Abstract
Growth factor independence 1 (Gfi1) is a transcriptional repressor essential for the function and development of many different hematopoietic lineages. The Gfi1 protein expression is regulated by the ubiquitin-proteasome system. In granulocytes, Gfi1 is rapidly degraded by the proteasome, while it is more stable in monocytes. How the ubiquitination and degradation of Gfi1 is regulated is unclear. Here, we show that the ubiquitin ligase Triad1 interacts with the DNA-binding domain of Gfi1. Unexpectedly, we found that Triad1 inhibited Gfi1 ubiquitination, resulting in a prolonged half-life. Down-regulation of endogenous Triad1 by siRNAs resulted in increased Gfi1 ubiquitination. In U937 cells, Triad1 caused an increase in endogenous Gfi1 protein levels and slowed cell proliferation in a similar manner when Gfi1 itself was expressed. A Triad1 mutant that lacks the Gfi1-binding domain did not affect Gfi1 levels and proliferation. Because neither proteasome-ubiquitin nor Triad1 ubiquitin ligase activity was required for the inhibition of Gfi1 ubiquitination, these data suggest that Triad1 competes for Gfi1 binding with as yet to be identified E3 ubiquitin ligases that do mark Gfi1 for proteasomal degradation. The finetuning of Gfi1 protein levels regulated by Triad1 defines an unexpected role for this protein in hematopoiesis.
Introduction
Hematopoiesis is characterized by the continuous generation of mature, functional blood cells from a pool of stem cells that reside in the bone marrow. To date, several transcription factors have been identified to play an essential role in hematopoiesis like PU.1,1,2 C/EBPα,3 and C/EBPϵ.4 The transcriptional repressor growth factor independence 1 (Gfi1) is also essential for proper hematopoiesis. Gfi1 was originally identified as a proviral insertion site resulting in IL-2–independent growth of T cells.5 The mammalian Gfi1 gene encodes a 55-kDa nuclear protein that contains 6 C-terminal C2-H2 zinc finger domains important for DNA binding and a characteristic N-terminal 20–amino acid SNAG domain essential for transcriptional repression. Overexpressed Gfi1 acts as a dominant oncogene and cooperates with known oncoproteins like Myc and Pim-1 in lymphoma development.6,7 In normal hematopoiesis, Gfi1 is essential for B- and T-cell development and regulates the self-renewal and long-term reconstituting potential of hematopoietic stem cells.8-10 Gfi1 was also shown to be important for lymphocyte-derived dendritic cell development.11 Furthermore, Gfi1 plays an essential role during granulocytic differentiation. Gfi1−/− knock-out mice are severely neutropenic due to a block in myeloid differentiation, which results in the accumulation of an atypical immature myeloid cell population.12,13 Recently, other essential Gfi1 functions outside the hematopoietic system also were identified, such as in the inner ear14,15 and in intestinal secretory lineage differentiation.16
As has been shown for other key regulators of hematopoietic differentiation such as AML1,17 C/EBPα,18 and RARα,19 it was recently shown that Gfi1 protein levels are controlled by the ubiquitin proteasome system (UPS).20,21 This system catalyzes the conjugation of ubiquitin to substrate molecules in a multienzyme process. First, the E1 enzyme activates free ubiquitin, a small peptide of 76 amino acids. The activated ubiquitin is subsequently transferred to an E2 ubiquitin–conjugating enzyme. In concert with the E2 enzyme, E3 ubiquitin ligases catalyze the covalent conjugation of ubiquitin to specific substrate proteins.22,23 Depending on the type of ubiquitin chain conjugated to the substrate, ubiquitination can regulate the activity or stability of substrate proteins.24 A crucial role of ubiquitin E3 ligases is the specific recognition of substrate proteins, which should be marked with ubiquitin. Therefore, the specificity of the UPS is mainly determined by the activity of E3 ubiquitin ligases.
Gfi1 protein levels are regulated by the UPS during myeloid differentiation.20 The Gfi1 protein is relatively stable in primary monocytes, while in mature granulocytes it is efficiently degraded by the 26S proteasome. This results in low Gfi1 protein levels in granulocytes, while mRNA levels are high. However, in primary monocytes, Gfi1 protein levels are much higher, while mRNA levels are significantly lower compared with granulocytes.20 Recent work showed that Gfi1 protein levels are also controlled by the UPS in neuronal cells.21 In these studies, it was shown that Ataxin-1 (Atx-1) interacts with Gfi1, resulting in enhanced degradation via the 26S proteasome. Mutations of Atx-1 in spinocerebellar ataxia type 1 result in decreased Gfi1 levels in neuronal cells, which is implicated in the onset of this neurodegenerative disease.21 However, no E3 ligase activity of Atx-1 has been described. While Gfi1 protein levels are regulated by the UPS in different cellular processes, the proteins involved are unknown. In this report, we show that Gfi1 interacts with the ubiquitin E3 ligase Triad1, and that Triad1 controls Gfi1 protein levels.
Materials and methods
Yeast 2-hybrid screening
The pretransformed matchmaker 2-hybrid system (Clontech, Palo Alto, CA) was used for yeast 2-hybrid analyses. Sequences encoding amino acids 109 to 493 of Triad1 were cloned in pGBD (Gal4 DNA-binding domain vector) as described,25 and the plasmid was transformed to yeast strain AH109. AH109 yeast cells were used to mate with the Y187 yeast strain that was pretransformed with a human bone marrow cDNA library in pACT2 (Clontech). Mating and yeast 2-hybrid screening was performed according to the manufacturer's instructions. Putative interactions in yeast were defined by growth of blue colonies within 12 days after plating the cells on leucine-, tryptophane-, adenine-, and histidine-deficient synthetic defined medium containing X-α-gal. Plasmids were isolated from positive yeast colonies using the plasmid mini kit (Qiagen, Valencia, CA), with an additional preincubation step with lysozyme. Isolated plasmid DNA was tested in rescreens to confirm the specificity of the interaction. The Gfi1B clone isolated from the yeast 2-hybrid screen was cloned in the pGBD vector to perform the reciprocal yeast 2-hybrid test with Triad1 in pGAD (Gal4 activation domain vector). For the fine mapping of protein-interacting domains, a panel of Triad1 deletion constructs was generated lacking either the entire TRIAD domain, the N- or C-terminal RING finger domains, or the DRIL domain using primers indicated in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) in sexual polymerase chain reaction (PCR) as described.26 Triad1 lacking the 2 C-terminal coiled-coil domains was generated by PCR using a primer introducing a stop codon (see Table S1 for primer sequences). The Triad1 deletion mutants were subsequently cloned in pGAD, and the integrity of all constructs was confirmed by sequence analysis.
Constructs, cell culture, and retroviral transduction
Triad1 and Gfi1 constructs were described earlier.20,25 For in vitro protein translation Gfi1B-Flag, Gfi1BΔzinc finger–Flag and Gfi1B-6 zinc finger–Flag kindly provided by Dr M. Osawa (Tsukaba, Japan) were used.27 Flag-Gfi1 and Gfi1-GFP were kindly provided by Dr T. Möröy, Montreal, QC. The retroviral Gfi1-IRES-GFP construct was kindly provided by Dr R. Dahl, Albuquerque, NM.28 Full-length Triad1 and the mutant lacking the C-terminal RING finger (Triad1dR2) were cloned into the pLZRS-IRES-ΔNGFR retroviral expression vector.29 The IRES (internal ribosomal entry site) allows for separate expression of Triad1 or its mutant and a truncated version of the nerve growth factor receptor (ΔNGFR) that lacks intracellular domains. The generation of retroviruses and retroviral transduction was performed as described.25,29 The percentage of GFP+ and ΔNGFR+ cells after transduction was monitored by fluorescence-activated cell-sorter (FACS) analysis (FC500; Beckman Coulter, Hialeah, FL). NGFR+ cells were sorted using magnetic-activated cell sorting (MACS) as described.29 COS-1 cells were cultured in Iscove modified Dulbecco medium (IMDM; Life Technologies, Gaithersburg, MD) supplemented with 10% fetal calf serum (FCS), penicillin (50 IU/mL), and streptomycin (50 μg/mL) (Life Technologies). HEK293 cells were cultured in Dulbecco modified Eagle medium (DMEM; Life Technologies) supplemented similar to that described for IMDM with the addition of 2 mM glutamine. Where indicated, cells were treated with 50 to 100 μg/mL cycloheximide (Sigma, St Louis, MO) for the indicated time. U937 cells were cultured in RPMI supplemented with 10% FCS and penicillin/streptomycin. Sf-9 and Hi-5 insect cells were grown at 27°C in a completely humidified atmosphere in TNM-FH medium (Sigma) supplemented as indicated for IMDM. For Sf-9 cells, 0.5% bovine serum albumin was added. Hi-5 cells were grown to 75% confluency and infected with baculovirus.
In vitro pull-down assays
His-tagged Triad1 was isolated from 4- to 5-day infected Hi-5 cells, which were lysed in RIPA buffer (PBS containing 1% Nonidet P-40 [NP-40], 0.5% natrium-deoxycholaat, 0.1% SDS, and complete protease inhibitor cocktail; Roche Diagnostics, Basel, Switzerland) followed by 2 hours incubation at 4°C with prewashed His-select beads (Sigma) in the presence of 5 mM imidazole (Sigma). Beads were washed 4 times with RIPA buffer supplemented with 10 mM imidazole. His-Triad1–loaded beads were incubated with 10 μL in vitro–translated, 35S methionine–labeled proteins for 16 hours at 4°C. The labeled proteins were produced using the rabbit reticulocyte lysate TnT Quick Coupled Transcription/Translation System (Promega, Madison, WI). After 4 wash steps with RIPA in the presence of 10 mM imidazole, proteins were eluted by the addition of 2× Laemli buffer and samples were heated for 5 minutes at 95°C. Samples were resolved by SDS-PAGE, and gels were fixed, treated with NAMP-100 (Amersham, Arlington Heights, IL), dried, and exposed.
Coimmunoprecipitation and Western blotting
COS-1 cells cultured in 10-cm dishes were transiently transfected as indicated using the calcium phosphate precipitation method. Cells were lysed 36 hours after transfection in RIPA buffer with complete protease inhibitors (Roche Diagnostics). GFP-tagged proteins were incubated with GFP antibody (Roche Diagnostics; clones 7.1 and 13.1) and protein A beads for 4 hours at 4°C with gentle rotation. Beads were washed 5 times with RIPA. Bound proteins were eluted in loading buffer by heating for 5 minutes at 95°C. Immunoprecipitates and protein lysates were separated on SDS-polyacrylamide gels, electrophoretically transferred onto polyvinylidenefluoride (PVDF) membranes (Bio-Rad, Hercules, CA), and blocked with 2% ELK (Campina, Woerden, the Netherlands) and 0.5% BSA (Sigma). Blots were probed with antibodies to Gfi1 (N20; Santa Cruz Biotechnology, Santa Cruz, CA), Flag (M2; Sigma), actin (AC40; Sigma), ubiquitin (6C1; Sigma), His-tag (penta-his; Qiagen), Triad1,25 or GFP (clones 7.1 and 13.1; Roche Diagnostics; used in all experiments except where indicated another GFP antibody was used [sc-8334; Santa Cruz Biotechnology]).
Transfection and in vivo ubiquitination assays
COS-1 cells grown in 10-cm dishes were transfected with 10 μg of His-tagged ubiquitin (His-Ub) and 10 μg of human Gfi1-GFP or Flag-Gfi1 expression constructs using calcium phosphate precipitation. For ubiquitination assays in which siRNAs were used, HEK293 cells were cotransfected with the indicated siRNAs with or without His-Ub and with or without GFP constructs using lipofectamine 2000 (Invitrogen, Carlsbad, CA). Where indicated, cells were sorted based on GFP positivity by FACS (Epics Altra; Beckman Coulter). The following siRNAs were used: Triad1 siRNA1, uugugaggaagaggaagaa; Triad1 siRNA2, aauugugaggaagaggaagaa; siRNA GFP, gcaagcugacccugaaguucau; a control siRNA, aggaguugguggcaauaau (designed against Sp1); and a control scrambled siRNA, aauugugaggaagaggaagaa (Allstar control; Qiagen). Where indicated, proteasome activity was inhibited by the addition of MG132 16 hours prior to harvesting cells. At 36 hours after transfection, cells were harvested for purification of His-tagged proteins. The cell pellet was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4, 10 mM imidazole, 10 mM β-mercaptoethanol, and complete protease inhibitor [Roche Diagnostics]) and incubated with His-select NTA beads (Sigma) for 4 hours at 4°C. The beads were washed 2 times with buffer A, 2 times with a mixture of 4 parts buffer A and 1 part of buffer B (50 mM Tris [pH 6.8] and 20 mM imidazole) and 2 times with buffer B. Bound proteins were eluted with 200 mM imidazole in Laemli buffer. The eluted proteins were analyzed by immunoblotting for the presence of Ub-conjugated Gfi1 using α-Flag, α-His, or α-Gfi1 antibodies. For measurements of Gfi1 stability by flow cytometry, HEK293 cells were transfected with Gfi1-GFP and Triad1 as indicated, and GFP intensities were measured 24 to 30 hours after transfection.
Results
Triad1 interacts with the zinc fingers of Gfi1 and Gfi1B
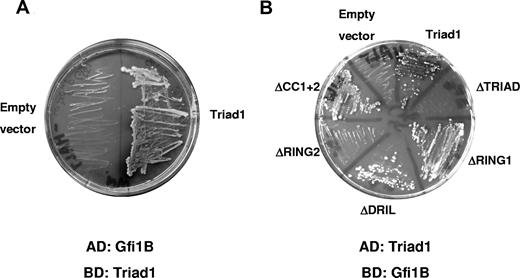
Triad1 is an E3 ubiquitin ligase implicated in myelopoiesis.25 To identify Triad1-binding partners, we searched for interacting proteins by screening a human bone marrow cDNA library in a yeast 2-hybrid assay using amino acids 109 to 493 of Triad1 as bait. This part of Triad1 comprises the TRIAD domain and the 2 coiled-coil regions.25 Candidate Triad1-interacting proteins were identified by screening 5.8 × 106 transformants on SD medium without adenine and histidine in the presence of X-α-gal. From blue-stained yeast colonies, pACT plasmid DNA was isolated and sequenced. One of the positive interacting proteins was identified as Gfi1B.30 The interacting clone spanned amino acids 91 to 330, containing 6 C2H2 zinc fingers of the DNA-binding domain. Gfi1B interacted specifically with Triad1 and not with the empty vector (Figure 1A). The interaction was validated by the reciprocal experiment using Gfi1B cloned in the pGBD bait vector and Triad1 in the pGAD prey vector (Figure 1B). To pinpoint the interacting domains, we tested the interaction of Gfi1B with various Triad1 deletion constructs. Deletion of the N-terminal RING, DRIL/IBR domain, or both coiled-coils of Triad1 did not influence the interaction. In contrast, deletion of the C-terminal RING finger abolished the observed interaction with Gfi1B, indicating that this domain of Triad1 is essential for the Gfi1B interaction (Figure 1B).
Gfi1B binds Triad1 in yeast 2-hybrid assays. (A) Yeast 2-hybrid screen with the Gfi1B clone that interacts with Triad1 (aa 109–493) and not with the empty vector control on double histidine and adenine selection medium containing X-α-gal. (B) Gfi1B was cloned into pGBD and tested for interaction in the reciprocal experiment with full-length Triad1 or deletion constructs in pGAD. The C-terminal RING finger (RING2) of Triad1 was the smallest essential domain for Gfi1B interaction. Used Triad1 mutants either lack the entire TRIAD domain (ΔTRIAD), the N- or C-terminal RING finger (respectively, ΔRING1 and ΔRING2), the DRIL domain (ΔDRIL), or both C-terminal coiled-coil regions (ΔCC1 + 2).
Gfi1B binds Triad1 in yeast 2-hybrid assays. (A) Yeast 2-hybrid screen with the Gfi1B clone that interacts with Triad1 (aa 109–493) and not with the empty vector control on double histidine and adenine selection medium containing X-α-gal. (B) Gfi1B was cloned into pGBD and tested for interaction in the reciprocal experiment with full-length Triad1 or deletion constructs in pGAD. The C-terminal RING finger (RING2) of Triad1 was the smallest essential domain for Gfi1B interaction. Used Triad1 mutants either lack the entire TRIAD domain (ΔTRIAD), the N- or C-terminal RING finger (respectively, ΔRING1 and ΔRING2), the DRIL domain (ΔDRIL), or both C-terminal coiled-coil regions (ΔCC1 + 2).
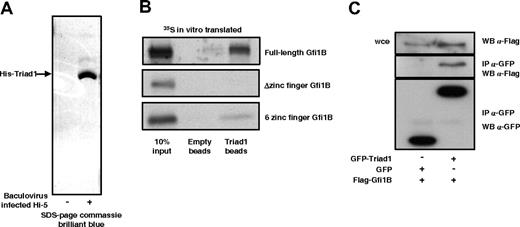
To confirm the Triad1 Gfi1B interaction with alternative methods, full-length His-tagged Triad1 was expressed in Hi-5 insect cells and purified with His-select beads (Figure 2A). Western blot analysis indicated that the purified band represented His-Triad1 (data not shown). The His-Triad1 beads were incubated in RIPA buffer with 35S-labeled in vitro–translated Gfi1B. As depicted in Figure 2B, Gfi1B interacted with Triad1 but not with the control beads. In vitro–translated fragments of Gfi1B were used to detect the part of Gfi1B responsible for interaction with Triad1. As expected, the Gfi1B zinc finger part interacted with Triad1, while the Gfi1B fragment containing the SNAG and non–zinc finger domain did not (Figure 2B). The interaction was further verified by coexpressing GFP or GFP-tagged Triad1 with N-terminally Flag-tagged Gfi1B in COS cells followed by GFP immunoprecipitation. The subsequent Flag staining (Figure 2C) showed that Gfi1B interacts with GFP-Triad1 and did not interact with GFP alone. Together, these data indicate that the zinc finger domain of Gfi1B interacts with the C-terminal RING finger of Triad1.
Gfi1B interacts with Triad1. (A) His-Triad1 was isolated from baculovirus-infected Hi-5 cells using His-Select beads. Proteins bound to beads were detected with SDS-PAGE followed by coomassie staining. A clear His-Triad1 band is visible, while no proteins can be detected when beads were incubated with uninfected Hi-5 cells. (B) In vitro–translated and 35S-labeled Gfi1B was incubated with empty beads or His-Triad1–loaded beads. Full-length Gfi1B and the 6–zinc finger domain interacted with Triad1, but not the Δzinc finger domain. (C) GFP-Triad1 or GFP was cotransfected with Flag-Gfi1B in COS cells. α-GFP–immunoprecipitated proteins (α-GFP clones 7.1 and 13.1; Roche Diagnostics) were immunoblotted and stained with a different α-GFP antibody (sc-8334; Santa Cruz Biotechnology) to check the immunopreciptiation. α-Flag staining showed that Gfi1B interacts with GFP-Triad1 but not with GFP alone. Whole-cell extract (wce) was blotted with α-Flag antibody to show equal Gfi1B expression.
Gfi1B interacts with Triad1. (A) His-Triad1 was isolated from baculovirus-infected Hi-5 cells using His-Select beads. Proteins bound to beads were detected with SDS-PAGE followed by coomassie staining. A clear His-Triad1 band is visible, while no proteins can be detected when beads were incubated with uninfected Hi-5 cells. (B) In vitro–translated and 35S-labeled Gfi1B was incubated with empty beads or His-Triad1–loaded beads. Full-length Gfi1B and the 6–zinc finger domain interacted with Triad1, but not the Δzinc finger domain. (C) GFP-Triad1 or GFP was cotransfected with Flag-Gfi1B in COS cells. α-GFP–immunoprecipitated proteins (α-GFP clones 7.1 and 13.1; Roche Diagnostics) were immunoblotted and stained with a different α-GFP antibody (sc-8334; Santa Cruz Biotechnology) to check the immunopreciptiation. α-Flag staining showed that Gfi1B interacts with GFP-Triad1 but not with GFP alone. Whole-cell extract (wce) was blotted with α-Flag antibody to show equal Gfi1B expression.
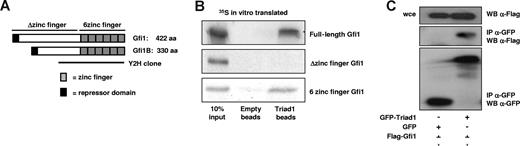
Gfi1B is a transcriptional repressor essential for proper erythroid and megakaryocytic differentiation and plays a role in T-cell activation.31,32 The Triad1-interacting zinc finger domain of Gfi1B is 97% identical to the DNA-binding domain of the Gfi1B homolog Gfi1 (Figure 3A).33 To test whether Gfi1 also interacts with Triad1, we performed in vitro pull-down experiments. This showed that 35S in vitro–translated Gfi1 also interacted with purified Triad1 on His-select beads via its zinc finger domain (Figure 3B). This interaction was verified by coimmunoprecipitating N-terminally Flag-tagged Gfi1 with GFP or GFP-Triad1 in transfected COS cell lysates. Gfi1 coimmunoprecipitated with GFP-Triad1, but not with GFP (Figure 3C). Expression constructs encoding only the zinc-finger domain of Gfi1, and Gfi1B could also be coimmunoprecipitated with Triad1 (data not shown). These experiments show that, similar to Gfi1B, the zinc fingers of Gfi1 interact with Triad1. Coimmunoprecipitation experiments showing endogenous interactions between Gfi1B and Gfi1 with Triad1 using lysates from U937 and other hematopoietic cell lines appeared negative, possibly due to the transient nature of interaction and low expression of the proteins (data not shown).
Gfi1 interacts with Triad1. (A) Schematic representation of Gfi1 and Gfi1B. The Triad1-interacting part of Gfi1B found in yeast 2-hybrid (indicated with the solid-line Y2H) contains the 6 zinc fingers that are 97% identical to Gfi1. (B) Like Gfi1B, full-length and the 6–zinc finger domain of Gfi1 also interacts with His-Triad1. No interaction was detected with the Δzinc finger domain of Gfi1. (C) GFP-Triad1 or GFP was cotransfected with Flag-Gfi1 in COS cells. α-Flag staining of whole-cell extract (wce) showed equal expression of Gfi1. α-Flag staining of the α-GFP immunoprecipitates showed that Gfi1 interacts with Triad1 but not with GFP alone. α-GFP staining (α-GFP sc-8334; Santa Cruz Biotechnology) of the immunoprecipitated proteins showed successful immunoprecipitation.
Gfi1 interacts with Triad1. (A) Schematic representation of Gfi1 and Gfi1B. The Triad1-interacting part of Gfi1B found in yeast 2-hybrid (indicated with the solid-line Y2H) contains the 6 zinc fingers that are 97% identical to Gfi1. (B) Like Gfi1B, full-length and the 6–zinc finger domain of Gfi1 also interacts with His-Triad1. No interaction was detected with the Δzinc finger domain of Gfi1. (C) GFP-Triad1 or GFP was cotransfected with Flag-Gfi1 in COS cells. α-Flag staining of whole-cell extract (wce) showed equal expression of Gfi1. α-Flag staining of the α-GFP immunoprecipitates showed that Gfi1 interacts with Triad1 but not with GFP alone. α-GFP staining (α-GFP sc-8334; Santa Cruz Biotechnology) of the immunoprecipitated proteins showed successful immunoprecipitation.
Triad1 inhibits the ubiquitination of Gfi1
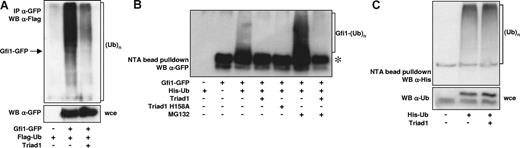
Since Triad1 functions as an ubiquitin E3 ligase in myelopoiesis,25 and Gfi1 was found to be ubiquitinated and degraded by the 26S proteasome in myeloid cells,20 we studied whether Triad1 ubiquitinates Gfi1. To this end, we transfected COS cells with Gfi1-GFP and Flag-Ub with or without Triad1. After GFP immunoprecipitation, the amount of ubiquitinated proteins bound to Gfi1 and ubiquitinated forms of Gfi1 itself were revealed by Flag staining in Western blot analysis. This resulted in a protein smear, including proteins with a lower molecular mass compared with Gfi1-GFP, probably representing ubiquitinated proteins bound to Gfi1. This indicates that Gfi1 is present in ubiquitinated protein complexes (Figure 4A). The intensity of the protein smear increases dramatically above the size of Gfi1-GFP, which suggests that Gfi1 itself is ubiquitinated. Remarkably, coexpression of Triad1 resulted in a severe reduction in the amount of ubiquitinated proteins.
Triad1 inhibits Gfi1 ubiquitination. (A) Gfi1-GFP and Flag-Ub were cotransfected with or without Triad1. Gfi1 was immunoprecipitated with α-GFP. Ubiquitinated Gfi1 or ubiquitinated proteins bound to Gfi1 were detected by α-Flag staining. The high molecular smear suggests that ubiquitinated proteins are bound to Gfi1, or that Gfi1 itself is ubiquitinated (→ marks molecular weight of unmodified Gfi1-GFP). The coexpression of Triad1 severely reduces the amount of ubiquitinated Gfi1 or ubiquitinated proteins bound to Gfi1. α-GFP staining of whole-cell extract (wce) was used to check for equal expression. (B) COS cells were transfected as indicated. Ubiquitinated proteins were isolated with His-select beads under denaturing conditions (6 M guanidium chloride). The high molecular smear represents polyubiquitinated forms of Gfi1. Coexpression of Triad1 or the H158A point mutant severely inhibited the ubiquitination of Gfi1. Incubation of cells with MG132 resulted in an accumulation of ubiquitinated forms of Gfi1, indicating that polyubiquitination of Gfi1 results in proteasomal degradation. Triad1 also inhibited the ubiquitination of Gfi1 in the presence of the proteasome inhibitor MG132. *Nonspecific interaction of Gfi1 with the His-select beads. (C) In vivo ubiquitination assay of endogenous proteins with His-Ub. Selection of ubiquitinated proteins (His-select beads) followed by an α-His staining showed that overall ubiquitination is not inhibited by Triad1. The α-ubiquitin staining of whole-cell extract (wce) in the bottom panel showed equal expression of His-tagged ubiquitin. The lower band is endogenous ubiquitin; the slower migrating band is His-Ub.
Triad1 inhibits Gfi1 ubiquitination. (A) Gfi1-GFP and Flag-Ub were cotransfected with or without Triad1. Gfi1 was immunoprecipitated with α-GFP. Ubiquitinated Gfi1 or ubiquitinated proteins bound to Gfi1 were detected by α-Flag staining. The high molecular smear suggests that ubiquitinated proteins are bound to Gfi1, or that Gfi1 itself is ubiquitinated (→ marks molecular weight of unmodified Gfi1-GFP). The coexpression of Triad1 severely reduces the amount of ubiquitinated Gfi1 or ubiquitinated proteins bound to Gfi1. α-GFP staining of whole-cell extract (wce) was used to check for equal expression. (B) COS cells were transfected as indicated. Ubiquitinated proteins were isolated with His-select beads under denaturing conditions (6 M guanidium chloride). The high molecular smear represents polyubiquitinated forms of Gfi1. Coexpression of Triad1 or the H158A point mutant severely inhibited the ubiquitination of Gfi1. Incubation of cells with MG132 resulted in an accumulation of ubiquitinated forms of Gfi1, indicating that polyubiquitination of Gfi1 results in proteasomal degradation. Triad1 also inhibited the ubiquitination of Gfi1 in the presence of the proteasome inhibitor MG132. *Nonspecific interaction of Gfi1 with the His-select beads. (C) In vivo ubiquitination assay of endogenous proteins with His-Ub. Selection of ubiquitinated proteins (His-select beads) followed by an α-His staining showed that overall ubiquitination is not inhibited by Triad1. The α-ubiquitin staining of whole-cell extract (wce) in the bottom panel showed equal expression of His-tagged ubiquitin. The lower band is endogenous ubiquitin; the slower migrating band is His-Ub.
We performed additional in vivo ubiquitination assays to further study the effect of Triad1 on Gfi1 ubiquitination. His-Ub was transfected with Gfi1-GFP with or without Triad1. Under denaturing conditions, ubiquitinated proteins were isolated and subsequently analyzed on Western blot. This showed that the amount of ubiquitinated Gfi1 was clearly diminished upon coexpression of Triad1 (Figure 4B). Also, the expression of the H158A Triad1 mutant, which cannot interact with the E2-conjugating enzyme UbcH7,25 diminished Gfi1 ubiquitination. This suggests that the inhibition of Gfi1 ubiquitination is independent of the E3 ligase activity of Triad1. Addition of the proteasome inhibitor MG132 resulted in a clear increase in ubiquitinated forms of Gfi1, indicating that ubiquitination of Gfi1 results in 26S proteasomal degradation. In the presence of MG132, Triad1 still inhibited the ubiquitination of Gfi1, indicating that proteasome activity is not necessary for the observed effect. Similar results were obtained when Flag-Gfi1 instead of Gfi1-GFP was used (Figure S1), thereby excluding any nonspecific effects in ubiquitination due to the GFP tag.
To rule out the possibility that the observed decrease of Gfi1 ubiquitination by Triad1 was caused by a nonspecific effect on total cellular ubiquitination, the effect of Triad1 on the total amount of ubiquitinated proteins was studied. To this end, His-Ub was transfected in COS cells with or without Triad1, followed by capture and staining of ubiquitinated proteins in Western blotting. As can be seen in Figure 4C, the total amount of ubiquitinated proteins was not altered upon coexpression of Triad1. Together, these data indicate that Triad1 specifically inhibits the ubiquitination of Gfi1.
Down-regulation of Triad1 levels results in an increase of Gfi1 ubiquitination
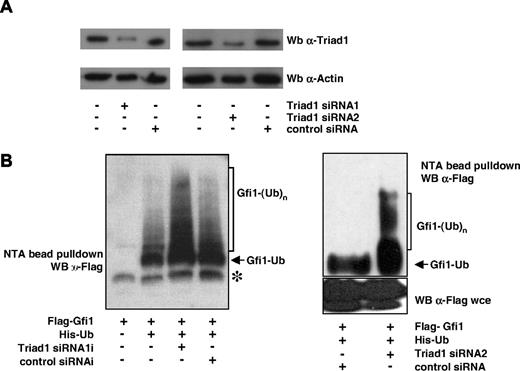
Synthetic siRNAs were developed to down-regulate endogenous Triad1 levels in HEK293 cells. The efficiency of these siRNAs was first tested by coexpressing them with GFP-Triad1. Two siRNAs designed to down-regulate Triad1 were equally efficient in down-regulating GFP-Triad1 as a control siRNA targeting GFP (Figure S2). In addition, coexpression of either construct with GFP followed by FACS sorting of transduced cells and staining for Triad1 indicated that both constructs resulted in down-regulation of endogenous Triad1 protein levels in HEK293 cells (Figure 5A). Subsequently, we tested whether Triad1 down-modulation affected Gfi1 ubiquitination. To study this, the Triad1 siRNAs were cotransfected with Flag-Gfi1 and His-Ub, followed by capture of ubiquitinated proteins and staining for Gfi1 in Western blot analysis. This showed that knocking down endogenous levels of Triad1 resulted in a clear increase in ubiquitinated forms of Gfi1 (Figure 5B). Control siRNAs (1 against Sp1; Figure 5B left panel; and a scrambled siRNA; Figure 5B right panel) had no effect on Gfi1 ubiquitination. These data show that decreasing endogenous Triad1 levels results in increased Gfi1 ubiquitination and indicate that limitation of essential cofactors is not the cause of the observed inhibition of Gfi1 ubiquitination when Triad1 is overexpressed (Figure 4). Together, these data indicate that Triad1 specifically inhibits the ubiquitination of Gfi1.
Inhibition of Triad1 expression results in increased Gfi1 ubiquitination. (A) HEK293 cells were not transfected or transfected with indicated siRNAs against Triad1 or a control siRNA (scrambled control; Qiagen) and GFP. At 48 hours after transfection, GFP+ cells were sorted by FACS and equal amounts of cells were lysed and stained for Triad1 expression, showing efficient down-modulation of endogenous Triad1. The same blots were probed with an actin antibody to demonstrate equal loading. (B) In vivo ubiquitination experiments in HEK293 cells showed that Triad1 siRNA expression resulted in increased polyubiquitination of Gfi1. Cotransfection with a control siRNA (left panel, siRNA against Sp1; right panel, scrambled siRNA; Qiagen) had no effect on Gfi1 ubiquitination. wce indicates whole-cell extract.
Inhibition of Triad1 expression results in increased Gfi1 ubiquitination. (A) HEK293 cells were not transfected or transfected with indicated siRNAs against Triad1 or a control siRNA (scrambled control; Qiagen) and GFP. At 48 hours after transfection, GFP+ cells were sorted by FACS and equal amounts of cells were lysed and stained for Triad1 expression, showing efficient down-modulation of endogenous Triad1. The same blots were probed with an actin antibody to demonstrate equal loading. (B) In vivo ubiquitination experiments in HEK293 cells showed that Triad1 siRNA expression resulted in increased polyubiquitination of Gfi1. Cotransfection with a control siRNA (left panel, siRNA against Sp1; right panel, scrambled siRNA; Qiagen) had no effect on Gfi1 ubiquitination. wce indicates whole-cell extract.
Triad1 inhibits Gfi1 protein turnover
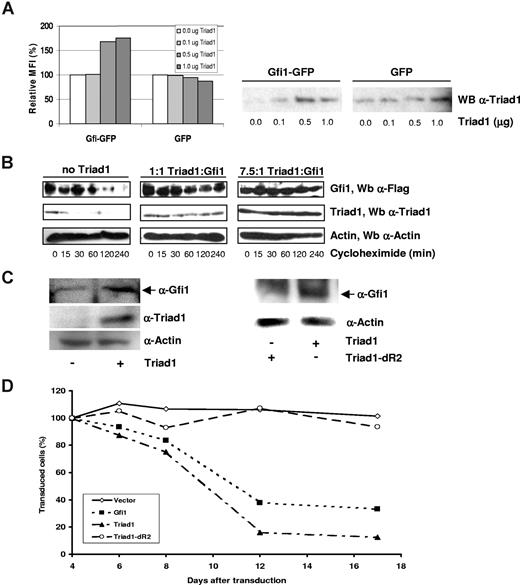
The ubiquitination of Gfi1, which results in proteasomal degradation, was inhibited by Triad1. Therefore, we studied the effect of Triad1 on Gfi1 protein levels. To address this question, we coexpressed Gfi1-GFP with different amounts of Triad1. Gfi1-GFP protein amounts were measured at the single-cell level by flow cytometry. Relative Gfi1-GFP protein levels were increased after Triad1 coexpression, while Triad1 had no effect on GFP alone (Figure 6A). The increase of Gfi1 protein levels caused by Triad1 suggests that Triad1 stabilizes Gfi1 by inhibiting its proteasomal degradation. To study the effect of Triad1 on Gfi1 stability, we transfected Flag-Gfi1 to COS cells with or without Triad1. The stability of Flag-Gfi1 protein was determined in the presence or absence of Triad1 after addition of the inhibitor of protein translation cycloheximide. In these experiments, we found that Triad1 resulted in a concentration-dependent increase in the half-life of Gfi1 (Figure 6B). To test the relevance of Gfi1 stabilization in hematopoiesis, we made use of U937 cells because we recently found that Gfi1 is efficiently targeted for proteasomal degradation in these cells.20 We generated a retroviral construct containing an expression cassette coding for Triad1 followed by an IRES and a truncated version of NGFR allowing for separate Triad1 and NGFR expression (“Constructs, cell culture, and retroviral transduction”). As control, we generated a similar construct containing Triad1 lacking coding sequences for the C-terminal RING finger (Triad1dR2) that are required for Gfi1 interaction. Both constructs were retrovirally transduced to U937 cells. NGFR+ cells were sorted, and Gfi1 protein levels were determined in Western blot analysis. This showed that Triad1 expression resulted in increased endogenous Gfi1 protein levels compared with nontransduced cells (Figure 6C). Expression of the Triad1dR2 mutant that lacked the Gfi1-binding domain did not cause an increase in Gfi1 protein levels.
Triad1 controls Gfi1 stability. (A) GFP or Gfi1-GFP was transfected with different amounts of Triad1 in HEK293 cells. At 24 hours after transfection, the Gfi1-GFP protein levels per cell were measured by flow cytometer. The mean fluorescence intensity (MFI) of transfected cells without Triad1 was set at 100%. The relative Gfi1-GFP protein levels increased when Triad1 was coexpressed, while Triad1 coexpression had no effect on the MFI of GFP transfected cells. Triad1 expression was checked in Western blot analysis (right panel). (B) HEK293 cells were transfected with 1 μg Flag-Gfi1 with or without Triad1 at indicated ratios. At 48 hours after transfection, cells were transferred to 6-well plates and allowed to adhere for 16 hours. Next, cells were treated with cycloheximide for the indicated time points. Total cell lysates were analyzed by Western blotting followed by an α-Flag staining. The Gfi1 half-life increased in a Triad1 concentration–dependent manner. The same samples were were reloaded for actin and Triad1 staining on 1 and the same blot to demonstrate equal loading and Triad1 expression. (C) U937 cells were transduced with Triad1- or Triad1dR2-encoding viruses. At 48 hours after transduction, cells were sorted based on NGFR expression using MACS. Equal amounts of NGFR+ cells were lysed and used in Western blot analysis to detect endogenous Gfi1 (N20 staining). To show equal loading, in the left panel, the samples were reloaded and stained for actin. The blot was reprobed with a Triad1 antibody to demonstrate Triad1 expression. In the right panel, the same blot was restained for actin to show equal loading. As a control to demonstrate retroviral Triad1 and Triad1dR2 expression, we transduced NIH3T3 cells followed by a Triad1 staining (Figure S3). (D) U937 cells were transduced with Gfi1, Triad1, Triad1dR2, or control viruses (encoding GFP and tNGFR). After transduction, cells were maintained as a mixed culture (containing transduced and nontransduced cells). At 4 days after transduction, the transduction efficiencies ranged from 6% to 67% based on either GFP positivity (Gfi1 and control vector) or NGFR positivity (Triad1 and Triad1dR2). At that time, the amount of GFP+ or NGFR+ cells was set at 100%. The relative amount of positive cells relative to this value was plotted in time. Both Gfi1+ and Triad1+ (based on GFP and NGFR positivity, respectively) cells were rapidly overgrown by nontransduced cells, while the relative amount of control and Triad1dR2+ cells remained stable during the culturing period. Gfi1 expression in Gfi1-transduced cells was confirmed at day 6 by Western blot analysis (Figure S4).
Triad1 controls Gfi1 stability. (A) GFP or Gfi1-GFP was transfected with different amounts of Triad1 in HEK293 cells. At 24 hours after transfection, the Gfi1-GFP protein levels per cell were measured by flow cytometer. The mean fluorescence intensity (MFI) of transfected cells without Triad1 was set at 100%. The relative Gfi1-GFP protein levels increased when Triad1 was coexpressed, while Triad1 coexpression had no effect on the MFI of GFP transfected cells. Triad1 expression was checked in Western blot analysis (right panel). (B) HEK293 cells were transfected with 1 μg Flag-Gfi1 with or without Triad1 at indicated ratios. At 48 hours after transfection, cells were transferred to 6-well plates and allowed to adhere for 16 hours. Next, cells were treated with cycloheximide for the indicated time points. Total cell lysates were analyzed by Western blotting followed by an α-Flag staining. The Gfi1 half-life increased in a Triad1 concentration–dependent manner. The same samples were were reloaded for actin and Triad1 staining on 1 and the same blot to demonstrate equal loading and Triad1 expression. (C) U937 cells were transduced with Triad1- or Triad1dR2-encoding viruses. At 48 hours after transduction, cells were sorted based on NGFR expression using MACS. Equal amounts of NGFR+ cells were lysed and used in Western blot analysis to detect endogenous Gfi1 (N20 staining). To show equal loading, in the left panel, the samples were reloaded and stained for actin. The blot was reprobed with a Triad1 antibody to demonstrate Triad1 expression. In the right panel, the same blot was restained for actin to show equal loading. As a control to demonstrate retroviral Triad1 and Triad1dR2 expression, we transduced NIH3T3 cells followed by a Triad1 staining (Figure S3). (D) U937 cells were transduced with Gfi1, Triad1, Triad1dR2, or control viruses (encoding GFP and tNGFR). After transduction, cells were maintained as a mixed culture (containing transduced and nontransduced cells). At 4 days after transduction, the transduction efficiencies ranged from 6% to 67% based on either GFP positivity (Gfi1 and control vector) or NGFR positivity (Triad1 and Triad1dR2). At that time, the amount of GFP+ or NGFR+ cells was set at 100%. The relative amount of positive cells relative to this value was plotted in time. Both Gfi1+ and Triad1+ (based on GFP and NGFR positivity, respectively) cells were rapidly overgrown by nontransduced cells, while the relative amount of control and Triad1dR2+ cells remained stable during the culturing period. Gfi1 expression in Gfi1-transduced cells was confirmed at day 6 by Western blot analysis (Figure S4).
Triad1 inhibits the proliferation of immature myeloid murine bone marrow cells and inhibits clonogenic growth of these cells in colony-forming unit granulocyte-macrophage (CFU-GM) assays.25 These effects could be mediated through stabilization of Gfi1. As Triad1 inhibits the proliferation of immature myeloid cells and stabilizes Gfi1 in U937 cells, we first tested whether forced Gfi1 expression inhibits proliferation of U937 cells. To this end, U937 cells were retrovirally transduced using a retroviral vector coding for Gfi1 and GFP.28 Proliferation assays using mixed cultures showed that Gfi1+ U937 cells were rapidly overgrown by nontransduced cells. Cells transduced with a control vector were not overgrown, but proliferated at a similar rate as nontransduced cells. Next, we tested whether Triad1 also inhibits proliferation and whether the Triad1 mutant that cannot bind to Gfi1 does not inhibit proliferation. Triad1-transduced U937 cells were overgrown by nontransduced cells in a similar rate as that of Gfi1+ cells. In contrast, Triad1dR2-transduced cells continued to proliferate at a similar rate as that of nontransduced cells. We conclude that Gfi1 is stabilized by Triad1 due to decreased ubiquitination and proteasomal degradation. The stabilization mediated by Triad1 depends on its Gfi1-interacting domain. Expression of both Triad1 and Gfi1 but not the Triad1dR2 mutant inhibits U937 cell growth, suggesting that Triad1 regulates the proliferation of immature myeloid blood cells by fine-tuning Gfi1 protein expression.
Discussion
Triad1 was recently reported as an ubiquitin E3 ligase implicated in hematopoiesis.25 Triad1 is the founding member of a subclass of RING finger E3 ligases.34,35 This subclass contains a TRIAD domain, which is characterized by a RING-DRIL/IBR-RING signature.34,35,37 Recent studies showed that the N-terminal RING finger of Triad1 interacts with the E2-conjugating enzyme UbcH7. UbcH7 mediates polyubiquitin chain formation linked via lysine-48 of ubiquitin. In line with this interaction, we showed that Triad1 functions as ubiquitin ligase and that it can catalyze the formation of polyubiquitin chains (linked via lysine-48) that mark proteins for proteasomal degradation.25 Triad1 is up-regulated upon monocytic and granulocytic differentiation, and Triad1 expression resulted in a severe reduction of clonogenic growth of primary murine bone marrow cells that could be partially rescued by proteasome inhibitors.25 Furthermore, the interaction with the E2-conjugating enzyme UbcH7 was essential for the observed Triad1 effect. In this report, we show that the C-terminal RING finger of Triad1 interacts with the zinc fingers of Gfi1 (Figure 3). Strikingly, we found that Triad1 inhibits the ubiquitination of Gfi1 (Figure 4). This indicates that Triad1 does not mark Gfi1 for proteasomal degradation and that other, currently unknown E3 ligases must exist that regulate Gfi1 turnover. Triad1 may inhibit the ubiquitination of Gfi1 by ubiquitinating these Gfi1 E3 ligases. Such a mechanism was recently described for the TRIAD E3 ligase Hoil-1 that interacts with SOCS6.38 Hoil-1 stabilizes the SOCS6 protein by mediating the ubiquitination and degradation of SOCS6-associated proteins.39 These associated proteins are normally responsible for the degradation of SOCS6. Our results show that the Triad1 H158A point mutant, which has lost its ability to bind with UbcH7,25 or wild-type Triad1 in the presence of proteasome inhibitors could still inhibit the ubiquitination of Gfi1 (Figure 4B). Together, these data strongly suggest that Triad1 inhibits the ubiquitination of Gfi1 independent of its E3 ligase activity. Such a model, in which an E3 ligase inhibits protein ubiquitination independent of its ligase activity, was recently described for the Von Hippel–Lindau E3 protein (pVHL). The pVHL ligase as well as a mutant lacking E3 ligase activity inhibited the ubiquitination of the tumor supressor p53 by disturbing the interaction between the ubiquitin ligase MDM2 and p53.40 As indicated, Triad1 inhibits Gfi1 ubiquitination in the presence of proteasome inhibitors and does not require UbcH7 to do this. This suggests that Triad1 competes for Gfi1 binding with other ubiquitin ligases that do mark Gfi1 for ubiquitination and subsequent proteasomal degradation. It also indicates that E3 ligases feature more functions than solely ubiquitinating proteins.
The Gfi1 protein levels are mainly regulated by the ubiquitin proteasome route during myelopoiesis.20 Gfi1 is efficiently degraded in granulocytes by the 26S proteasome. However, in CD14+ monocytes it is more stable due to diminished proteasomal degradation, resulting in an accumulation of Gfi1 protein levels.20 Triad1 protein levels are induced during terminal monocytic differentiation.25 Thus, Triad1 may inhibit the ubiquitination of Gfi1 in mature monocytes, which could result in the accumulation of Gfi1 protein levels. Indeed, forced expression of Triad1 in U937 cells resulted in an increase in endogenous Gfi1 protein levels (Figure 6C). In addition, Triad1 inhibited U937 cell expansion in a similar manner as Gfi1, while a Triad1 mutant lacking the Gfi1-interacting domain (Triad1dR2) did not (Figure 6D). Together, these data suggest that Triad1 may control myeloid proliferation through regulation of Gfi1 protein levels. However, in granulocytes, Triad1 is also significantly expressed although Gfi1 protein levels are low.20,25 A possible reason why Triad1 does not protect Gfi1 for degradation in mature granulocytes is the fact that the proteins may not colocalize. Triad1 localizes to specific subnuclear DAPI dull regions (euchromatin) in the nucleus of granulocytes.25 Recent studies showed that the Gfi1 paralog Gfi1B localizes to heterochromatin.41 If Gfi1 localizes to the same compartment as Gfi1B, this implies that Triad1 and Gfi1 are located in different subcellular structures in mature granulocytes, which may hamper the inhibitory effects of Triad1 on Gfi1 ubiquitination. Recently, it was also shown that Atx-1 influences Gfi1 protein stability in neuronal cells.21 In contrast to Triad1, Atx-1 destabilized Gfi1 by enhancing its degradation via the proteasome, although it is unclear whether Atx-1 functions as E3 ubiquitin ligase. Upon granulocytic differentiation, Atx-1 levels are induced.42 We found that Atx-1 mRNA expression levels are 4-fold higher in granulocytes compared with monocytes (Figure S5). The higher Atx-1 expression in granulocytes might overcome Triad1-mediated inhibition of Gfi1 ubiquitination, while Triad1 can stabilize Gfi1 in monocytes due to lower Atx-1 expression.
In this report, we have described that the ubiquitin E3 ligase Triad1 can stabilize Gfi1, which results in an accumulation of Gfi1 protein levels. This suggests that Triad1 is an important posttranscriptional regulator of the transcriptional repressor Gfi1, and in this way may regulate Gfi1 function. Because Triad1 and Gfi1 are not only expressed in the hematopoietic compartment, these effects may also play a role in the nervous system,14,21 lung development,43 and intestine.16 To better understand how Triad1 and Atx-1 affect the ubiquitination and protein stability of Gfi1, it will be important to identify the ubiquitin E3 ligase(s) that mark Gfi1 for proteasomal degradation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank Dr T. Möröy, Dr M. Osawa, and Dr R. Dahl for constructs, and Wouter Hofkens and Marleen Wienholts for technical assistance.
This work was supported by grants from the Dutch Cancer Society project code KUN2001–2395 (J.A.F.M., L.v.E.), the Royal Netherlands Academy of Sciences KNAW (Koninklijke Nederlandse Academie voor Wetenschappen; J.H.J.), and the Vanderes Foundation (B.A.V.d.R.).
Authorship
Contribution: J.A.F.M. and B.A.V.d.R. wrote the paper; J.A.F.M. and L.T.v.d.M. designed and performed research and analyzed data; L.v.E. performed research; S.v.R. and W.W. performed yeast-2-hybrid studies; and T.d.W., J.H.J., and B.A.V.d.R. analyzed data and designed the studies.
J.A.F.M. and L.T.v.d.M. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bert A. Van der Reijden, Central Hematology Laboratory, Molecular Hemato-Oncology Unit, Radboud University Nijmegen Medical Center, Nijmegen Center for Molecular Life Sciences, PO Box 9101, 6500 HB Nijmegen, the Netherlands; e-mail: b.vanderreijden@chl.umcn.nl.