT signaling, cells were stimulated with 50 ng/mL recombinant human G-CSF (Amgen) at 37°C. For analysis of Ras/MAPK signaling, cells were incubated in DME supplemented with 0.5% FCS (vol/vol) for 8 hours at 37°C with 10% CO2 in air, followed by stimulation with 50 ng/mL recombinant human G-CSF. At each time point, cells were lysed in RIPA buffer (1% Triton X-100; 0.1% SDS; 1% sodium deoxycholate; 150 mM NaCl; 10 mM Tris-HCl, pH 7.5; 0.01% [wt/vol] sodium azide) supplemented with 2 mM sodium vanadate, 10 mM sodium fluoride, 1 mM PMSF, and complete protease inhibitor (Roche). Total protein (20 μg) was electrophoresed in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, electrophoretically transferred onto PVDF membrane, and immunoblotted with antibodies against phospho-STAT3 (Tyr705; Cell Signaling, Beverly, MA), STAT3 (Santa Cruz Biotechnology), phospho-p44/p42 (Cell Signaling), p44/p42 (Cell Signaling), or an in-house polyclonal antibody raised against SOCS3.
Hematologic analysis
Mice were bled from the retro-orbital sinus and peripheral cell counts were performed using an ADVIA 120 automated cell analyzer (Bayer, Leverkusen, Germany). Differential counts and morphologic analyses were performed manually on May-Grünwald-Giemsa–stained peripheral blood smears or cytocentrifuge preparations of spleen or bone marrow suspensions. For progenitor cell assays, bone marrow cells at a final concentration of 25 000 cells/mL or spleen cells (50 000 cells/mL) were added to triplicate 36-mm–diameter bacterial plates in 1 mL of semisolid medium (DME with 20% selected BCS and 0.3% agar [wt/vol]) with cytokines at the following final concentrations: 100 ng/mL recombinant murine stem cell factor (SCF; produced in Pichia pastoralis), 10 ng/mL G-CSF, 10 ng/mL recombinant murine granulocyte-macrophage colony-stimulating factor (GM-CSF; produced in murine erythroleukemia cells), 10 ng/mL recombinant murine macrophage colony-stimulating factor (M-CSF; Cetus, Berkeley, CA), 10 ng/mL murine interleukin 3 (IL-3; Peprotech, Rocky Hill, NJ), 100 ng/mL recombinant human IL-6 (a gift from Richard Simpson of the Ludwig Institute for Cancer Research, Melbourne, Australia), and 100 ng/mL recombinant human IL-11 (a gift from Genetics Institute, Andover, MA). Cells were incubated at 37°C in 10% CO2 in air for 7 days before plates were fixed, floated onto glass slides, dried, and stained with acetylcholinesterase, Luxol fast blue, and hematoxylin. Total and differential counts were performed by microscopic examination of the stained culture slides. To determine the cellular content of colonies, cultures were resuspended in phosphate-buffered saline, triturated to create single-cell suspensions, and counted using a hemocytometer. Peripheral blood progenitors were enumerated by culturing 10 μL (for G-CSF–treated animals) or 30 μL (for saline-treated animals) of blood as described in this paragraph for 7 days, fixed, stained, and counted at × 40 magnification.43 The relative contribution of different cell populations to peripheral blood, peritoneal cells, spleen, bone marrow, lymph node, and thymus were assessed by flow cytometry. Single-cell suspensions were created from each organ in phosphate-buffered saline supplemented with 2% FCS and 0.02% (wt/vol) sodium azide. Cell suspensions were stained with either phycoerythrin- or fluorescein isothiocyanate–conjugated antibodies against Mac1, Gr1, B220, CD4, CD8, CD71, Ter119, and IgM (Pharmingen, San Diego, CA). Dead cells were excluded by propidium iodide uptake. Stained suspensions were analyzed on an LSR flow cytometer (Becton Dickinson, Mountain View, CA) and data were compiled using FloJo software (v.6.4.7, Treestar, Ashland, OR). Proliferation assays were performed using bone marrow cells after red cell lysis. Cells (105) were placed into wells of a 96-well plate in DME supplemented with 10% FCS with saline/BSA, G-CSF, or IL-3 and incubated at 37°C in 10% CO2 in air for 48 hours. Cells were pulsed with 1 μCi (0.037 MBq) [3H]-thymidine for 8 hours, harvested onto Inotech (Berkeley, CA) glass filters, and counted in a TopCount NXT Microplate Scintillation Counter (Packard, Meriden, CT). Six replicates for each concentration were assayed and the result was averaged. To generate radiation chimeras, C57BL/6.SJL (Ly5.1) mice were lethally irradiated (11 Gy, split dose) and injected with 106 donor bone marrow cells via the tail vein. Two months after transplantation, reconstitution was assessed using flow cytometry of peripheral blood with antibodies against Ly5.1 and Ly5.2 (Pharmingen). Only mice with greater than 80% reconstitution by donor cells were used in further experiments.
In vivo response to G-CSF
mBSA/IL-1–induced arthritis
Induction of acute arthritis was performed as described.45 Briefly, adult mice were anesthetized and injected intra-articularly with 10 μL of a 20-mg/mL solution methylated bovine serum albumin (mBSA; endotoxin level, < 1 pg/μg; Sigma-Aldrich) into knee joints. Mice were injected with 20 μL of 12.5 μg/mL recombinant human IL-1β (endotoxin level, < 0.1 ng/μg; PeproTech) in the footpad. The IL-1 injection was repeated for the next 2 days. Mice were killed on day 7 following intra-articular mBSA injection, and the knee joints were removed and processed as described previously.45 Severity of arthritis was assessed using 5 histologic features of joint disease (exudate, synovitis, pannus, cartilage, and bone destruction), each graded for severity from 0 (normal) to 5 (severe) by an investigator blinded to the experimental groups, as described previously.45 The mean histologic severity score for an experimental group (maximum of 25) was calculated by averaging the sum of the 5 histologic features, with each feature scored at 4 section depths. For histological images, a Zeiss (Camperdown, Australia) Axioplan 2 microscrope was used. Fixed tissues were paraffin sectioned and stained with hematoxylin and eosin and viewed with 10×/0.30 air objective. Images were acquired with a Zeiss Axiocam camera and Axiovision software version 3.1. Image labelling was made using Freehand software version 11.0.2.
Results
Generation of SOCS3ΔSB/ΔSB mice
Gene targeting was used to create mice expressing SOCS3 protein lacking the SOCS box (hereafter referred to as SOCS3ΔSB/ΔSB) as outlined in Figure 1A. Exon 2 of SOCS3 was replaced by an exon in which the sequence encoding the SOCS box (nucleotides 573 to 693) was deleted in frame. The resulting SOCS3 protein therefore lacked the 40 C-terminal amino acids that encode the SOCS box but contained the native stop codon, 3′ untranslated region, and polyadenylation signal sequence. Correct homologous recombination and cre recombinase–mediated excision of the neomycin selection cassette were confirmed by Southern blotting (Figure 1B). RNA was prepared from livers of WT, SOCS3ΔSB/+, and SOCS3ΔSB/ΔSB mice treated with IL-6 60 minutes prior to the time that they were killed, and SOCS3 transcripts were amplified by RT-PCR. Using primers spanning the sequence encoding the SOCS box, only the shorter transcript, representing the SOCS box–deleted form, was detectable in SOCS3ΔSB/ΔSB mice (Figure 1C). SOCS3 protein in IL-6–stimulated liver lysates was detected by immunoprecipitation and subsequent immunoblot (Figure 1D). Full-length SOCS3 protein was present in WT and SOCS3ΔSB/+ livers, whereas in SOCS3ΔSB/+ and SOCS3ΔSB/ΔSB mice a shorter 20-kDa band, representing SOCS box–deleted SOCS3 protein (SOCS3ΔSB), was evident.
SOCS3ΔSB/ΔSB mice are viable and do not develop spontaneous inflammatory disease
Unlike SOCS3-null mice, which die at midgestation due to placental insufficiency,12,14 intercrosses of SOCS3ΔSB/+ mice gave rise to expected numbers of SOCS3ΔSB/ΔSB offspring. The pups were of normal size, and histologic examination of the placentae of embryonic day–17 pups did not reveal any abnormalities (data not shown). The weight and appearance of neonatal pups and of 8- to 12-week-old SOCS3ΔSB/ΔSB mice did not differ from WT littermates, and the mice were fertile and nursed normally. Full blood, gross, and histologic examination of 8- to 12-week-old SOCS3ΔSB/ΔSB mice was normal, with the exception of spleen weight, which was increased (Table 1). Flow-cytometric analysis of cells from lymph node, spleen, thymus, bone marrow, and peritoneal cavity using a panel of specific monoclonal antibodies to myeloid, B-cell, T-cell, and erythroid antigens did not detect any differences between WT and SOCS3ΔSB/ΔSB mice (data not shown). A cohort of SOCS3ΔSB/ΔSB mice was followed to 16 months of age. In contrast to mice with a hematopoietic and endothelial cell–specific deletion of SOCS3 or mice with deletion of SOCS3 and LIF,10,13 both of which developed neutrophilia and chronic inflammatory lesions with advancing age, SOCS3ΔSB/ΔSB mice remained healthy although they developed neutrophilia (WT 3.7 × 103/mL, n = 5; SOCS3ΔSB/ΔSB 9.3 × 103/mL, n = 10; P < .01). Aside from splenomegaly, gross and microscopic analysis was normal, with no evidence of chronic inflammatory lesions (data not shown).
Altered STAT activation and G-CSF responsiveness in SOCS3ΔSB/ΔSB bone marrow cells
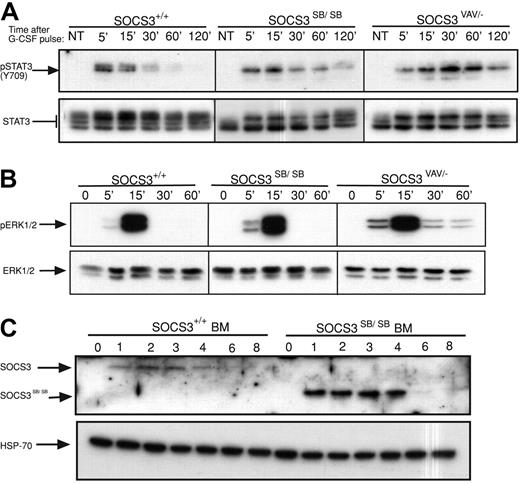
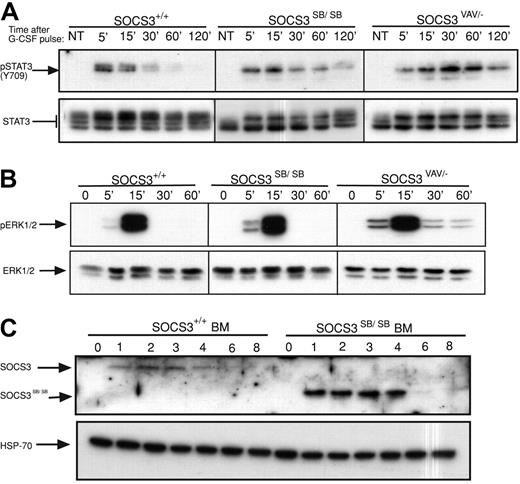
Previous analysis of mice with deletion of SOCS3 in hematopoietic and endothelial cells (SOCS3ΔVAV/−, SOCS3ΔTIE2/−) or in neutrophils (SOCS3ΔLYSM/−) revealed that SOCS3 is a major physiologic regulator of G-CSF signaling.10,15 Therefore, the time course of activation of STAT3 in primary bone marrow cells from WT or SOCS3ΔSB/ΔSB mice in response to a pulse of G-CSF (50 ng/mL) was compared. Prolonged STAT3 activation was observed in SOCS3ΔSB/ΔSB bone marrow cells. This alteration in the kinetics of STAT3 activation was not as dramatic as that seen in bone marrow lacking SOCS3 (SOCS3ΔVAV/−; Figure 2A; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Activation of ERK1/2 was greater and was prolonged in SOCS3ΔVAV/− cells but not in SOCS3ΔSB/ΔSB cells (Figure 2B; Figure S1). To detect SOCS3 and SOCS3ΔSB in bone marrow, whole-cell lysates were stimulated with G-CSF for 1 to 8 hours in the presence of the proteasomal inhibitor MG132. Compared with WT SOCS3, the level of SOCS3ΔSB protein was higher (Figure 2C). This difference was observed despite the presence of a proteasomal inhibitor, indicating that a nonproteasomal but SOCS box–mediated mechanism is involved in degradation of SOCS3. The increase in SOCS3ΔSB protein in G-CSF–stimulated bone marrow lysates may be due to sustained elevation of STAT3, leading to increased transcription of SOCS3. Alternatively, the half-life of SOCS3ΔSB may be altered.
Removal of the SOCS3 SOCS box alters the regulation of the JAK/STAT pathway in vivo. (A) Activation of STAT3 in response to G-CSF. Bone marrow cells from SOCS3+/+ and SOCS3ΔSB/ΔSB or SOCS3ΔVAV/− mice were left untreated (NT) or stimulated with 50 ng/mL G-CSF for 15 minutes, washed, then lysed at intervals between 5 minutes and 120 minutes. Lysates were analyzed by immunoblot using antibodies specific for phospho-STAT3 or total STAT3. (B) Activation of ERK in response to G-CSF. Bone marrow cells from SOCS3+/+ and SOCS3ΔSB/ΔSB or SOCS3ΔVAV/− mice were left unstimulated (0 time point) or stimulated with 50 ng/mL G-CSF for intervals between 5 minutes and 60 minutes. Lysates were analyzed by immunoblot using antibodies specific for phospho-ERK or total ERK. (C) Induction of WT and SOCS box–deleted SOCS3 protein in SOCS3+/+ and SOCS3ΔSB/ΔSB bone marrow cells. Cells were left unstimulated (0 time point) or incubated in the presence of 10 ng/mL G-CSF and 10 μM MG132 for 1 to 8 hours and analyzed by immunoblot using antibodies specific for SOCS3 or HSP-70. BM indicates bone marrow.
Removal of the SOCS3 SOCS box alters the regulation of the JAK/STAT pathway in vivo. (A) Activation of STAT3 in response to G-CSF. Bone marrow cells from SOCS3+/+ and SOCS3ΔSB/ΔSB or SOCS3ΔVAV/− mice were left untreated (NT) or stimulated with 50 ng/mL G-CSF for 15 minutes, washed, then lysed at intervals between 5 minutes and 120 minutes. Lysates were analyzed by immunoblot using antibodies specific for phospho-STAT3 or total STAT3. (B) Activation of ERK in response to G-CSF. Bone marrow cells from SOCS3+/+ and SOCS3ΔSB/ΔSB or SOCS3ΔVAV/− mice were left unstimulated (0 time point) or stimulated with 50 ng/mL G-CSF for intervals between 5 minutes and 60 minutes. Lysates were analyzed by immunoblot using antibodies specific for phospho-ERK or total ERK. (C) Induction of WT and SOCS box–deleted SOCS3 protein in SOCS3+/+ and SOCS3ΔSB/ΔSB bone marrow cells. Cells were left unstimulated (0 time point) or incubated in the presence of 10 ng/mL G-CSF and 10 μM MG132 for 1 to 8 hours and analyzed by immunoblot using antibodies specific for SOCS3 or HSP-70. BM indicates bone marrow.
Growth of SOCS3ΔSB/ΔSB bone marrow cells in semisolid agar cultures in response to a range of cytokines was compared with that of bone marrow from WT mice. In SOCS3ΔSB/ΔSB cultures, when maximally stimulated by GM-CSF, M-CSF, IL-3, or SCF, the frequency of myeloid progenitors was normal, however there was a selective increase in colony-forming cells (CFCs) in response to G-CSF (WT, 9 ± 3; SOCS3ΔSB/ΔSB, 16 ± 1; P < .05 both per 25 000 cells plated; triplicate cultures from 3 mice of each genotype) or IL-6 (WT, 11 ± 5; SOCS3ΔSB/ΔSB, 18 ± 3; P < .05 both per 25 000 cells plated; triplicate cultures from 3 mice of each genotype). The increase in number and size of G-CSF–responsive and IL-6–responsive CFCs in SOCS3ΔSB/ΔSB bone marrow was intermediate compared with that seen in SOCS3ΔVAV/− bone marrow (Figure 3A-B). In addition to this quantitative difference, 2 qualitative differences between SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− were observed. Firstly, as observed previously, G-CSF–responsive SOCS3ΔVAV/− progenitor cells gave rise to abnormal numbers of macrophage and granulocyte-macrophage colonies,10 but this phenomenon was not observed in SOCS3ΔSB/ΔSB bone marrow cultures (data not shown). Secondly, synergy between SCF and IL-6 was not observed in SOCS3ΔVAV/− bone marrow cultures, whereas in SOCS3ΔSB/ΔSB bone marrow cultures, synergy between SCF and IL-6 was preserved (Figure 3C-D).
SOCS3ΔSB/ΔSB bone marrow cells are hyperresponsive to G-CSF. (A) Bone marrow cells (25 000) from WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures with cytokines as indicated and colonies were counted after 7 days. Results represent the mean (± SD) from at least 3 mice of each genotype. (B) To ascertain colony size, cultures were plucked, resuspended, and counted. Results represent the mean (± SD) for at least 3 mice of each genotype. (C) To assess synergy between SCF and other cytokines, 25 000 bone marrow cells from SOCS3+/+, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures stimulated by SCF and/or additional cytokines as indicated and colonies were counted after 7 days. Results for each cytokine are represented as groups containing WT, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− data for the single cytokine, SCF, and the combination of the 2. Results represent the mean (± SD) from at least 3 mice of each genotype. Synergy between SCF and IL-6 was not detected in cultures of SOCS3ΔVAV/− bone marrow cells. (D) Average colony size. Results represent the mean (± SD) for at least 3 mice for each genotype. Again, synergy between SCF and IL-6 was not observed in cultures of SOCS3ΔVAV/− cells. (E-F) Bone marrow cells were stimulated with G-CSF (E) or IL-3 (F) and proliferative activity was assessed after 48 hours by 3[H]-thymidine incorporation. Results represent the mean (± SD) for 2 mice per genotype; analysis of each mouse included 6 replicates (*P < .05; **P < .01).
SOCS3ΔSB/ΔSB bone marrow cells are hyperresponsive to G-CSF. (A) Bone marrow cells (25 000) from WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures with cytokines as indicated and colonies were counted after 7 days. Results represent the mean (± SD) from at least 3 mice of each genotype. (B) To ascertain colony size, cultures were plucked, resuspended, and counted. Results represent the mean (± SD) for at least 3 mice of each genotype. (C) To assess synergy between SCF and other cytokines, 25 000 bone marrow cells from SOCS3+/+, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures stimulated by SCF and/or additional cytokines as indicated and colonies were counted after 7 days. Results for each cytokine are represented as groups containing WT, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− data for the single cytokine, SCF, and the combination of the 2. Results represent the mean (± SD) from at least 3 mice of each genotype. Synergy between SCF and IL-6 was not detected in cultures of SOCS3ΔVAV/− bone marrow cells. (D) Average colony size. Results represent the mean (± SD) for at least 3 mice for each genotype. Again, synergy between SCF and IL-6 was not observed in cultures of SOCS3ΔVAV/− cells. (E-F) Bone marrow cells were stimulated with G-CSF (E) or IL-3 (F) and proliferative activity was assessed after 48 hours by 3[H]-thymidine incorporation. Results represent the mean (± SD) for 2 mice per genotype; analysis of each mouse included 6 replicates (*P < .05; **P < .01).
Additional evidence of an enhanced in vitro response to G-CSF was demonstrated by measuring the proliferation of bone marrow cells from WT, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− mice cultured in increasing concentrations of G-CSF or IL-3. Proliferation of cells of all 3 genotypes in response to IL-3 was similar, whereas SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− cells were hyperresponsive to G-CSF (Figure 3E-F). Taken together, these results demonstrate that the deletion of the SOCS3 SOCS box renders bone marrow cells hyperresponsive to G-CSF but that the magnitude of the response is less than that observed when full-length SOCS3 protein is absent. In addition, qualitative aspects of SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− bone marrow cultures differed.
The SOCS box of SOCS3 is required for negative regulation of “emergency granulopoiesis”
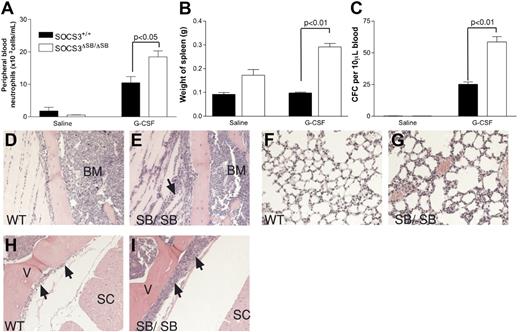
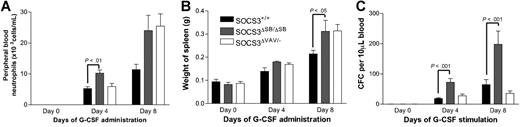
tk;4Under normal circumstances, circulating levels of G-CSF are low; however, during insults such as bacterial infection, circulating levels of G-CSF increase dramatically. This “emergency granulopoiesis” can be mimicked in vivo by injecting mice twice daily with pharmacologic doses of G-CSF. SOCS3 has previously been shown to be important in regulating G-CSF signaling during emergency granulopoiesis.10 WT and SOCS3ΔSB/ΔSB mice were injected intraperitoneally twice daily with 2.5 μg G-CSF or endotoxin-free diluent for 5 days, and peripheral blood neutrophil counts, spleen weight, and blood progenitor cell mobilization were measured. Compared with WT mice, all 3 parameters were significantly increased in the SOCS3ΔSB/ΔSB mice (Figure 4A-C). Histologic examination of tissues from the treated mice revealed increased neutrophilic infiltration of tissues in treated SOCS3ΔSB/ΔSB mice, particularly muscle, lung, and intrathecal space (Figure 4D-I).
Increased progenitor cell mobilization in SOCS3ΔSB/ΔSB mice in response to G-CSF stimulation in vivo. SOCS3+/+ and SOCS3ΔSB/ΔSB mice (4 per group) were injected intraperitoneally twice daily with either 2.5 μg/kg G-CSF or endotoxin-free saline/BSA vehicle for 5 days. Mice were killed on day 6. Results are means (± SD). (A) Peripheral blood neutrophil count. (B) Spleen weight. (C) Peripheral blood colony-forming cells. Peripheral blood (10 μL for G-CSF–treated, 30 μL for vehicle-treated) from all mice was cultured in triplicate in the presence of SCF/IL-3 for 7 days, fixed and stained, and the number of colonies was counted at × 40 magnification. Histologic examination showed increased neutrophil infiltration into the following: (D,E) Parasternal muscles. The arrow indicates neutrophils between the muscle fibers. (E,F) Lung. Note the thickening of the alveolar walls, due to increased numbers of neutrophils, in SOCS3ΔSB/ΔSB mice. (H,I) Intrathecal space. A transverse section through the spinal column at the level of the thoracic vertebrae reveals increased numbers of intrathecal granulocytes in the SOCS3ΔSB/ΔSB mice. BM indicates bone marrow; V, vertebral column; and SC, spinal cord.
Increased progenitor cell mobilization in SOCS3ΔSB/ΔSB mice in response to G-CSF stimulation in vivo. SOCS3+/+ and SOCS3ΔSB/ΔSB mice (4 per group) were injected intraperitoneally twice daily with either 2.5 μg/kg G-CSF or endotoxin-free saline/BSA vehicle for 5 days. Mice were killed on day 6. Results are means (± SD). (A) Peripheral blood neutrophil count. (B) Spleen weight. (C) Peripheral blood colony-forming cells. Peripheral blood (10 μL for G-CSF–treated, 30 μL for vehicle-treated) from all mice was cultured in triplicate in the presence of SCF/IL-3 for 7 days, fixed and stained, and the number of colonies was counted at × 40 magnification. Histologic examination showed increased neutrophil infiltration into the following: (D,E) Parasternal muscles. The arrow indicates neutrophils between the muscle fibers. (E,F) Lung. Note the thickening of the alveolar walls, due to increased numbers of neutrophils, in SOCS3ΔSB/ΔSB mice. (H,I) Intrathecal space. A transverse section through the spinal column at the level of the thoracic vertebrae reveals increased numbers of intrathecal granulocytes in the SOCS3ΔSB/ΔSB mice. BM indicates bone marrow; V, vertebral column; and SC, spinal cord.
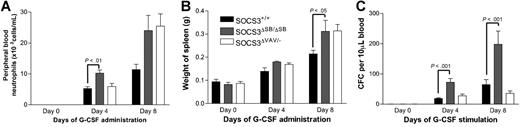
The SOCS3ΔSB/ΔSB mice showed increased responsiveness to G-CSF compared with WT mice. However, the phenotype was milder than that observed when G-CSF was administered to SOCS3ΔVAV/− mice.10 To directly compare the responsiveness of SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− bone marrow cells and to establish whether the altered responsiveness is intrinsic to the hematopoietic system, lethally irradiated WT C57BL/6.SJL (Ly5.1) mice were reconstituted with WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− bone marrow cells. Ten to 12 weeks later, mice reconstituted with more than 80% donor cells were treated for 4 or 8 days with G-CSF. Following 8 days of treatment, an increase in spleen weight among all groups was observed, with significantly larger spleens observed in mice receiving SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− bone marrow compared with WT bone marrow (Figure 5A). The number of progenitor cells in the peripheral blood after 4 or 8 days of G-CSF treatment was significantly increased in mice that received SOCS3ΔSB/ΔSB bone marrow compared with mice receiving WT bone marrow. At day 4, peripheral blood neutrophils were also significantly higher in the SOCS3ΔSB/ΔSB mice (Figure 5B-C). In previous studies,10 we observed that mice reconstituted with SOCS3ΔVAV/− bone marrow had enhanced responsiveness to G-CSF and it was unclear why this cohort responded poorly in this experiment. It may reflect differences in the level of reconstitution in the 2 experiments. In keeping with previous studies, there was increased neutrophil infiltration in liver, muscle, and lung of G-CSF–treated SOCS3ΔVAV/− bone marrow chimeras compared with controls. The extent of neutrophil infiltration in G-CSF–treated SOCS3ΔSB/ΔSB chimeras was intermediate between WT and SOCS3ΔVAV/− chimeras (data not shown).
Comparison of the response of SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− bone marrow cells to administration of G-CSF in vivo using radiation chimeras. C57BL/6.SJL (Ly5.1) mice reconstituted with SOCS3+/+, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− bone marrow cells (4 of each genotype per group) were injected intraperitoneally twice daily with either 2.5 μg/kg G-CSF or saline/BSA vehicle for 4 or 8 days. Mice were killed on days 0, 4, or 8. (A) Peripheral blood neutrophil count. (B) Spleen weight. (C) Peripheral blood colony-forming cells. Peripheral blood (10 μL for G-CSF–treated, 30 μL for vehicle-treated) from all mice was cultured in triplicate in the presence of SCF/IL-3 for 7 days, fixed, and stained, and the number of colonies was counted at × 40 magnification. Results are means (± SD).
Comparison of the response of SOCS3ΔSB/ΔSB and SOCS3ΔVAV/− bone marrow cells to administration of G-CSF in vivo using radiation chimeras. C57BL/6.SJL (Ly5.1) mice reconstituted with SOCS3+/+, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− bone marrow cells (4 of each genotype per group) were injected intraperitoneally twice daily with either 2.5 μg/kg G-CSF or saline/BSA vehicle for 4 or 8 days. Mice were killed on days 0, 4, or 8. (A) Peripheral blood neutrophil count. (B) Spleen weight. (C) Peripheral blood colony-forming cells. Peripheral blood (10 μL for G-CSF–treated, 30 μL for vehicle-treated) from all mice was cultured in triplicate in the presence of SCF/IL-3 for 7 days, fixed, and stained, and the number of colonies was counted at × 40 magnification. Results are means (± SD).
SOCS3ΔSB/ΔSB mice show abnormal responses in an arthritis disease model
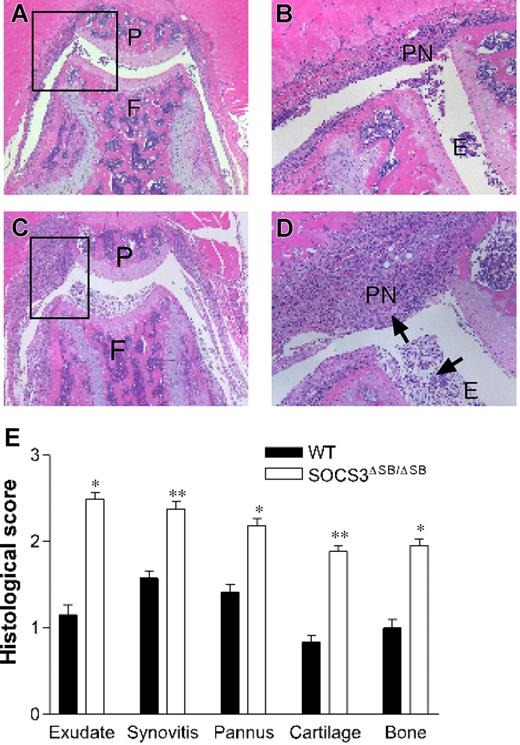
Acute inflammation elicits a complex web of interacting positive and negative regulatory signals mediated by cytokines, chemokines, and intracellular signaling pathways.46 To test the in vivo requirement for the SOCS box of SOCS3 in the response to inflammation, we examined the response of SOCS3ΔSB/ΔSB mice to acute, IL-1–dependent inflammatory arthritis.45 This model is dependent on G-CSF.47 WT and SOCS3ΔSB/ΔSB mice were injected intra-articularly with mBSA, followed by 3 daily subcutaneous injections of IL-1. Mice were killed on day 7 and knee joints were processed for histology. Joint pathology was not observed in either group in the absence of mBSA injection. In comparison with WT mice, SOCS3ΔSB/ΔSB mice developed exacerbated arthritis with markedly increased inflammatory infiltrate in the knee joint (Figure 6A-D). Exudate, synovitis, pannus, bone, and cartilage destruction were all enhanced in SOCS3ΔSB/ΔSB mice, as graded histologically.45 The overall disease score, as measured by the sum of individual histologic scores for the 5 features examined, was increased significantly in SOCS3ΔSB/ΔSB mice (Figure 6E).
SOCS3ΔSB/ΔSB mice exhibit exacerbated mBSA/IL-1–induced acute arthritis. (A-D) Acute inflammatory arthritis was induced by intra-articular injection of mBSA into the knee joint followed by 3 daily subcutaneous injections of IL-1. Mice were killed on day 7. Frontal hematoxylin and eosin–stained sections through knee joints from arthritic WT (A-B) and SOCS3ΔSB/ΔSB (C-D) mice. E indicates exudate; P, patella; PN, pannus; F, femur; and arrows, increased exudate and inflammatory cells, predominantly neutrophils, in the joint space of treated SOCS3ΔSB/ΔSB mice. (E) Joint sections were graded for 5 features of inflammatory arthritis, each on a scale of 0 (normal) to 5 (severe) by an investigator blinded to the experimental groups. Results are shown as the mean (± SD) for 5 joints per group (*P < .05; **P < .01).
SOCS3ΔSB/ΔSB mice exhibit exacerbated mBSA/IL-1–induced acute arthritis. (A-D) Acute inflammatory arthritis was induced by intra-articular injection of mBSA into the knee joint followed by 3 daily subcutaneous injections of IL-1. Mice were killed on day 7. Frontal hematoxylin and eosin–stained sections through knee joints from arthritic WT (A-B) and SOCS3ΔSB/ΔSB (C-D) mice. E indicates exudate; P, patella; PN, pannus; F, femur; and arrows, increased exudate and inflammatory cells, predominantly neutrophils, in the joint space of treated SOCS3ΔSB/ΔSB mice. (E) Joint sections were graded for 5 features of inflammatory arthritis, each on a scale of 0 (normal) to 5 (severe) by an investigator blinded to the experimental groups. Results are shown as the mean (± SD) for 5 joints per group (*P < .05; **P < .01).
Discussion
SOCS3ΔSB/ΔSB mice, which produce a truncated SOCS3 protein that lacks the C-terminal SOCS box, exhibit exaggerated responses to G-CSF administration and to mBSA/IL-1–induced acute arthritis, a G-CSF–dependent process. This is the first in vivo evidence that SOCS3 inhibits cytokine signaling by 2 mechanisms: the kinase inhibitory region/SH2 domain and the SOCS box.
In vitro cultures revealed both quantitative and qualitative differences between SOCS3ΔSB/ΔSB, SOCS3-null (SOCS3ΔVAV/−), and WT bone marrow cells. In SOCS3ΔSB/ΔSB and SOCS3-null bone marrow, the number of CFCs responsive to G-CSF or IL-6 was increased, as was colony size, and the cells showed increased proliferation in response to G-CSF. The intermediate phenotype of SOCS3ΔSB/ΔSB bone marrow compared with SOCS3-null bone marrow mirrored the quantitative difference in the duration of STAT3 activation in the cells. There were also qualitative differences in the response of SOCS3ΔSB/ΔSB and SOCS3-null bone marrow cells. Whereas colonies arising in SOCS3-null bone marrow cultures stimulated with G-CSF contained macrophages in addition to granulocytes, this skewing of differentiation was not observed in SOCS3ΔSB/ΔSB cultures. In addition, synergy between SCF and IL-6 was lost in SOCS3-null bone marrow cultures but was preserved in SOCS3ΔSB/ΔSB cultures. These qualitative differences may reflect a requirement for a different threshold of STAT3 activation for these phenomena or alterations in other pathways, such as the ERK/Ras/MAPK pathway (Figure 2B), in SOCS3-null cells. In SOCS3-null ES cells, alterations in ERK/Ras/MAPK signaling were shown to direct cell differentiation.48
Mice lacking SOCS3 die around embryonic day 13 due to a placental defect resulting from dysregulated LIF signaling in the placenta.12-14 In contrast, SOCS3ΔSB/ΔSB pups are born in normal numbers and the architecture of the SOCS3ΔSB/ΔSB placentae is normal, indicating that the SOCS box is not required to negatively regulate LIF signaling in the placenta or that the level of regulation is sufficient for normal placentation. The role of SOCS3 during adult life has been studied using mouse strains with conditional deletions in various tissues. Mice with a hematopoietic and endothelial cell–specific deletion of SOCS3 were born viable and healthy but from 17 weeks of age developed a fatal illness characterized by inflammation in the pleural and peritoneal cavities, neutrophil leukocytosis, and infiltration of the liver and lungs by hematopoietic cells of all lineages.10 This phenotype was not observed in SOCS3ΔSB/ΔSB mice, which remained healthy to 16 months of age.
STAT3 tyrosine phosphorylation is prolonged in G-CSF–stimulated SOCS3ΔSB/ΔSB bone marrow cells. Previous mutagenesis studies using overexpression systems found that SOCS3ΔSB can inhibit JAK/STAT signaling as effectively as WT protein.39 This, together with our observation that ERK1/2 phosphorylation is not altered in G-CSF–stimulated SOCS3ΔSB/ΔSB bone marrow, suggests that the interaction of endogenous SOCS3ΔSB with tyrosine 728 of the receptor is preserved. The current models for the mechanism of SOCS3 protein function include direct inhibition of JAK/STAT signaling, regulation of signal transduction adaptor proteins, as well as targeting of signal transduction molecules for degradation through the ECS complex.49-51 The best studied example of the latter mechanism is the targeting of the TELJAK2 oncogenic fusion protein by SOCS1.30-32 It is likely that the altered kinetics of STAT3 activation in response to G-CSF stimulation is due to disruption of SOCS box–mediated degradation of the receptor-activated JAK/STAT complex via the proteasome. Our data also suggest that the SOCS3 SOCS box regulates SOCS3 levels.
In the steady-state, SOCS3ΔSB/ΔSB mice had normal peripheral blood parameters and remained healthy. However, the response of the SOCS3ΔSB/ΔSB mice to G-CSF administration, a treatment that mimics emergency granulopoiesis and therapeutic use of G-CSF, was exaggerated with significantly greater increases in spleen weight, peripheral blood neutrophil counts, and progenitor cell mobilization. Histologic examination revealed increased neutrophil infiltration of tissues. These differences persisted in mice that had received transplants of WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− bone marrow, demonstrating that the altered responsiveness is intrinsic to hematopoietic cells.
Recently, the SOCS3 protein was shown to ubiquitinate the G-CSFR on a membrane proximal lysine, thereby triggering routing of the receptor to lysosomes.52 This process was SOCS box dependent. It is possible that G-CSFR routing may be altered in SOCS3ΔSB/ΔSB mice and that this contributes to the altered in vivo and in vitro responsiveness of these mice to G-CSF.
The realization that cytokine circuits cue the termination of their own signal through the induction of SOCS proteins points to these proteins as candidate players in the initiation and/or propagation of chronic inflammatory diseases. Rheumatoid arthritis is an autoimmune disease characterized by inflammation, synovial hyperplasia, neoangiogenesis, and progressive destruction of cartilage and bone. The etiology is complex, and inflammatory cytokines, including G-CSF, play a central role.47 To model acute inflammation, we used the mBSA/IL-1 model of acute inflammatory arthritis. IL-1 is a major regulator of rheumatoid arthritis and systemic injection of IL-1 in mice converts the transient CD4+ T-lymphocyte–dependent inflammatory reaction to mBSA into a florid monoarthritis. Synovial macrophages, neutrophils, G-CSF, and IL-6 are important mediators of the pathology in the mBSA/IL-1 model.45,47,53 In SOCS3ΔVAV/− mice, mBSA/IL-1–dependent arthritis was markedly exacerbated. The arthritis was characterized by increased numbers of neutrophils in the inflamed synovium, most likely due to hyperresponsiveness to G-CSF and IL-6. SOCS3 was also shown to be an important regulator of CD4+ T lymphocytes and of macrophage and osteoclast generation.54 Adenovirus-mediated transfer of SOCS3 into the ankle joints of mice with collagen-induced arthritis has been shown to reduce the severity of disease.55 In keeping with these data, we found that SOCS3ΔSB/ΔSB mice exhibited an increased inflammatory joint response in the mBSA/IL-1 arthritis model. The model is dependent on G-CSF, and the exacerbated arthritis seen in SOCS3ΔSB/ΔSB mice is most likely due to their enhanced responsiveness to G-CSF. Our experiments have not identified the critical cell types responsible for the increase in acute inflammation observed in SOCS3ΔSB/ΔSB joints and cell transfer, and lineage-specific deletion experiments will be required to dissect this.
Recently, intracellular delivery of SOCS3 has been trialed as an anti-inflammatory agent. Exogenous delivery of SOCS3 to mice increased survival after lipopolysaccharide-induced endotoxic shock and attenuated liver apoptosis and hemorrhagic necrosis.56,57 These data highlight a therapeutic role for SOCS3 in tempering signaling in the immune system by inhibiting inflammatory cytokine circuits in autoimmune and other inflammatory diseases. Increased understanding of the role of the SOCS box in vivo will aid in the design of effective therapies for clinically important G-CSF–dependent processes.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jeff Babon and Sandra Nicholson for helpful discussions. We are grateful for the excellent technical assistance of Sandra Mifsud, Ladina DiRago, Lynne Hartley, Janelle Lochland, and Evelyn Trounson. Dannielle Bennett is thanked for her caring animal husbandry.
This work was supported by grants from the National Institute of Health (CA-22556) and the National Health and Medical Research Council of Australia (Program grants 257500, 461219).
National Institutes of Health
Authorship
Contribution: K.B., P.E., S.R., D.M., T.A.W., A.W.R., and L.R. designed and performed research and analyzed data; I.P.W., D.J.H., N.A.N., and W.S.A. designed research and analyzed data; K.B. and L.R. wrote the paper; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lorraine Robb, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Pde, Parkville 3050, Victoria, Australia; e-mail: robb@wehi.edu.au.

![Figure 3. SOCS3ΔSB/ΔSB bone marrow cells are hyperresponsive to G-CSF. (A) Bone marrow cells (25 000) from WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures with cytokines as indicated and colonies were counted after 7 days. Results represent the mean (± SD) from at least 3 mice of each genotype. (B) To ascertain colony size, cultures were plucked, resuspended, and counted. Results represent the mean (± SD) for at least 3 mice of each genotype. (C) To assess synergy between SCF and other cytokines, 25 000 bone marrow cells from SOCS3+/+, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures stimulated by SCF and/or additional cytokines as indicated and colonies were counted after 7 days. Results for each cytokine are represented as groups containing WT, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− data for the single cytokine, SCF, and the combination of the 2. Results represent the mean (± SD) from at least 3 mice of each genotype. Synergy between SCF and IL-6 was not detected in cultures of SOCS3ΔVAV/− bone marrow cells. (D) Average colony size. Results represent the mean (± SD) for at least 3 mice for each genotype. Again, synergy between SCF and IL-6 was not observed in cultures of SOCS3ΔVAV/− cells. (E-F) Bone marrow cells were stimulated with G-CSF (E) or IL-3 (F) and proliferative activity was assessed after 48 hours by 3[H]-thymidine incorporation. Results represent the mean (± SD) for 2 mice per genotype; analysis of each mouse included 6 replicates (*P < .05; **P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/5/10.1182_blood-2007-03-079178/6/m_zh80170706380003.jpeg?Expires=1768160425&Signature=q~ddXJYnNdNsnh8LjwJxt8XsEknNUencweYj8T2JwDQcBolgL2IeIaH06rQVSZOMuFjuN3rtCZ-v8jU2XBYaI-OAyFVvCJLsFHq0-hHtZOZBBtFdD0~X51gK2feyy9Hdxhq~XB8B80ZcUimG5A1MVVr32lSXxK463qGa~6kHCqcfGSQqI21SyMib~4MkptsDk-QPHw21YS3E94Pnbi5KvhX8t0RowbsNjep7PsPRrSCTcrDn~8TPdu6HSE3tBL5WgFQn4auRYawyK52yXoYdZRzmIAiDDpsW1ZvnBLB5atuhaz4b3iUXa4hxuW2wB38Gw8ldSfG2MfmWP0-7nH6khw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



![Figure 3. SOCS3ΔSB/ΔSB bone marrow cells are hyperresponsive to G-CSF. (A) Bone marrow cells (25 000) from WT, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures with cytokines as indicated and colonies were counted after 7 days. Results represent the mean (± SD) from at least 3 mice of each genotype. (B) To ascertain colony size, cultures were plucked, resuspended, and counted. Results represent the mean (± SD) for at least 3 mice of each genotype. (C) To assess synergy between SCF and other cytokines, 25 000 bone marrow cells from SOCS3+/+, SOCS3ΔSB/ΔSB, or SOCS3ΔVAV/− mice were plated in semisolid agar cultures stimulated by SCF and/or additional cytokines as indicated and colonies were counted after 7 days. Results for each cytokine are represented as groups containing WT, SOCS3ΔSB/ΔSB, and SOCS3ΔVAV/− data for the single cytokine, SCF, and the combination of the 2. Results represent the mean (± SD) from at least 3 mice of each genotype. Synergy between SCF and IL-6 was not detected in cultures of SOCS3ΔVAV/− bone marrow cells. (D) Average colony size. Results represent the mean (± SD) for at least 3 mice for each genotype. Again, synergy between SCF and IL-6 was not observed in cultures of SOCS3ΔVAV/− cells. (E-F) Bone marrow cells were stimulated with G-CSF (E) or IL-3 (F) and proliferative activity was assessed after 48 hours by 3[H]-thymidine incorporation. Results represent the mean (± SD) for 2 mice per genotype; analysis of each mouse included 6 replicates (*P < .05; **P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/5/10.1182_blood-2007-03-079178/6/m_zh80170706380003.jpeg?Expires=1768285026&Signature=mczJd7IPDliKDJQ4EudJm5lcsakBjoZByR-lkuv~EdX307mdYWMC2bDPmnQz2oPkJVUIKQswb2p7syMSVuNLslgn96W2RiXn700cNxzbaglZ8-ShA4sHxH5xmdPi~gbLVu-IyYR~wo-M74qAo1IJgAnJh0KcQv8-GB6A-PxDvOKJ6ajDU2De7o2du5DvyYc9jMYAa5B7jEnvHk2JDrVzU-3K01v4GC80MBE61nP4j13h3bmnfVxuho10W8vDJqtmnsRrnvLKQ8oRT16ztrUFAqpeLBzoJGJ8YuBGUiYD7W9YJOhgoMyfLrNXSciTdGVA2q2VAZ46o1~O9BbjRuPoWQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)