Abstract
Spontaneous Rh phenotype alteration interferes with pretransfusion and prenatal blood group examinations and may potentially indicate hematologic disease. In this study, the molecular background of this biologic phenomenon was investigated. In 9 patients (3 with hematologic disease), routine RhD typing showed a mixture of D-positive and D-negative red cells not attributable to transfusion or hematopoietic stem-cell transplantation. In all patients, congenital and acquired chimerism was excluded by microsatellite analysis. In contrast to D-positive red cells, D-negative subpopulations were also negative for C or E in patients genotyped CcDdee or ccDdEe, respectively, which suggested the presence of erythrocyte precursors with an apparent homozygous cde/cde or hemizygous cde/— genotype. Except for one patient with additional Fyb antigen anomaly, no other blood group systems were affected. RH genotyping of single erythropoietic burst-forming units, combined with microsatellite analysis of blood, different tissues, sorted blood cell subsets, and erythropoietic burst-forming units, indicated myeloid lineage–restricted loss of heterozygosity (LOH) of variable chromosome 1 stretches encompassing the RHD/RHCE gene loci. Fluorescent in situ hybridization studies indicated that LOH was caused by either somatic recombination or deletion. Therefore, most cases of spontaneous Rh phenotype splitting appear to be due to hematopoietic mosaicism based on LOH on chromosome 1.
Introduction
The Rh blood group antigens are exposed on the RhD (CD240D) and RhCcEe (CD240CE) polypeptides encoded by the closely linked RHD and RHCE genes, respectively, on the short arm of chromosome 1 (1p36.2-p34).1 Especially the highly immunogenic D (RH1) antigen carried by the RhD polypeptide is of clinical relevance for transfusion medicine and prenatal management, because anti-D alloimmunization in D-negative individuals may be responsible for severe hemolytic transfusion reactions and is a major cause of hemolytic disease of the newborn.2 In whites, a deletion of the whole RHD gene (RHd) is the usual basis for an RhD-negative phenotype.3 To minimize the risk associated with exposure of D-negative individuals to D-positive red blood cells (RBCs), unambiguous RhD typing results are a prerequisite of pretransfusion and prenatal investigations
Occasionally, in some individuals the simultaneous presence of D-positive and D-negative RBCs is identified by a mixed-field agglutination pattern in routine RhD typing that is most frequently due to previous D-mismatched RBC transfusions or hematopoietic stem-cell transplantation (HSCT). However, in certain cases, a mixed RhD phenotype is observed to occur spontaneously even in the absence of transfusion or transplantation. Known reasons for such findings include hematopoietic or whole-body chimerism (presence of cells from 2 or more zygotes), mosaicism (presence of cells of different phenotypes derived from one zygote), and somatic RHD mutation.4–7 Recently, also mosaicism involving a cell line with uniparental isodisomy for chromosome 1 was described as a reason for ambiguous RhD typing.8
Many individuals with spontaneous Rh phenotype splitting or progressive Rh antigen loss were found to suffer from hematologic disease, such as acute or chronic myelogenous leukemia, myeloproliferative disease, or myelodysplastic syndrome, in the majority of cases without detectable cytogenetic abnormalities of chromosome 1.2,9–15 However, spontaneous appearance of D-negative RBCs was also observed in originally D-positive healthy subjects or patients with nonhematologic disease.11,12,16,17 A potentially serious risk is posed by blood donors with mixtures of D-positive and D-negative RBC subsets erroneously typed D-negative,18 as their RBC units were demonstrated to induce alloanti-D in D-negative recipients.19
In this study, the molecular background of spontaneous Rh phenotype splitting in 9 patients with hematologic as well as nonhematologic diseases was investigated. In this series of patients, thus far the largest, hematopoietic mosaicism was identified as a principal reason for RBC phenotype anomalies. For the first time, molecular genetic in vivo evidence is provided to suggest loss of heterozygosity (LOH) along variable chromosome 1 stretches encompassing the RHD/RHCE gene loci as the most prevalent mechanism for Rh mosaicism. The clinical significance of this phenomenon with regard to anti-D alloimmunization and as potential marker for hematologic disease is discussed.
Patients, materials, and methods
Patients
Nine patients (all white; 4 male, 5 female; median age, 63 years) without a recent history of transfusion or HSCT showing mixed-field agglutination in routine serologic RhD typing were included into this study. After institutional review board approval by the Clinic of Blood Group Serology, Medical University of Vienna, informed consent was obtained in accordance with the Declaration of Helsinki. Patients' characteristics are shown in Table 1.
Serologic blood group typing
Serologic blood group typing, antierythrocyte antibody screening, and direct antiglobulin testing were carried out by gel centrifugation technique using DiaMed (Cressier, Switzerland) reagents and equipment, as described previously.20 For Duffy (Fy, CD234) blood group typing, polyclonal human anti-Fya and anti-Fyb (Immucor, Rödermark, Germany) concentrated by adsorption onto antigen-positive group O RBCs followed by acid elution (DiaCidel; DiaMed) were used in the indirect antiglobulin test in gel cards (DiaMed). Partial D identification was performed by using 2 commercially available monoclonal anti-D antibody panels (D-Screen; Diagast, Loos, France; and ID-Partial RhD-Typing Set; DiaMed).
Red cell flow cytometry
Absolute D antigen densities of the D-positive RBC subpopulations from patients and of control RBC samples were determined by flow cytometry (FACSCalibur with CellQuest acquisition software; BD Biosciences, San Jose, CA) after indirect immunofluorescence staining with 11 different monoclonal anti-D antibodies exactly as described.21 Adequate fluorescence marker placements allowed for the quantification of D-positive RBC fractions of patient blood samples.
For separate analysis of D-positive and D-negative RBC subpopulations with respect to expression of Rh antigens, Rh complex molecules (LW and Rh-associated glycoprotein), and Fyb antigen, a double-color immunofluorescence approach was used. First, RBCs were incubated with human polyclonal anti-C (Molter, Neckargemünd, Germany), anti-c (DiaMed), anti-e (Molter), anti-Fyb (Immucor), or human monoclonal anti-E (clone 907; Diagast). All polyclonal sera had been prepared for use by repeat adsorption onto antigen-positive group O RBCs (Ccddee, ccddee, and CcDdee RBCs with anti-C, anti-c, and anti-e, respectively, and Fy[a-b+] RBCs with anti-Fyb), washing, and subsequent acid elution.22 Alternatively, murine monoclonal antibodies against LW (clone BS46, provided by Manfred Ernst, Biotest, Dreieich, Germany), Rh-associated glycoprotein (RhAG; 2D10, donated by Leo de Jong, Sanquin, Amsterdam, the Netherlands), or total Rh polypeptide (using anti-Rh recognizing both RhD and RhCcEe polypeptides; Bric69, International Blood Group Reference Laboratory [IBGRL], Bristol, United Kingdom) were used. After a washing step, R-phycoerythrin (PE)–conjugated goat anti–human IgG F(ab′)2 (Immunotech, Marseille, France) or rabbit anti–mouse IgG F(ab′)2 (Dako Cytomation, Glostrup, Denmark) was used as secondary reagent.23 After washing, RBC samples were additionally labeled with fluorescein isothiocyanate (FITC)–conjugated human monoclonal anti-D (clone Brad3, IBGRL), washed, and subjected to flow cytometric analysis.
Sorting of nucleated cell subsets from peripheral blood
Cell subsets of anticoagulated blood samples were quantified by 4-color FACSCalibur flow cytometry after isotonic RBC lysis with ammonium chloride buffer and dual platform cell staining.24 For cell sorting, 3-laser FACSAria (BD Biosciences) equipment with DIVA software (BD Biosciences) was used. Two vials with 106 leukocytes each were prepared per blood sample, and sorted using the following fluorescent dye and murine monoclonal antibody combinations: Syto41 (Molecular Probes, Eugene, OR), anti-CD8 FITC (clone DK25; Dako Cytomation), anti-CD56 PE (clone NCAM16.2; BD Biosciences), anti-CD3 PE Texas Red (clone UCHT1; PharMingen, San Diego, CA), anti-CD45 peridin chlorophyll protein (PerCP; clone 2D1; BD), anti-CD4 PE-cyanin7 (clone SK3; BD), anti-CD71 allophycocyanine (APC; clone L01.1; BD Biosciences), and anti-CD14 APC-cyanin7 (clone MoP9; BD) for CD4+, CD8+, natural killer cells, and normoblasts; and Syto41, anti-CD15 FITC (clone C3D1; Dako Cytomation), anti-CD33 PE (clone P67.6; BD), anti-CD45RA PE Texas Red (clone 2H4; Coulter, Miami, FL), anti-CD45 PerCP, anti-CD34 PE-cyanin7 (clone 8B12; BD), and anti-CD19 APC (clone SJ25C1; BD) for CD34+ cells, monocytes, granulocytes, and B cells. The purity of sorted cell subsets generally exceeded 98%.
Erythropoietic burst-forming unit cultures
Cultures for erythropoietic burst-forming units (BFU-Es) were performed exactly as described elsewhere.25 After cultivation for 2 to 3 weeks, all colonies were scored according to standard criteria.26 Single BFU-E colonies were picked using sterile Pasteur pipettes and resuspended in 100 μL PBS in 0.5-mL microcentrifuge tubes for subsequent DNA isolation.
DNA isolation
Genomic DNA from anticoagulated blood was extracted with the GenoPrep Cartridge B 350 on a GenoM-6 instrument (GenoVision, Vienna, Austria), with Chelex27 or with Nucleon BACC2 reagents (Amersham, Buckinghamshire, United Kingdom). DNA isolation from single BFU-E colonies was performed with BACC2 reagents, with adjusted reagent volumes (100 μL buffer B, 25 μL sodium perchlorate, 100 μL chloroform, and 30 μL resin suspension). Precipitated DNA from BFU-Es was resuspended in 12 μL water. DNA from buccal swabs, hair samples, and sorted peripheral blood cells was extracted with Chelex; DNA from finger nails, with the Qiamp DNA Mini Kit (Qiagen, Valencia, CA).
Molecular blood group genotyping
For RHD and RHCE genotyping, testing for variant RHD alleles and RHD zygosity of all patient and all RH genotype–matched control blood samples, commercially available typing kits (CDE, RHd; Innotrain, Kronberg, Germany) were used employing polymerase chain reaction (PCR) technique with sequence-specific priming (SSP).21 Duffy genotyping of patient 5 was performed as described.28
The C, c, and e alleles of RHCE were detected from DNA isolated from single BFU-E colonies with sequence-specific monoplex real-time PCR using primers, probes, and real-time PCR reagents as previously described,29 with a modification of the cycle protocol for increased sensitivity. Because very low DNA concentrations were observed, the internal control was replaced by an external real-time PCR using primers and probe specific for the single copy gene beta globin on the same PCR plate.30 Primers and probes for RHD exons 5 (patient 6) and 7 (patients 2 and 3) were used as recently described.31 Reaction conditions were 95°C for 10 minutes, 15 2-step cycles with 64°C for 40 seconds and 94°C for 10 seconds followed by 35 2-step cycles with 60°C for 40 seconds and 94°C for 10 seconds. An ABI 7300 Sequence Detection System (Applied Biosystems, Foster City, CA) was used. BFU-E samples of patient 7 were genotyped by PCR-SSP using primers and reaction conditions for RHD (exon 7), RHCE*E, and RHCE*e, as described previously.32,33 Since the amount of DNA per BFU-E colony was expected to be very low, only 3 specificities per sample were tested in a 35-cycle PCR protocol. Positive amplification was generated with 2 separate primers in each PCR, giving rise to a 434-bp human growth hormone product as described previously.32
Microsatellite analysis
To test for congenital or acquired chimerism, multiplex-PCR of 15 highly polymorphic autosomal short tandem repeat loci (D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818, FGA) and amelogenin was performed on DNA samples from blood using the AmpFlSTR IDentifiler PCR Amplification Kit (Applied Biosystems).
DNA samples from whole blood, nucleated blood cell subsets, fingernails, eyebrows, buccal swabs, or BFU-E colonies were studied with a series of polymorphic dinucleotide microsatellite marker systems located on chromosome 1 with human mapping primers D1S468, D1S507, D1S2697, D1S2644, D1S199, D1S2864, D1S233, D1S2890, D1S2635, and D1S2836 (Applied Biosystems) according to the manufacturer's instructions In case of samples with low DNA concentration, the sample volume was increased up to 6-fold and the number of cycles were increased by 3. In patient 5, 8 additional markers (D1S2841, D1S206, D1S252, D1S498, D1S218, D1S238, D1S249, D1S2833) were investigated. The instrument platforms Geneamp PCR System 9700 and ABI Prism 310 Genetic Analyser (Applied Biosystems) were used for amplification and further analysis.
Fluorescent in situ hybridization analyses
Dual-color fluorescent in situ hybridization (FISH) analyses on methanol/acetic acid–fixed peripheral blood cells obtained from 6 patients (1, 2, 3, 5, 6, and 9) were performed as described.34 P1-based artificial chromosome (PAC) clones RP11-335G20 (accession no. AL928711; kindly provided by Mariano Rocchi, Insitituto di Genetica, Bari, Italy) and RP11-316M1 (Research Genetics, Huntsville, AL) that encompass the RHD/RHCE and AF1q gene loci, respectively, were used. Additional analyses were conducted in patient 6 with probes specific for the 1q and 1p subtelomeres as well as chromosome 1 and 7 centromeres. At least 200 cells per patient were scored and the signal patterns recorded separately for segmented and round nuclei.
Results
Qualitative and quantitative properties of D-positive and D-negative red cell subsets
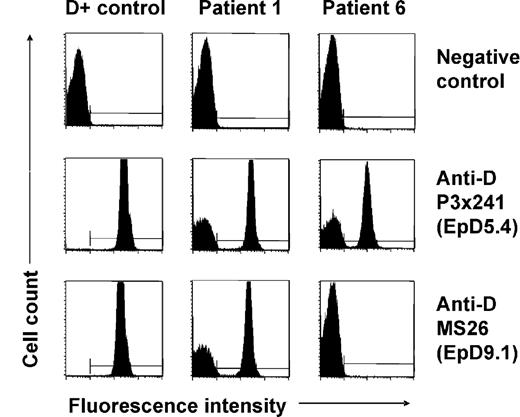
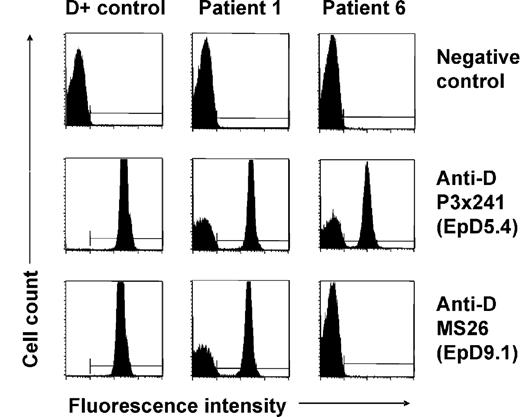
None of the patients (Table 1) with spontaneous mixed RhD phenotype identified by routine RhD typing had a history of transfusion or HSCT. In 5 patients (patients 1, 2, 3, 7, and 8), previous RhD typing performed at least 19 months before recognition of D antigen loss had shown normal RhD-positive phenotypes; in the other 4 patients no earlier typing data were available. At initial presentation, mixed-field agglutination was evident using different monoclonal anti-D reagents in plate, tube, as well as gel centrifugation technique. Flow cytometry with indirect immunofluorescence staining with 11 different monoclonal anti-D antibodies confirmed the simultaneous presence of D-positive and D-negative RBCs in all patients. Representative results for 2 anti-D clones (P3 × 241 and MS26) are shown in Figure 1. Interestingly, patient 6 displayed signs of D epitope loss, as some but not all of the anti-D clones were reactive with his D-positive RBC subsets (Figure 1). Analysis of the D epitope expression pattern35 as well as RHD/RHCE genotyping led to the recognition of an inherited partial RhD variant, D category IV type 4 (genotype CcDvardee), in this patient. All other patients showed normal RHD-positive genotypes (7 CcDdee and 1 ccDdEe, Table 1). The patients were all heterozygous for the presence of a hybrid Rhesus box and, therefore, apparently hemizygous for RHD. The individual percentages of D-positive RBCs and the absolute D antigen densities of these subsets as determined by flow cytometry are shown in Table 1. All D antigen densities were found to be equivalent to RH genotype–matched controls.
Flow cytometric analysis of red cell D antigen expression of patients with spontaneous mixed RhD phenotype. Representative immunofluorescence histograms of RBCs of a D-positive (CcDdee) control and of patients 1 and 6 indirectly stained with 2 different monoclonal anti-D antibodies (P3x241 and MS26) are shown. Note D-positive and D-negative RBC subpopulations in patient 1. Similar results were obtained with further 9 monoclonal anti-D antibodies, and samples of patients 2, 3, 4, 5, 7, and 9. In comparison, a proportion of RBCs of patient 6 was reactive only with some but not all anti-D clones (see nonreactivity with clone MS26) indicating the absence of certain D epitopes (EpD) from his D-positive cells; the D epitope expression pattern was characteristic of a D category IV blood group variant.35
Flow cytometric analysis of red cell D antigen expression of patients with spontaneous mixed RhD phenotype. Representative immunofluorescence histograms of RBCs of a D-positive (CcDdee) control and of patients 1 and 6 indirectly stained with 2 different monoclonal anti-D antibodies (P3x241 and MS26) are shown. Note D-positive and D-negative RBC subpopulations in patient 1. Similar results were obtained with further 9 monoclonal anti-D antibodies, and samples of patients 2, 3, 4, 5, 7, and 9. In comparison, a proportion of RBCs of patient 6 was reactive only with some but not all anti-D clones (see nonreactivity with clone MS26) indicating the absence of certain D epitopes (EpD) from his D-positive cells; the D epitope expression pattern was characteristic of a D category IV blood group variant.35
None of the patients had irregular antierythrocytic antibodies or a positive direct antiglobulin test. The median transfusion-free follow-up of the studied patients was 33 months (range, 5 to 65 months). On repeat serologic re-evaluation, 5 patients presented a stable mixed-field agglutination pattern over time, 2 patients exhibited progressively diminishing proportions of D-positive RBCs, and 2 showed complete D antigen loss during their observation period (Table 1).
Rh antigen and Rh complex expression profiles suggest an apparently Rh haplotype–dependent phenomenon
Extended blood group phenotyping using gel centrifugation technique additionally argued against transfusion or HSCT as cause for the ambiguous RhD typing results: in 8 patients, mixed-field agglutination was restricted to Rh system antigens, whereas typing of other blood group antigens yielded unequivocal results (Table 1). Only in patient 5, a mixed Fyb (FY2) phenotype was also noticed (Table 1; Figure 2B), despite her apparently normal FY*A/FY*B genotype. This is remarkable because not only the RH but also the Duffy blood group-encoding DARC gene is located on chromosome 1 (1q22-q23).36 Importantly, the RhD phenotype splitting was generally accompanied by mixed C (RH2) or E (RH3) antigen typing results in CcDdee and ccDdEe individuals, respectively (Table 1).
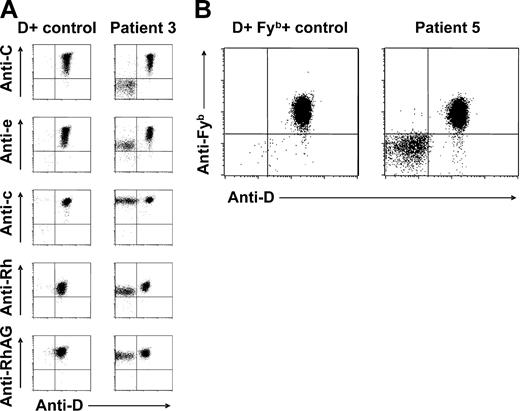
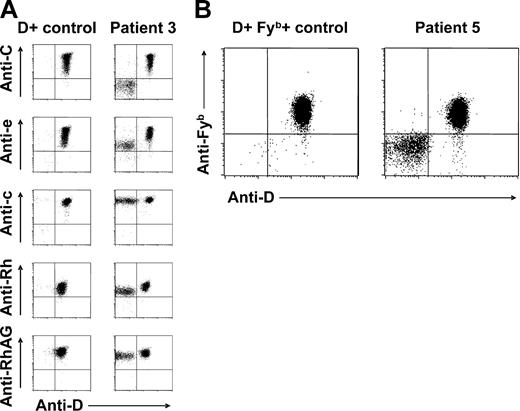
Red cell Rh antigen, Rh complex molecule, and Fyb antigen expression of patients with spontaneous mixed RhD phenotype. (A) Double-color flow cytometric analyses of Rh antigen D, C, e, and c, total Rh protein, and RhAG expression of RBC samples from patient 3 and a D-positive (CcDdee) control are shown. Note presence of C+c+D+e+ and C−c+D−e+ RBC subpopulations. Similar results were obtained with samples from patients 1, 2, 4, and 6. (B) Double-color flow cytometric analysis of D and Fyb antigen expression of patient 5 and a CcDdee, Fy(a+b+) control. Only the D-positive red cell subset of this patient is also Fyb positive.
Red cell Rh antigen, Rh complex molecule, and Fyb antigen expression of patients with spontaneous mixed RhD phenotype. (A) Double-color flow cytometric analyses of Rh antigen D, C, e, and c, total Rh protein, and RhAG expression of RBC samples from patient 3 and a D-positive (CcDdee) control are shown. Note presence of C+c+D+e+ and C−c+D−e+ RBC subpopulations. Similar results were obtained with samples from patients 1, 2, 4, and 6. (B) Double-color flow cytometric analysis of D and Fyb antigen expression of patient 5 and a CcDdee, Fy(a+b+) control. Only the D-positive red cell subset of this patient is also Fyb positive.
According to double-color flow cytometry, the D-positive RBC subpopulations were also positive for C or E, whereas D-negative RBC subsets were generally negative for C or E, in CcDdee or ccDdEe individuals, respectively (Figure 2A). In all studied patients, D-negative RBCs were found to express c (RH4) and e (RH5) antigens (Figure 2A). In patient 5, only D-positive but not D-negative RBCs were positive for the Fyb antigen (Figure 2B), whereas all RBCs typed Fya positive (data not shown). In all patients, all RBCs expressed RhAG (CD241) (Figure 2A), excluding a possible regulatory role of this molecule in Rh phenotype splitting.37 In light of the known strong haplotypic association of both C and E with D,2 these findings were suggestive of an underlying RHD/RHCE haplotype-specific phenomenon: patients genotyped CcDdee had a mixture of C+c+D+E−e+ and C−c+D−E−e+ RBCs apparently deriving from 2 different stem-cell lines with CDe/cde and cde/cde (or possibly cde/—) genotypes, respectively. Analogously, patient 7 (ccDdEe) had 2 circulating RBC subsets with Rh antigen expression compatible with cDE/cde and cde/cde (or cde/—) genotypes.
Exclusion of congenital or acquired chimerism
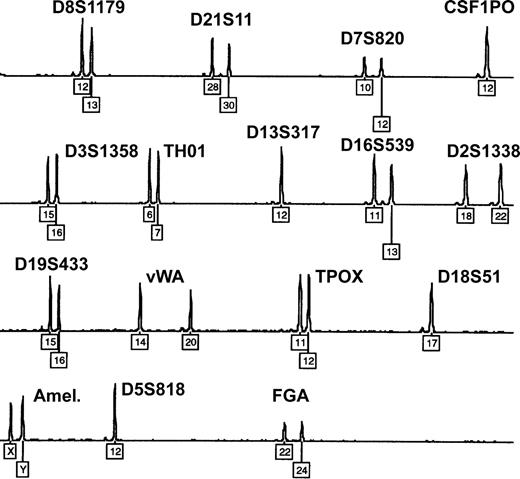
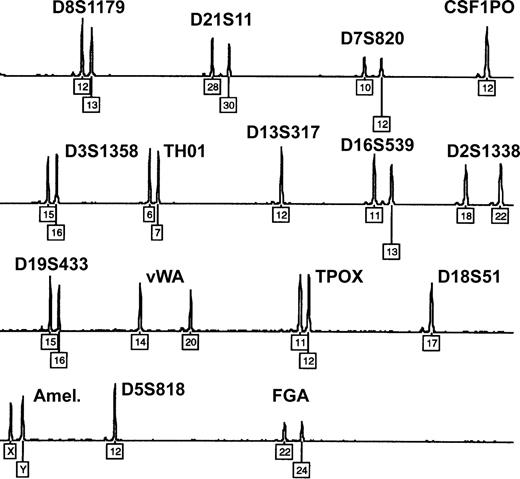
Twin, dispermic, or artificial chimerism is known as possible reason for mixed blood group phenotypes.4,38 However, all 9 patients denied having a twin, and none had a history of transplantation. Moreover, microsatellite analysis of whole-blood DNA samples (15 different loci located on chromosomes 2-8, 11-13, 16, 18, 19, and 21) ruled out congenital or acquired chimerism in each of the studied patients: exclusively homozygous or well-balanced heterozygous allelic peaks were found, with a maximum of 2 alleles present at each locus (Figure 3). In case of chimerism, a mixed DNA profile showing up to 3 or 4 alleles and extremely unbalanced heterozygous peaks would be expected.39
Exclusion of congenital or acquired chimerism by microsatellite marker analysis. Representative electropherogram showing the blood DNA profile of patient 1 after multiplex-PCR of 15 highly polymorphic autosomal short tandem repeat loci and amelogenin (Amel.). Numbers denote allelic designations of individual loci. No additional allelic peaks and only well-balanced heterozygous peaks are observed. Similar results were obtained with samples from the other 8 patients.
Exclusion of congenital or acquired chimerism by microsatellite marker analysis. Representative electropherogram showing the blood DNA profile of patient 1 after multiplex-PCR of 15 highly polymorphic autosomal short tandem repeat loci and amelogenin (Amel.). Numbers denote allelic designations of individual loci. No additional allelic peaks and only well-balanced heterozygous peaks are observed. Similar results were obtained with samples from the other 8 patients.
Demonstration of RH genotype splitting by molecular analysis of single erythroid progenitor cells
In patients 2, 3, 6, and 7, the collection of separate BFU-E colonies allowed for DNA isolation from single erythroid progenitor cells. Real-time PCR genotyping for RHD, RHCE*C, RHCE*c, and RHCE*e was performed in CcDdee patients, and PCR-SSP genotyping for RHD, RHCE*E, and RHCE*e in the ccDdEe patient. BFU-E DNA samples negative for RHD also typed negative for RHCE*C and RHCE*E in CcDdee and ccDdEe individuals, respectively, but were generally positive for RHCE*e (and confirmed RHCE*c positive in CcDdee patients). On the other hand, RHD-positive samples of patient 7 (ccDdEe) were also positive for RHCE*E and RHCE*e. In CcDdee patients, all RHD-positive samples typed positive also for RHCE*C, RHCE*c, and RHCE*e (Table 2). These findings support the notion of 2 RBC lines, one of “complete” Rh phenotype encoded by 2 parental RH haplotypes (CDe/cde or cDE/cde) and a second with an “incomplete” Rh phenotype encoded by the RHD-negative parental RH haplotype only (homozygous cde/cde or hemizygous cde/—).
Rh phenotype splitting associated with loss of heterozygosity on chromosome 1
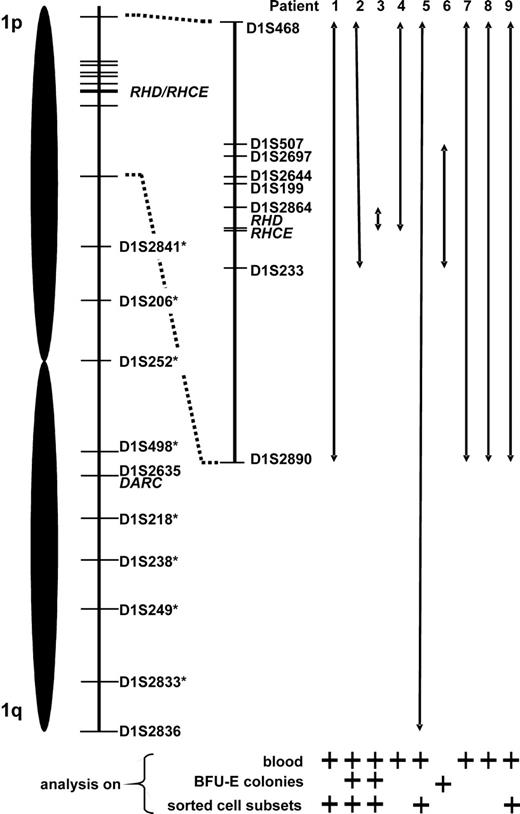
DNA samples extracted from whole blood and different tissues (buccal swabs, finger nails, eyebrows) were subjected to analysis of 10 microsatellite markers coded for by chromosome 1. At some microsatellite markers flanking the RHD/RHCE loci, remarkably low peak height ratios of the 2 heterozygous peaks (height of lower peak divided by height of higher allele) were observed in whole blood DNA samples, which contrasted to generally normal values obtained with hair samples (Table 3). This phenomenon could be explained by a mixture of heterozygous and apparently homozygous (possibly hemizygous) peaks, which had only one allele (high peak) in common. This common allele appeared to be much higher than the unshared allele (low peak) of the heterozygous cell line and resulted in a strong peak imbalance and a low peak height ratio. Judging from the peak height ratios of blood compared with hair samples, the extent of LOH on chromosome 1 differed among patients (Table 3).40 In patient 3 it was confined to the D1S2864 locus located 2.8 Mb telomeric of RHD, in patients 1, 7, 8, and 9 it encompassed all informative loci on the short arm of chromosome 1 (spanning an interval of at least 54.1 Mb, from 21.9 Mb telomeric to 32.2 Mb centromeric of RHD), in patient 4 it included all loci telomeric of RHD (at least 21.9 Mb), whereas in patient 2 the alteration included also the D1S233 locus centromeric of RHD (at least 7.7 Mb). In patient 5, the unusual peak imbalance apparently spanned all informative chromosome 1 loci affecting also the DARC locus on the long arm of this chromosome (mixed Fyb phenotype of this subject, Table 1). Affection of the entire chromosome 1 in this patient was confirmed by analyzing additional 8 microsatellite markers (D1S2841, D1S206, D1S252, D1S498, D1S218, D1S238, D1S249, D1S2833 at a distance of 79.26, 99.95, 117.36, 149.57, 172.77, 186.41, 203.98, and 228.81 Mb, respectively, from the p-telomeric end): all but 2 (D1S252 and D1S238) were informative and showed analogous results (data not shown). Importantly, the analysis of whole blood DNA samples of patients 7 and 8 with progressive loss of D expression revealed decreasing peak height ratios at all affected microsatellite loci over time (Table 3), demonstrating the analytical power of the applied technique.
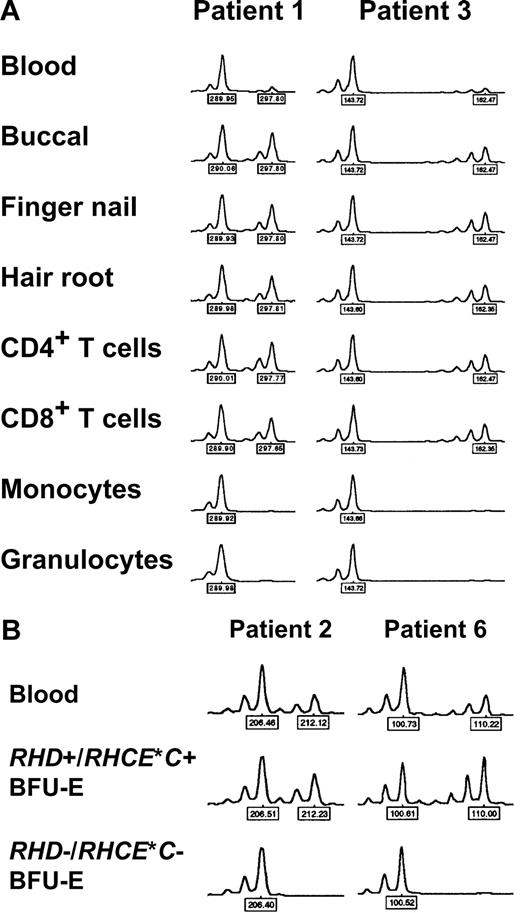
Detection of myeloid lineage–restricted loss of heterozygosity on chromosome 1
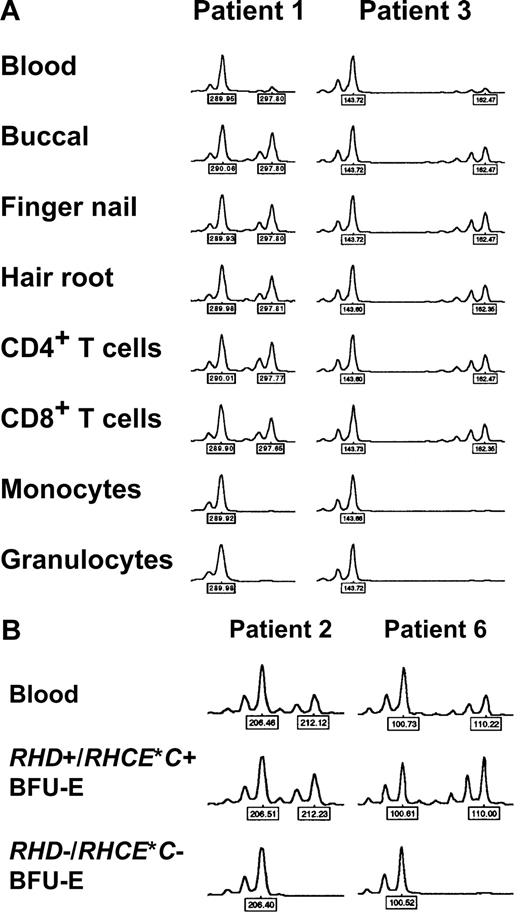
In 5 patients, DNA samples from different peripheral blood cell subsets were investigated. In 4 cases (patients 1, 2, 3, and 9), a clear-cut difference between myeloid and lymphoid cells was evident, with extremely low relative peak height ratios obtained with myeloid cells but not lymphocytes (Figure 4A; Table 4). Since RBCs derive from myeloid progenitor cells, these findings could explain loss of Rh expression in a proportion of mature RBCs via loss of one RH haplotype (CDe or cDE, depending on the individual patient's genotype) in a percentage of stem cells. In contrast, in patient 5 no difference between myeloid and lymphocyte subsets was observed; in addition, relative peak imbalances could be detected all over chromosome 1 (Table 4) as already shown by analyzing DNA of blood from this patient (Table 3).
Loss of heterozygosity (LOH) on chromosome 1 in patients with spontaneous Rh phenotype splitting. (A) Myeloid lineage–restricted LOH on chromosome 1. Representative electropherograms of the D1S2697 and D1S2864 microsatellite markers with DNA samples from blood, different tissues, and sorted blood cell subsets of patient 1 and 3, respectively, are shown. Note pronounced peak imbalance in blood, and apparent LOH in myeloid cell subsets. Similar results were obtained with samples of patients 1, 2, and 9 and further microsatellite markers (as indicated by ‡ in Table 3). (B) LOH on chromosome 1 in RHD−/RHCE*C− (only RHCE*c+/RHCE*e+) but not in RHD+/RHCE*C+ (and additionally RHCE*c+/RHCE*e+) erythropoietic blast-forming units (BFU-Es). Representative electropherograms of the D1S233 and D1S199 microsatellite markers with DNA samples from blood and single BFU-Es of patient 2 and 6, respectively, are shown. Similar observations were made with samples of patients 2, 3, and 6 and with other microsatellite markers (all markers showing LOH as given in Table 5). Peak heights represent fluorescence intensity; numbers denote relative fragment size (bp).
Loss of heterozygosity (LOH) on chromosome 1 in patients with spontaneous Rh phenotype splitting. (A) Myeloid lineage–restricted LOH on chromosome 1. Representative electropherograms of the D1S2697 and D1S2864 microsatellite markers with DNA samples from blood, different tissues, and sorted blood cell subsets of patient 1 and 3, respectively, are shown. Note pronounced peak imbalance in blood, and apparent LOH in myeloid cell subsets. Similar results were obtained with samples of patients 1, 2, and 9 and further microsatellite markers (as indicated by ‡ in Table 3). (B) LOH on chromosome 1 in RHD−/RHCE*C− (only RHCE*c+/RHCE*e+) but not in RHD+/RHCE*C+ (and additionally RHCE*c+/RHCE*e+) erythropoietic blast-forming units (BFU-Es). Representative electropherograms of the D1S233 and D1S199 microsatellite markers with DNA samples from blood and single BFU-Es of patient 2 and 6, respectively, are shown. Similar observations were made with samples of patients 2, 3, and 6 and with other microsatellite markers (all markers showing LOH as given in Table 5). Peak heights represent fluorescence intensity; numbers denote relative fragment size (bp).
Loss of heterozygosity on chromosome 1 in RHD-negative but not RHD-positive erythropoietic blast-forming units
DNA samples extracted from BFU-E colonies of 3 CcDdee patients were further investigated. Because of the limited DNA quantities, only selected informative microsatellite loci were studied. In all studied BFU-E colonies genotyped RHD+/RHCE*C+/RHCE*c+/RHCE*e+, exclusively well-balanced heterozygous peaks were found, indicative of normal unmixed DNA profiles. In contrast, in RHD−/RHCE*C− but RHCE*c+/RHCE*e+ colonies, LOH was evident by the presence of only one allele (Figure 4B) at exactly those microsatellite markers with low peak height ratios in whole blood (Table 3). Informative loci with heterozygosity in only RHCE*c+/RHCE*e+ BFU-E colonies confined the region of LOH on chromosome 1 (Table 5). These data indicated a causal relationship between the RH haplotype loss detected by real-time PCR and the LOH at chromosome 1 segments very probablyencompassing the RHD/RHCE loci. Hence, LOH on chromosome 1 appeared to be responsible for the obvious haplotype-dependent Rh antigen deficiency of a proportion of RBCs. Figure 5 summarizes the proposed minimum extension of partial LOH on chromosome 1 in individual patients.
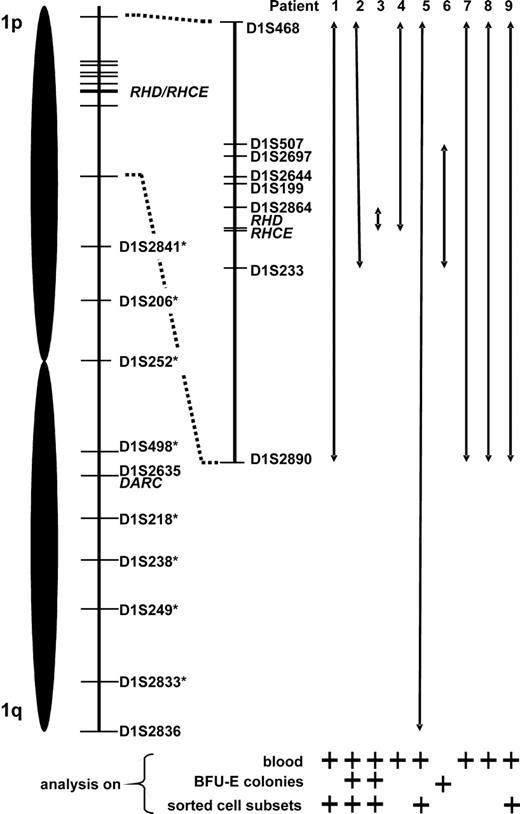
Proposed minimum expansion of loss of heterozygosity (LOH) on chromosome 1. Relative positions of the studied microsatellite markers as well as of the RHD/RHCE and DARC gene loci are depicted. Chromosome 1 stretches apparently affected by LOH are represented by double arrows. This data compilation is based on results of microsatellite analysis of blood, erythropoietic blast-forming units (BFU-Es), and sorted blood cell subsets; for each patient, lines of evidence are specified below the arrows. In patient 5, LOH appeared to span the entire chromosome 1 (markers with an asterisk were analyzed only in patient 5).
Proposed minimum expansion of loss of heterozygosity (LOH) on chromosome 1. Relative positions of the studied microsatellite markers as well as of the RHD/RHCE and DARC gene loci are depicted. Chromosome 1 stretches apparently affected by LOH are represented by double arrows. This data compilation is based on results of microsatellite analysis of blood, erythropoietic blast-forming units (BFU-Es), and sorted blood cell subsets; for each patient, lines of evidence are specified below the arrows. In patient 5, LOH appeared to span the entire chromosome 1 (markers with an asterisk were analyzed only in patient 5).
Spontaneous Rh phenotype splitting caused by either somatic recombination-associated duplication or deletion
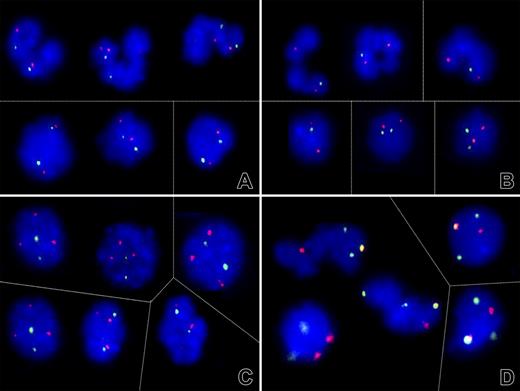
To determine whether the D-negative cell clone resulted from a simple hemizygous deletion or a more complex somatic recombination-associated duplication of the cde haplotype, dual-color FISH analyses on fixed peripheral blood cells obtained from 6 patients (1, 2, 3, 5, 6, and 9) were performed. In 5 of 6 cases, FISH analysis confirmed the diploid presence of the RH loci in all cells (Figure 6A), whereas in patient 3 (with the smallest region of LOH) only one signal was found in 88% segmented and 14% round nuclei (Figure 6B). These results are consistent with the view that somatic recombination-associated loss of the D-positive and duplication of the D-negative allele is more common than a simple deletion alone. The incidental detection of 3 AF1q signals in 75% of nuclei from patient 6 (who suffered from MDS that subsequently transformed into AML) prompted further investigations, which revealed also 3 chromosome 1 (green) and 2 chromosome 7 (red) centromeres with one colocalization (Figure 6C-D). Together with the 2 1p and 3 1q subtelomere signals (data not shown), this FISH pattern defines a der(1,7)(q10;p10), a nonrandom chromosome rearrangement that is highly specific for a particular subset of neoplastic myeloid disorders.41,42 Although these findings corroborate a potential biologic link between the phenomenon described herein and the recently documented acquired partial uniparental disomies in AML, the small homozygous stretch surrounding the RHD/RHCE gene region in this instance also implied that its underlying somatic recombination event was not directly linked with the one generating the chromosomal abnormality.41,43,44
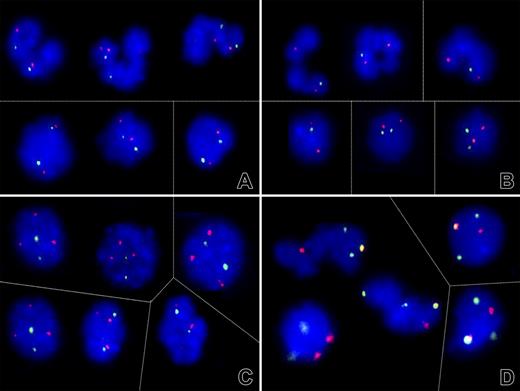
Evidence for different mechanisms of loss of heterozygosity on chromosome 1. (A-C) Dual-color fluorescent in situ hybridization signal patterns of selected cell nuclei obtained with PAC clones that encompass the RHD/RHCE (FITC, green) and, as a control, AF1q gene sequences (Cy3, red). (A) As shown here for patient 9, both the segmented (top) and round (bottom) nuclei in 4 of 6 analyzed cases contained 2 signals each and, thus, 2 RH gene loci. (B) The lack of one green signal in the majority of segmented nuclei and a minority of round nuclei of patient 3, on the other hand, confirmed a monoallelic deletion of at least the RHD/RHCE gene locus. (C) The 2 green RH and 3 red AF1q signal pattern in patient 6 prompted further investigations that (D) confirmed the presence of 3 chromosome 1 (green) and 2 chromosome 7 (red) centromeres with one colocalization (orange). This pattern reflects the presence of a der (1;7)(q10;p10), a highly specific chromosome abnormality in particular subtypes of myeloid malignancies. Thin lines separate individual images of representative cell nuclei. Signals were recorded after overnight incubation at 37°C. Images were visualized using a Zeiss Axioplan microscope equipped with 100×/1.45 alpha Plan-Fluar lens (Carl Zeiss, Heidelberg, Germany). Images were acquired using a Photometrics (Tucson, AZ) charge-coupled device camera, and IPLAB software (VYSIS, Stuttgart, Germany), and were processed using PowerPoint (Microsoft, Redmond, WA). Original magnification × 1000. Thin lines separate individual images of representative nuclei.
Evidence for different mechanisms of loss of heterozygosity on chromosome 1. (A-C) Dual-color fluorescent in situ hybridization signal patterns of selected cell nuclei obtained with PAC clones that encompass the RHD/RHCE (FITC, green) and, as a control, AF1q gene sequences (Cy3, red). (A) As shown here for patient 9, both the segmented (top) and round (bottom) nuclei in 4 of 6 analyzed cases contained 2 signals each and, thus, 2 RH gene loci. (B) The lack of one green signal in the majority of segmented nuclei and a minority of round nuclei of patient 3, on the other hand, confirmed a monoallelic deletion of at least the RHD/RHCE gene locus. (C) The 2 green RH and 3 red AF1q signal pattern in patient 6 prompted further investigations that (D) confirmed the presence of 3 chromosome 1 (green) and 2 chromosome 7 (red) centromeres with one colocalization (orange). This pattern reflects the presence of a der (1;7)(q10;p10), a highly specific chromosome abnormality in particular subtypes of myeloid malignancies. Thin lines separate individual images of representative cell nuclei. Signals were recorded after overnight incubation at 37°C. Images were visualized using a Zeiss Axioplan microscope equipped with 100×/1.45 alpha Plan-Fluar lens (Carl Zeiss, Heidelberg, Germany). Images were acquired using a Photometrics (Tucson, AZ) charge-coupled device camera, and IPLAB software (VYSIS, Stuttgart, Germany), and were processed using PowerPoint (Microsoft, Redmond, WA). Original magnification × 1000. Thin lines separate individual images of representative nuclei.
Rh antigen and Rh complex molecule expression levels of D-negative red cell subpopulations reflect the mechanisms of loss of heterozygosity
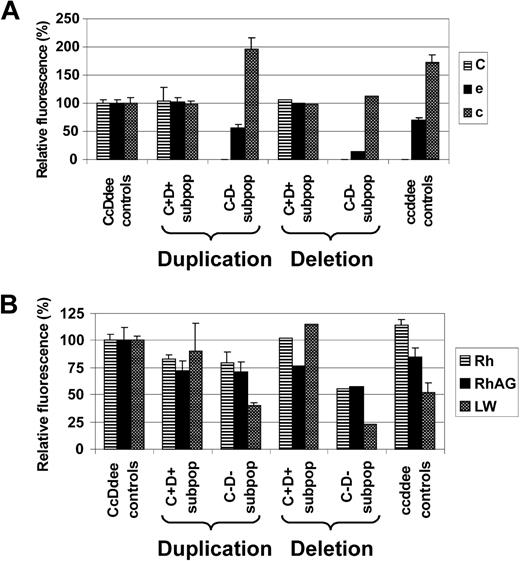
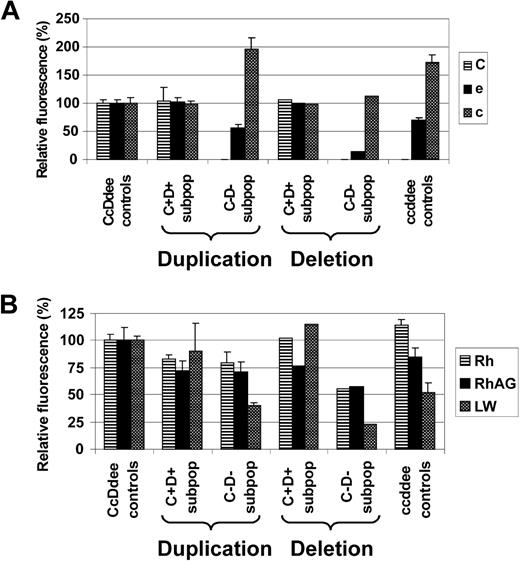
The apparent mode of LOH as evidenced by FISH (RH duplication versus monoallelic RH deletion) seemed to influence Rh antigen and Rh complex molecule expression of the affected RBC subpopulations. While C+D+ RBC subpopulations of CcDdee patients 1, 2, 3, and 6 expressed Rh antigens C, e, and c at levels comparable with CcDdee controls, only C−D− RBC subsets from patients with apparent somatic recombination-associated RH duplication (patients 1, 2, 6) displayed equal e and c levels, compared with ccddee controls. In contrast, the C−D− RBC subpopulation of patient 3 affected by monoallelic RH deletion showed markedly diminished levels of e and c, compared with ccddee controls (Figure 7A). Moreover, the C−D− RBC fraction of patient 3 had also reduced expression of total Rh protein and the Rh complex molecules RhAG and LW (CD242),45 compared with ccddee controls or the respective subpopulations from patients with RH duplication (Figure 7B). These findings emphasized that the C−D− RBC subsets of patients affected by LOH were derived from stem cells of ccddee or hemizygous cde genotype consequent to somatic recombination-associated RH (cde) duplication or RH (CDe) deletion, respectively.
Rh antigen and Rh complex expression levels of red cell subsets from patients apparently affected by either somatic recombination-associated RH duplication or deletion. Expression of (A) Rh antigens C, e, and c, and of (B) total Rh protein, RhAG, and LW of C+D+ and C−D− RBC subpopulations (C+D+ subpop and C−D− subpop) of CcDdee patients with either somatic recombination-associated RH duplication (duplication: patients 1, 2, and 6) or monoallelic RH deletion (resulting in RH hemizygosity; deletion: patient 3) is shown. For comparison, values of control CcDdee (n = 3) and ccddee (n = 3) RBC samples are depicted. Monoallelic RH deletion in patient 3 is reflected by markedly reduced e and c expression of his C−D− subpopulation, compared with ccddee controls. In contrast, C−D− subpopulations from patients with RH duplication express e and c levels similar to ccddee controls. Moreover, the partial RH hemizygosity of patient 3 was also associated with particularly low expression of total Rh protein, RhAG, and LW of his C−D− subset. Data are shown as the averaged mean fluorescence intensities relative to values of CcDdee control RBCs set as 100%. Error bars represent SD.
Rh antigen and Rh complex expression levels of red cell subsets from patients apparently affected by either somatic recombination-associated RH duplication or deletion. Expression of (A) Rh antigens C, e, and c, and of (B) total Rh protein, RhAG, and LW of C+D+ and C−D− RBC subpopulations (C+D+ subpop and C−D− subpop) of CcDdee patients with either somatic recombination-associated RH duplication (duplication: patients 1, 2, and 6) or monoallelic RH deletion (resulting in RH hemizygosity; deletion: patient 3) is shown. For comparison, values of control CcDdee (n = 3) and ccddee (n = 3) RBC samples are depicted. Monoallelic RH deletion in patient 3 is reflected by markedly reduced e and c expression of his C−D− subpopulation, compared with ccddee controls. In contrast, C−D− subpopulations from patients with RH duplication express e and c levels similar to ccddee controls. Moreover, the partial RH hemizygosity of patient 3 was also associated with particularly low expression of total Rh protein, RhAG, and LW of his C−D− subset. Data are shown as the averaged mean fluorescence intensities relative to values of CcDdee control RBCs set as 100%. Error bars represent SD.
Discussion
In this study, the puzzling phenomenon of spontaneous coexistence of D-positive and D-negative RBC subpopulations was investigated. In a representative number of 9 patients with mixed Rh phenotype, the prevailing molecular background of this condition was elucidated. Rh mosaicism involving a hematopoietic stem-cell line with LOH on chromosome 1 was demonstrated to be the key mechanism for the observed RBC phenotype anomalies.
Frequent causes of mixed Rh phenotype, such as RBC transfusion or HSCT, had been ruled out in these patients, which was also confirmed by the fact that Rh and in one case also Fy but not other blood group antigens were affected. In addition, none of the patients displayed congenital or acquired chimerism,4 as evidenced by microsatellite analysis. This result was remarkable, as spontaneous chimerism had been reported to occur quite frequently.5,46
The immunohematologic properties of the D-positive and D-negative RBC subpopulations indicated loss of one complete RH haplotype (CDe or cDE, depending on the individual RH genotype) in the D-negative fraction. This was corroborated by genotyping of BFU-E colonies derived from single erythroid progenitor cells, showing RHD+/RHCE*C+/RHCE*c+/RHCE*e+ (or RHD+/RHCE*E+/RHCE*e+ in the ccDdEe patient) as well as solely RHCE*c+/RHCE*e+ (or only RHCE*e+ in patient 7) results in samples from individual patients. Moreover, microsatellite analysis of chromosome 1 demonstrated LOH on variable chromosome 1 stretches encompassing the RHD/RHCE gene loci in DNA samples from blood, in marked contrast to other tissues. Most importantly, this molecular alteration was confirmed by microsatellite analysis of single BFU-E colonies, showing a remarkable pattern: without exception, BFU-E colonies solely positive for RHCE*c and RHCE*e displayed LOH on variable stretches of chromosome 1, which, in contrast, was not the case for RHD+/RHCE*C+/RHCE*c+/RHCE*e+ colonies. Therefore, Rh phenotype splitting was ascribed to RH haplotype elimination in stem cell lines affected by LOH on chromosome 1.
Possible mechanisms for LOH include somatic recombination, deletion, gene conversion, and chromosome loss with or without duplication of the remaining chromosome. However, allelic loss in vitro occurred predominantly by somatic recombination and chromosome loss with duplication of the remaining chromosome,47 whereas LOH in vivo was due mainly to somatic recombination.48 According to FISH analysis, Rh mosaicism based on LOH was also caused mainly by homologous recombination between the chromatides of chromosome 1 prior to mitosis (acquired partial uniparental disomy), explaining the lack of detectable cytogenetic abnormalities in most individuals with this condition.2 Deletion of a rather short chromosome 1 stretch including the RH locus appeared to be responsible for LOH only in patient 3. The results of quantitative Rh antigen expression studies corroborated both the real-time PCR and the FISH data. Of interest, in 4 of 5 further investigated patients, such LOH was found to be lineage specific, occurring in a percentage of myeloid cell subsets but not in lymphocytic compartments or other tissues. No such lineage restriction was found in patient 5, and unusual peak imbalances were seen at all of her informative chromosome 1 microsatellite markers. Importantly, in this patient not only a mixed Rh but also Fy phenotype were found, which supported the view that both the short and the long arms of chromosome 1 were affected by LOH. Two cases with similar Rh and Fy phenotype splitting had been reported years ago, without demonstrable cytogenetic abnormalities.16,49 In patient 5, the combined Rh and Fy mosaicism could be explained by loss of one chromosome 1 and concomitant duplication of the remaining chromosome in a pluripotent hematopoietic stem-cell clone.47
Isolated Rh mosaicism based on LOH on chromosome 1 is not necessarily associated with negative effects. In many cases, it could represent simply an index for genetic alteration acquired in the course of normal aging. In fact, the median age of the patients of this study was 63 years, and serologic screening for Rh phenotype splitting among apparently healthy persons 60 years or older had revealed a considerable frequency of at least 1 in 500.17 Importantly, Rh mosaicism in apparently healthy subjects may have clinically relevant consequences,18 as already minor proportions of D-positive RBCs in blood units from donors erroneously typed D-negative were demonstrated to incite anti-D immunization in D-negative recipients.19
Loss of Rh antigens, after ABO,50 is the second most commonly reported blood group alteration associated with hematologic disease.15 In this study, one third of the patients suffered from different hematologic diseases, whereas the rest of the study cohort appeared hematologically healthy within even extended observation periods (in comparison, only 1.8% of all patients who receive blood grouping tests at the Division of Blood Group Serology in Vienna suffer from hematologic diseases). Currently, it is not known at which rate Rh mosaicism accompanies or precedes overt bone marrow disease, and therefore its significance as a possible marker for disease is uncertain. Judging from the literature, Rh mosaicism appears to be preferably recognized in patients suffering from hematologic disease.2,9–15,51 Moreover, in a number of cases, re-establishment of the normal unmixed Rh phenotype upon remission of hematologic disease, and Rh antigen loss upon relapsing or progressive disease, documented the dependence of Rh mosaicism on pathological stem-cell clones.2,13,14,51 However, varying proportions of RBC subsets of different phenotypes over time, as also observed in some of the patients of this study, are not an exclusive feature of Rh mosaicism but were also reported for hematopoietic chimerism.4,5 Moreover, LOH is a major mechanism for inactivation of tumor-suppressor genes, and some of these genes are believed to reside on the short arm of chromosome 1, next to the RH locus.52 Because of the aforementioned circumstances, it seems generally advisable to perform diagnostic steps to exclude hematologic disease in individuals with spontaneous blood group phenotype splitting.
In conclusion, all patients of this study showed Rh mosaicism based on LOH along variable chromosome 1 stretches encompassing the RHD/RHCE gene loci. Hence, this alteration seems to represent by far the most common molecular background of spontaneous Rh phenotype splitting. Further studies providing reliable frequency data of Rh mosaicism could aid in the interpretation of this biologic phenomenon and shed light on the long-term outcome with respect to the possible development of hematologic disease.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Grant 10991 from the Österreichische Nationalbank (G.F.K.), and a grant from the Faculty of Health Sciences, Copenhagen University in Denmark (F.B.C.).
The authors acknowledge the expert technical assistance of Margit König, Kornelia Gerhartl, Eva Jäger, and Bianca Doninger, and the support of Dr Johannes Radek and Dr Michael Schuck in sample acquisition.
Authorship
Contribution: G.F.K., D.S., W.R.M., and C.G. designed research; all authors performed research and collected data; G.F.K., E.-M.D., and C.G. contributed analytical tools, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Günther F. Körmöczi, Department of Blood Group Serology and Transfusion Medicine, Medical University of Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria; e-mail: guenther.koermoeczi@meduniwien.ac.at.