Abstract
The aims of our study were to verify whether it was possible to generate in vitro, from different adult human tissues, a population of cells that behaved, in culture, as multipotent stem cells and if these latter shared common properties. To this purpose, we grew and cloned finite cell lines obtained from adult human liver, heart, and bone marrow and named them human multipotent adult stem cells (hMASCs). Cloned hMASCs, obtained from the 3 different tissues, expressed the pluripotent state–specific transcription factors Oct-4, NANOG, and REX1, displayed telomerase activity, and exhibited a wide range of differentiation potential, as shown both at a morphologic and functional level. hMASCs maintained a human diploid DNA content, and shared a common gene expression signature, compared with several somatic cell lines and irrespectively of the tissue of isolation. In particular, the pathways regulating stem cell self-renewal/maintenance, such as Wnt, Hedgehog, and Notch, were transcriptionally active. Our findings demonstrate that we have optimized an in vitro protocol to generate and expand cells from multiple organs that could be induced to acquire morphologic and functional features of mature cells even embryologically not related to the tissue of origin.
Introduction
The presently accumulated evidence indicates that adult bone marrow (BM) contains at least 2 populations of stem cells: hematopoietic stem cells (HSCs) and mesenchymal stem cells (MSCs), responsible for the generation of the BM microenvironment.1
Intriguingly, several reports have demonstrated the ability of MSCs to differentiate toward derivatives of germ layers other than mesoderm.2-6 Although it is still unclear whether widely multipotent cells do exist in vivo and if they play a significant role in tissue repair and turnover, the ability to generate in vitro cells that, under defined culture conditions, display a very high developmental plasticity is nonetheless of important clinical relevance.
Until now, the most convincing evidence, although debated,7 of the possibility to grow in culture a population of widely multipotent cells in humans has been obtained only for BM,8 while a similar feature has been just postulated for other adult human tissues.9
We therefore planned to verify if human multipotent adult stem cells (hMASCs) could be produced from other adult human organs on top of BM, and we used this latter as a control/reference tissue.
By systematically using a highly reproducible method, we were able to grow in culture cell lines from adult human liver, heart, and BM. These cell lines, once cloned at single-cell level, maintained the in vitro properties of parental lines, including the capability to differentiate into morphologically mature and functionally competent cells, even of tissues embryologically not related to the one of origin.
Finally, we performed a comparative in vitro analysis on hMASCs originated from the 3 different sources with respect to immunophenotype, growth kinetics, specific transcriptional settings, telomerase activity, and global gene expression profile. Altogether the obtained results indicate that cardiac- and liver-derived hMASC cultures do not differ significantly from the BM-derived ones, and, in particular, we demonstrated that the respective single-cell–derived clones maintained the ability to differentiate along some derivatives of the 3 germ layers as defined by immunohistochemical, molecular, and functional assays.
Materials and methods
Human samples were collected after informed consent was obtained in accordance with the Declaration of Helsinki and with approval by the Independent Ethics Committee of the University of Udine, Udine, Italy.
Tissue donors
The principal characteristics of tissue donors are summarized in Table 1 and Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell isolation and culture
Cells less than 30 μm in diameter were isolated from human livers and hearts through collagenase digestion (Worthington, Lakewood, NJ) followed by filtration (Filcons; Dako, Carpinteria, CA). Peripheral blood and BM mononuclear cells were isolated through density gradient centrifugation (Biocoll; Biochrom, Berlin, Germany) (Document S1).
Primary culture.
Freshly isolated cells (1.5 × 106) were plated in 100-mm dishes (Corning BV life sciences, Corning, NY) in Mesencult (StemCell Technologies, Vancouver, BC).
Subcultures.
When colonies that developed in primary culture reached confluence (after 2-3 weeks), cells were detached by 0.25% trypsin-EDTA (Sigma-Aldrich, St Louis, MO) and subcultured onto human fibronectin (Sigma-Aldrich)–coated 100-mm dishes in a proliferation medium composed as follows2 : 60% low-glucose DMEM (Invitrogen, Carlsbad, CA), 40% MCDB-201, 1 mg/mL linoleic acid–BSA, 10−9 M dexamethasone, 10−4 M ascorbic acid-2 phosphate, 1 × insulin-transferrin-sodium selenite (all from Sigma-Aldrich), 2% fetal bovine serum (Invitrogen or StemCell Technologies), 10 ng/mL hPDGF-BB, 10 ng/mL hEGF (both from Peprotech EC, London, United Kingdom). Cells were seeded at the density of 2 × 103 cells/cm2. Medium was replaced every 4 days. After 3 to 4 population doublings (corresponding to about 1 week in culture), cells were detached with 0.25% trypsin-EDTA (Sigma-Aldrich) and split onto fibronectin-coated dishes in proliferation medium. Passages were performed keeping constant seeding density and population doublings (3-4) before further passaging. Passages were numbered starting from P1, corresponding to the first subculture onto fibronectin-coated dishes in proliferation medium.
Single-cell cloning
Twelve cell lines (n = 4 BM derived, n = 6 liver derived, and n = 2 heart derived) at the second passage (P2) in proliferation medium were cloned at single-cell level. A high-speed cell sorter (MoFlo; Dako) was used to seed single cells into 96-well plates coated either with MS5 murine feeder layer (n = 5790 wells) or with fibronectin (n = 5680 wells). CFSE (carboxy-fluorescein-diacetate-succinimidyl-ester; Molecular Probes/Invitrogen, Carlsbad, CA) was used to exclude improperly seeded wells and to determine sorting efficiency. Of the 576 proliferating clones, 277 were expanded up to 5 × 105 cells (72 from liver, 107 from heart, and 98 from BM), before either freezing or differentiating them. Twenty selected clones were expanded up to 107 cells (Supplemental Methods).
Induction and assessment of multilineage differentiation
P3-polyclonal cell lines (n = 33) and single-cell–derived clones (n = 185) were exposed to standardized differentiation-inducing conditions10-12 for 5 to 112 days to induce their differentiation into osteoblasts, myocytes, endothelial cells, hepatocytes, neurons, and glial cells. Subsequently, cells were analyzed by immunofluorescence, histochemistry, and reverse-transcription–polymerase chain reaction (RT-PCR) for the expression of lineage-specific markers (Supplemental Methods). The functional competence of cells differentiated along a neurogenic lineage was assessed by electrophysiological analyses and dosing their glutamate release in culture supernatant (Supplemental Methods). Functionally competent myocytes were detected by the presence of spontaneous calcium transients, as evaluated by FLUO-4 imaging (Molecular Probes/Invitrogen) and by spontaneously beating cells (Supplemental Methods). Active calcium deposition sites were identified by tetracycline (Sigma-Aldrich) incorporation into osteogenic areas of differentiating cultures. The active uptake of acetylated low-density lipoprotein (LDL) was used as a marker of endothelial cell function. Hepatocyte function was assessed dosing albumin in culture supernatants, evaluating the activity of the phenobarbital-regulated cytochrome-CYP2B6 (pentoxyresorufin O-dealkylation [PROD] assay; Molecular Probes/Invitrogen), and detecting glycogen by periodic acid Schiff staining (PAS) and PAS after diastase digestion (Document S1). In a first experimental set, we evaluated the differentiation potential of 141 clones to single lineages of differentiation, while, in a second group of experiments, we established the percentage of multipotent, bipotent, and unipotent single-cell–derived clones (n = 44). Clones were scored as multipotent when able to generate at least one derivative of each germ layer.
Flow cytometry, immunofluorescence, and FISH
Immunofluorescence and fluorescent in situ hybridization (FISH) analyses were performed on 4% buffered paraformaldehyde or methanol/acetone fixed P3 cells, while fluorescence-activated cell sorting (FACS) analysis was performed on P3 cells detached from the culture substrate through a short incubation in a trypsin-EDTA solution (Sigma-Aldrich). Staining was performed using either properly conjugated primary antibodies or unconjugated primary antibodies followed by incubation with conjugated secondary antibodies. Intracellular stainings were performed after a permeabilization step using the Intrastain Fixation and Permeabilization kit (Dako), following the manufacturer's instructions. FISH was performed using X- and Y-chromosome probes (Vysis, Des Plaines, IL), following the manufacturer's instructions. DNA content was determined on ethanol-fixed, propidium iodide–stained cells.
RT-PCR analysis
DNA and RNA were extracted from 5 × 105 P3-hMASCs grown in expansion or differentiation media.
mRNA was reverse-transcribed and cDNA amplified using the appropriate primers (Document S1 and Table S1).
Telomeric repeat amplification protocol (TRAP) assay
The detection of telomerase activity was performed using the TRAPeze kit (Chemicon International, Temecula, CA) following the manufacturer's instructions.
Karyotyping
Metaphase spreads were prepared from single-cell–derived clones cultured in expanding medium for 72 hours. Chromosome analysis was performed according to standard procedures using Q-banding by fluorescence with guinacrine dye (QFQ) techniques at 400-band resolution.
Soft agar assay
Subconfluent P3 cultures were harvested, suspended in 0.5% agarose at a concentration of 5 × 103/mL, and seeded in 35-mm plates containing a basal layer of 1% agarose. Colonies were counted after 2 weeks under a phase-contrast microscope.
Microarray analysis
Total RNAs from P3-hMASCs were extracted using TRIZOL reagent (Invitrogen) according to the manufacturer's recommendations, and subjected to DNase I treatment (Promega, Madison, WI).
After RNA quality control, microarray cDNA targets were prepared by following the 2-step indirect fluorescent labeling procedure starting from 10 μg total RNA.13
Each sample was labeled using the Cy5 fluorophore and hybridized to the LNCIB 18K features cDNA microarray slides along with the reference RNA labeled with the Cy3 fluorophore14 (Document S1).
Results
Cell growth characteristics and immunophenotype of hMASCs obtained in vitro from adult bone marrow, liver, and heart
By the systematic use of the cell expansion protocol, finite cell lines were obtained from 27 BMs, 55 hearts, and 17 human livers (Table 1). No cell line could be obtained from 8 different peripheral blood samples. Age, sex, and organ pathology did not impair the possibility to generate hMASC cultures. In fact, cell lines could be obtained from end-stage samples (ie, end-stage heart failure), mildly injured samples (steatotic livers), and nonpathological samples (donor atria and BMs).
BM-derived (number of tested cell lines obtained from distinct individuals [n] = 11), cardiac-derived (n = 20), and liver-derived (n = 5) cell lines showed a similar colony-forming efficiency (15 ± 5, 18 ± 11, and 10 ± 5 clones/105 plated cells, respectively).
BM-derived (n = 4), cardiac-derived (n = 10), and liver-derived (n = 3) cell lines exhibited similar growth kinetics in culture with the cell population doubling times (calculated at P3) of 43 (± 7), 46 (± 19), and 50 (± 21) hours, respectively (Figure S1A). From the growth curve analysis, we observed that the maximum cell expansion was achieved with a seeding density of 1000 cells/cm2 (Figure S1B), and when below 500 cells/cm2 the cells grew poorly and eventually died.
Although finite, expanded cell lines were able to proliferate above 40 population doublings (PDs).
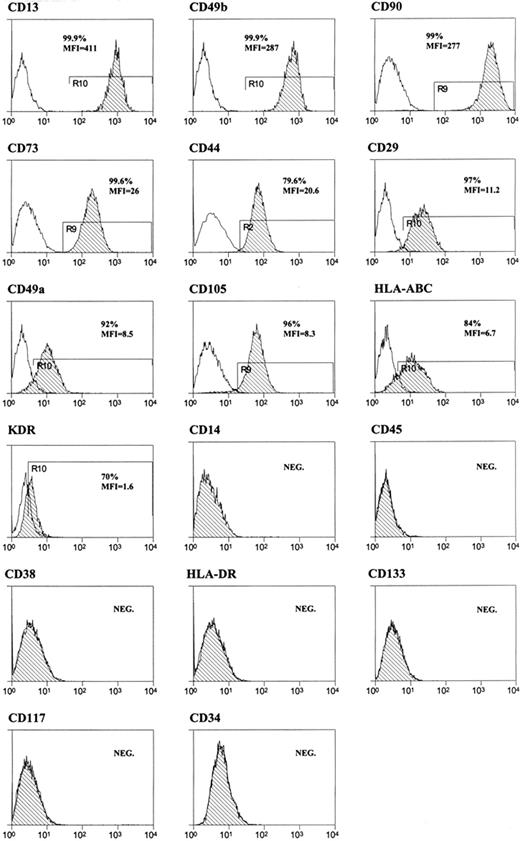
Cell surface immunophenotype of cell lines (BM, n = 9; heart, n = 18; liver, n = 8) growing in expansion medium was evaluated by flow cytometry at P3. The obtained antigenic pattern was very similar to the one described for mesenchymal stem cells15 (Figure 1).
FACS analysis of hMASCs. Representative flow cytometry histograms of multipotent cell populations. Plots show isotype control IgG-staining profile (open histogram) versus specific antibody staining profile (dash histogram). All the tested cell lines displayed a similar immunophenotype, being on an average (1) the cell fraction highly expressing CD13, CD49b, or CD90: 97% (± 7%), 84% (± 14%), and 79% (± 20%), respectively, and (2) the cell fraction expressing low levels of CD73, CD44, HLA-ABC, CD29, CD105, KDR, or CD49a: 93% (± 10%), 91% (± 17%), 85% (± 6%), 79% (± 15%), 75% (± 16%), 72% (± 14%), and 70% (± 9%), respectively. The vast majority of the cell population (> 99%) was, instead, negative for CD14, CD45, CD38, HLA-DR, CD133, CD117, and CD34.
FACS analysis of hMASCs. Representative flow cytometry histograms of multipotent cell populations. Plots show isotype control IgG-staining profile (open histogram) versus specific antibody staining profile (dash histogram). All the tested cell lines displayed a similar immunophenotype, being on an average (1) the cell fraction highly expressing CD13, CD49b, or CD90: 97% (± 7%), 84% (± 14%), and 79% (± 20%), respectively, and (2) the cell fraction expressing low levels of CD73, CD44, HLA-ABC, CD29, CD105, KDR, or CD49a: 93% (± 10%), 91% (± 17%), 85% (± 6%), 79% (± 15%), 75% (± 16%), 72% (± 14%), and 70% (± 9%), respectively. The vast majority of the cell population (> 99%) was, instead, negative for CD14, CD45, CD38, HLA-DR, CD133, CD117, and CD34.
No major differences were detected among the cell lines derived from the 3 different sources (Table S2).
Analysis of stem cell phenotype and properties
We first studied the expression of ABCG2 and MDR1, proteins involved in the generation of the side population.18 A small fraction of cells growing in expansion medium expressed high levels of ABCG2 (1.55% ± 0.57%) and MDR1 (0.93% ± 0.37%) (Figure S2).
We then assessed the expression of pluripotent state–specific transcription factor genes and differentiation-related genes.
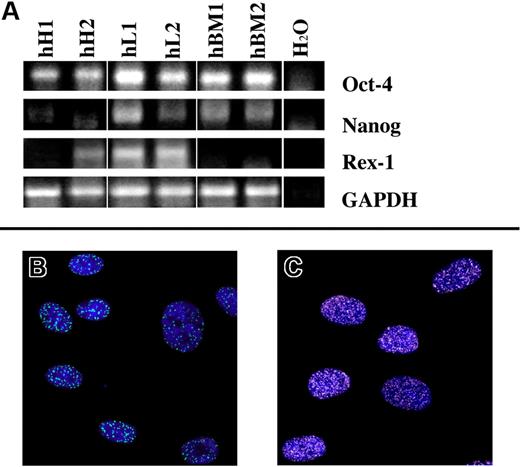
With respect to the first set of genes, we analyzed (n = 12) Oct-4, REX1, and NANOG.19,20 Most of the tested cell lines in expansion medium did express these markers at mRNA level (Figure 2A).21 Noteworthy, oct-4 and nanog proteins were detected by immunofluorescence in the 74% (± 14%) and in 40% (± 14%) of the cells, respectively (Figure 2B,C).
Pluripotent state–specific transcription factor (Oct-4, NANOG, and REX-1) expression. (A) Human heart-derived (hH1 and hH2), liver-derived (hL1 and hL2) and bone marrow–derived (hBM1 and hBM2) cells express, at mRNA level, at least 2 of the tested markers. Green dots (A488; B), and white dots (A488; C) represent the nuclear expression of oct-4 and nanog proteins, respectively. Nuclei are depicted by the blue fluorescence of DAPI staining. Image acquisition was carried out by a Confocal Laser Microscope (Leica TCS-SP2; Leica Microsystems, Milano, Italy), using a 63 × oil immersion objective (numeric aperture: 1.40). Adobe Photoshop software was used to combine RGB channels, overlay the images, and adjust contrast (Adobe, San Jose, CA).
Pluripotent state–specific transcription factor (Oct-4, NANOG, and REX-1) expression. (A) Human heart-derived (hH1 and hH2), liver-derived (hL1 and hL2) and bone marrow–derived (hBM1 and hBM2) cells express, at mRNA level, at least 2 of the tested markers. Green dots (A488; B), and white dots (A488; C) represent the nuclear expression of oct-4 and nanog proteins, respectively. Nuclei are depicted by the blue fluorescence of DAPI staining. Image acquisition was carried out by a Confocal Laser Microscope (Leica TCS-SP2; Leica Microsystems, Milano, Italy), using a 63 × oil immersion objective (numeric aperture: 1.40). Adobe Photoshop software was used to combine RGB channels, overlay the images, and adjust contrast (Adobe, San Jose, CA).
Regarding commitment related genes, we performed a FACS analysis to investigate the expression of BM-specific (GATA2, TAL1; n = 5), cardiac-specific (GATA4, GATA2, Nkx2.5; n = 11), and hepato-specific (GATA4, HNF4-alpha; n = 6) transcription factors in hMASCs. Only a minority of cells expressed the tested markers (Figure S3A,B). RT-PCR analysis was performed to validate cytofluorimetric results and to extend the analysis to other markers such as PU.1 and myocardin (MYOCD) (Figure S3C).
Several reports have demonstrated the role of telomeric length and telomerase activity in stem cell self-renewing, ageing, and mobilization processes.22,23
In order to verify whether cells growing under our experimental conditions possessed telomerase activity, a telomeric repeat amplification protocol assay was performed (n = 17) and we observed that cells at P3 displayed this activity (Figure 3A,B).
Telomeric repeat amplification protocol (TRAP) assay. (A) Telomerase activity products of human heart-derived (hH1, hH2, and hH3), liver-derived (hL1, hL2, and hL3), and bone marrow–derived (hBM1, hBM2, and hBM3) multipotent cells display 6-bp periodicity. Cells treated with RNase (-i) were used as negative control. TSR8 lane represents the telomerase quantitation control template. Developed autoradiographic film was acquired by SnapScan 1236 (Agfa, Mortsel, Belgium) connected to a Macintosh G3 Computer (Apple, Cupertino, CA) equipped with ScanWise software (v1.2.1; Agfa). (B) Quantitation of multipotent cell line telomerase activity. Each unit of total product generated (TPG) corresponds to the number of TS primers (in 1 × 10−3 amole or 600 molecules) extended with at least 4 telomeric repeats by telomerase. Results are presented as mean and standard deviation. The differences among cell lines did not reach statistical significance.
Telomeric repeat amplification protocol (TRAP) assay. (A) Telomerase activity products of human heart-derived (hH1, hH2, and hH3), liver-derived (hL1, hL2, and hL3), and bone marrow–derived (hBM1, hBM2, and hBM3) multipotent cells display 6-bp periodicity. Cells treated with RNase (-i) were used as negative control. TSR8 lane represents the telomerase quantitation control template. Developed autoradiographic film was acquired by SnapScan 1236 (Agfa, Mortsel, Belgium) connected to a Macintosh G3 Computer (Apple, Cupertino, CA) equipped with ScanWise software (v1.2.1; Agfa). (B) Quantitation of multipotent cell line telomerase activity. Each unit of total product generated (TPG) corresponds to the number of TS primers (in 1 × 10−3 amole or 600 molecules) extended with at least 4 telomeric repeats by telomerase. Results are presented as mean and standard deviation. The differences among cell lines did not reach statistical significance.
Altogether, these data indicate that cells, displaying low levels of commitment, expressing pluripotent state–specific markers, and possessing telomerase activity can be obtained in culture from adult human tissues.
Single-cell cloning and multilineage differentiation potential
Clues on the multipotency of the stem cell lines generated from the 3 different tissues came from our preliminary results on multilineage differentiation assays (n = 33) of noncloned cells. In fact, almost all of the tested polyclonal cell lines were able to differentiate in at least one derivative of each germ layer (Figures S4,S5).
However, the most critical parameter in the definition of stem cells is their clonality, which is the ability of single undifferentiated cells to self-renew, proliferate, and differentiate to produce mature progeny cells.24 Therefore, hMASC multipotency was tested at a clonal level in order to exclude that the wide differentiation capacity, displayed by the finite cell lines, had to be attributed to the presence, in culture, of multiple unipotent stem cells.
We thus sorted single cells from 12 distinct growing cultures, obtained from the 3 different organs, either on a preirradiated murine stromal layer (MS5; BM, n = 2; liver, n = 2; and heart, n = 1) or on fibronectin-coated wells (BM, n = 2; liver, n = 4; and heart, n = 1), in culture medium enriched with 10% FBS. In both cases, we were able to generate clones with similar efficiency (17% ± 11% vs 20% ± 8%).
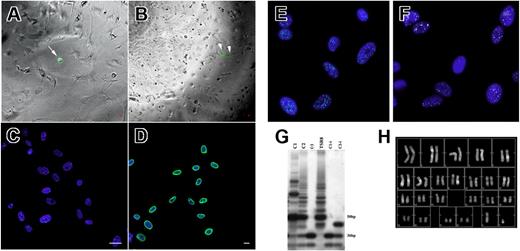
We used CFSE labeling to assess the efficiency of the sorting process and to exclude wells improperly seeded with more than one cell. The fraction of wells containing a single cell was 56% (± 9%), while we never identified a single well seeded with more than one cell. Few days after sorting, the presence of doublets of labeled cells was also documented (Figure 4A,B).
Single-cell sorting. (A) CFSE-labeled (green fluorescence, arrow) single cell seeded on an irradiated murine feeder layer (phase contrast). (B) Doublet of CFSE-labeled cells originated from a single cell 4 days after sorting (arrowheads). (C) X (spectrum green fluorescence) and Y (spectrum orange fluorescence) human chromosomes revealed in hMASC nuclei (blue fluorescence) by FISH analysis. (D) Antihuman lamin A/C antibody decorates (A488, green fluorescence) decorates the nuclei of expanded clones. Blue fluorescence of DAPI identifies nuclei. Scale bars represent 10 μm. (E) Epifluorescence image of oct4 protein expression (A488; green fluorescence) in a hMASC single-cell clone. (F) Epifluorescence image of nanog protein expression (A488; white fluorescence) in a hMASC single-cell clone. Nuclei are depicted by the blue fluorescence of DAPI. (G) Telomerase activity products of a hMASC single-cell clone (C) display 6-bp periodicity. Cells treated with RNase (Ci) were used as negative control. TSR8 lane represents the telomerase quantitation control template. (H) Representative karyotype of single-cell–derived expanded clones. (A,B,E,F) Epifluorescence and phase contrast images were obtained using a live cell imaging dedicated system consisting of a Leica DMI 6000B microscope connected to a Leica DFC350FX camera (Leica Microsystems) equipped with a 5 × dry objective (numeric aperture: 0.12; B), a 10 × dry objective (numeric aperture: 0.25; A), and a 63 × oil immersion objective (numeric aperture: 1.4; E,F). (C,D) Image acquisition was carried out, at room temperature (RT), by a confocal laser microscope (Leica TCS-SP2), using a 40 × oil immersion objective (numeric aperture: 1.25). Adobe Photoshop software was used to combine RGB channels, overlay the images, and adjust contrast. (G) Developed autoradiographic film was acquired by SnapScan 1236 (Agfa) connected to a Macintosh G3 Computer (Apple) equipped with ScanWise software (v1.2.1; Agfa).
Single-cell sorting. (A) CFSE-labeled (green fluorescence, arrow) single cell seeded on an irradiated murine feeder layer (phase contrast). (B) Doublet of CFSE-labeled cells originated from a single cell 4 days after sorting (arrowheads). (C) X (spectrum green fluorescence) and Y (spectrum orange fluorescence) human chromosomes revealed in hMASC nuclei (blue fluorescence) by FISH analysis. (D) Antihuman lamin A/C antibody decorates (A488, green fluorescence) decorates the nuclei of expanded clones. Blue fluorescence of DAPI identifies nuclei. Scale bars represent 10 μm. (E) Epifluorescence image of oct4 protein expression (A488; green fluorescence) in a hMASC single-cell clone. (F) Epifluorescence image of nanog protein expression (A488; white fluorescence) in a hMASC single-cell clone. Nuclei are depicted by the blue fluorescence of DAPI. (G) Telomerase activity products of a hMASC single-cell clone (C) display 6-bp periodicity. Cells treated with RNase (Ci) were used as negative control. TSR8 lane represents the telomerase quantitation control template. (H) Representative karyotype of single-cell–derived expanded clones. (A,B,E,F) Epifluorescence and phase contrast images were obtained using a live cell imaging dedicated system consisting of a Leica DMI 6000B microscope connected to a Leica DFC350FX camera (Leica Microsystems) equipped with a 5 × dry objective (numeric aperture: 0.12; B), a 10 × dry objective (numeric aperture: 0.25; A), and a 63 × oil immersion objective (numeric aperture: 1.4; E,F). (C,D) Image acquisition was carried out, at room temperature (RT), by a confocal laser microscope (Leica TCS-SP2), using a 40 × oil immersion objective (numeric aperture: 1.25). Adobe Photoshop software was used to combine RGB channels, overlay the images, and adjust contrast. (G) Developed autoradiographic film was acquired by SnapScan 1236 (Agfa) connected to a Macintosh G3 Computer (Apple) equipped with ScanWise software (v1.2.1; Agfa).
In order to perform additional analyses, selected clones (n = 20) were expanded to more than 107 cells. The human origin of the expanded clones was confirmed performing both FISH analyses for human X- and Y-chromosomes and staining for human nuclear lamin A/C (Figure 4C,D).
In order to exclude that hMASCs could have fused with MS5 cells generating hybrids, DNA content, karyotype, and the presence of human-specific DNA sequences were assessed, demonstrating that expanded clones were of both human origin and diploid (Figures 4H,S6, and data not shown). The karyotype analysis excluded the presence of chromosomal aberrations as well. Moreover, hMASC cell lines (n = 10) did not grow in soft agar and, when injected subcutaneously in nonobese diabetic–severe combined immunodeficient (Nod-Scid) mice (n = 3), they did not form neoplasia.
We next evaluated whether single-cell clones maintained a stable immunophenotype or if this procedure selected cells with an antigenic pattern radically different from parental lines. FACS analysis showed that the immunophenotype of the expanded clones was similar to that described for polyclonal hMASCs (Figure S7). Moreover, expanded clones continued to express Oct-4 and NANOG and to display telomerase activity (Figure 4E-G).
Finally, we evaluated the multipotency of single-cell–derived clones. For this purpose, first we induced differentiation of BM-derived (n = 59), heart-derived (n = 63), and liver-derived (n = 63) hMASC clones using methods previously described,2,9,11,12,25,26 then we analyzed the acquisition of functional and molecular evidences of differentiation.
Neuroectodermic differentiation.
Cells exposed for one week to EGF and b-FGF displayed a remarkable morphologic change. The majority of the cells that maintained a flattened morphology were positive for the glial fibrillary acidic protein (GFAP; Figures 5J,S8A,D), while neuronlike cells expressed NeuD, a neuron-specific transcription factor (Figure S8B,E), beta3 tubulin arrayed in filaments and bundles (Figure 5A,J,N), neuron-specific enolase (Figure S8C,F), tyrosine hydroxylase (Figure 5B), and acetylcholintransferase (Figure 5C). Interestingly, GFAP and beta3-tubulin were coexpressed in a fraction of differentiating cells, consistent with recent findings27 (Figure 5J).
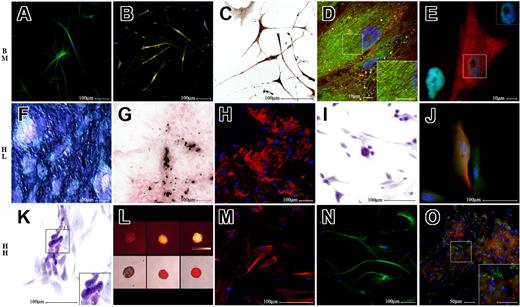
Multilineage differentiation of human single-cell–derived clones. Differentiation of bone marrow (BM)–, liver (HL)– and heart (HH)–derived single-cell clones (SCCs). The top row of figures refers to a BM-derived SCC, the middle row refers to HL-derived SCC, while the bottom row refers to HH-derived SCC. The first 3 pictures of each row illustrate multiple demonstrations of the differentiation into an ectodermic derivative (neurons, top row), a mesodermic derivative (osteoblasts, middle row), and an endodermic derivative (hepatocytes, bottom row). The last 2 pictures of each row demonstrate that each clone was also able to differentiate into derivatives of the other 2 germ layers. Specifically, panel A illustrates beta3 tubulin staining in green (A488), and panel B demonstrates tyrosine hydroxylase staining in yellow (A488). Panel C demonstrates a positive immunohistochemistry staining for acetylcholine transferase. Panel D shows α-actinin (A555, red fluorescence) and ryanodine receptor (BODIPY, green fluorescence) stainings; the insert box shows at higher magnification interdigitating α-actinin and ryanodine receptor positivities. Panel E illustrates GATA4 (A488, green fluorescence in nuclei) and cytokeratin (A555, red fluorescence) labeling; the insert box documents that cytokeratin-positive cells are also GATA4 positive. Panel F reveals a positive cytochemical reaction for alkaline phosphatase activity. Panel G documents a positive von Kossa cytochemical reaction. Panel H shows the red fluorescence of tetracyclines incorporated in calcification sites. Panel I shows in purple a positive PAS reaction. Panel J documents in green GFAP (A488) and in red beta3 tubulin–positive cells (A555). Panel K reveals PAS positivity of differentiated cells. Panel L shows, in the top row, in pseudocolors (Q-LUT, scale bar) resorufin fluorescence of cell aggregates exposed to pentoxyresorufin (PR), and in the bottom row, overlay of phase contrast and red fluorescence images of the same cells; from the left to the right are depicted the following: negative control, cells exposed to PR only, and cells exposed to phenobarbital and PR. Panel M illustrates cytokeratin staining (A555, red fluorescence). Panel N shows beta3 tubulin staining in green (A488). Panel O shows von Willebrand factor in green (A488) and DiI-labeled acetylated LDL in red; insert box is a higher magnification of the marked field. DAPI was used in all fluorescence images to label nuclei in blue. (E,J,L,M) Epifluorescence and phase contrast images obtained using a live cell imaging dedicated system consisting of a Leica DMI 6000B microscope connected to a Leica DFC350FX camera (Leica Microsystems), equipped with a 63× immersion oil (numeric aperture: 1.4; E), a 40× dry (numeric aperture: 0.6; J,M), and a 10× dry (numeric aperture: 0.25; L) objective. (B,D,H,N,O) Image acquisition was carried out by a confocal laser microscope (Leica TCS-SP2), using a 63× immersion oil objective (numeric aperture: 1.40; D) and a 20× dry objectives (numeric aperture: 0.50; A,B,H,N,O). (C,F,G,I,K) Bright field images were captured using an Olympus AX70 microscope connected to an Olympus DP50 camera (Olympus, Italy). A 20× dry objective (numeric aperture: 0.70; F,G) and a 40× dry objective (numeric aperture: 0.95; C,I,K) were used for this purpose. Color temperature: 5400°K. Adobe Photoshop software was used to compose, overlay the images and to adjust contrast (Adobe).
Multilineage differentiation of human single-cell–derived clones. Differentiation of bone marrow (BM)–, liver (HL)– and heart (HH)–derived single-cell clones (SCCs). The top row of figures refers to a BM-derived SCC, the middle row refers to HL-derived SCC, while the bottom row refers to HH-derived SCC. The first 3 pictures of each row illustrate multiple demonstrations of the differentiation into an ectodermic derivative (neurons, top row), a mesodermic derivative (osteoblasts, middle row), and an endodermic derivative (hepatocytes, bottom row). The last 2 pictures of each row demonstrate that each clone was also able to differentiate into derivatives of the other 2 germ layers. Specifically, panel A illustrates beta3 tubulin staining in green (A488), and panel B demonstrates tyrosine hydroxylase staining in yellow (A488). Panel C demonstrates a positive immunohistochemistry staining for acetylcholine transferase. Panel D shows α-actinin (A555, red fluorescence) and ryanodine receptor (BODIPY, green fluorescence) stainings; the insert box shows at higher magnification interdigitating α-actinin and ryanodine receptor positivities. Panel E illustrates GATA4 (A488, green fluorescence in nuclei) and cytokeratin (A555, red fluorescence) labeling; the insert box documents that cytokeratin-positive cells are also GATA4 positive. Panel F reveals a positive cytochemical reaction for alkaline phosphatase activity. Panel G documents a positive von Kossa cytochemical reaction. Panel H shows the red fluorescence of tetracyclines incorporated in calcification sites. Panel I shows in purple a positive PAS reaction. Panel J documents in green GFAP (A488) and in red beta3 tubulin–positive cells (A555). Panel K reveals PAS positivity of differentiated cells. Panel L shows, in the top row, in pseudocolors (Q-LUT, scale bar) resorufin fluorescence of cell aggregates exposed to pentoxyresorufin (PR), and in the bottom row, overlay of phase contrast and red fluorescence images of the same cells; from the left to the right are depicted the following: negative control, cells exposed to PR only, and cells exposed to phenobarbital and PR. Panel M illustrates cytokeratin staining (A555, red fluorescence). Panel N shows beta3 tubulin staining in green (A488). Panel O shows von Willebrand factor in green (A488) and DiI-labeled acetylated LDL in red; insert box is a higher magnification of the marked field. DAPI was used in all fluorescence images to label nuclei in blue. (E,J,L,M) Epifluorescence and phase contrast images obtained using a live cell imaging dedicated system consisting of a Leica DMI 6000B microscope connected to a Leica DFC350FX camera (Leica Microsystems), equipped with a 63× immersion oil (numeric aperture: 1.4; E), a 40× dry (numeric aperture: 0.6; J,M), and a 10× dry (numeric aperture: 0.25; L) objective. (B,D,H,N,O) Image acquisition was carried out by a confocal laser microscope (Leica TCS-SP2), using a 63× immersion oil objective (numeric aperture: 1.40; D) and a 20× dry objectives (numeric aperture: 0.50; A,B,H,N,O). (C,F,G,I,K) Bright field images were captured using an Olympus AX70 microscope connected to an Olympus DP50 camera (Olympus, Italy). A 20× dry objective (numeric aperture: 0.70; F,G) and a 40× dry objective (numeric aperture: 0.95; C,I,K) were used for this purpose. Color temperature: 5400°K. Adobe Photoshop software was used to compose, overlay the images and to adjust contrast (Adobe).
Whole-cell patch-clamp recordings assessed the level of functional competence achieved by the cells. Twenty-nine differentiated hMASCs and 26 undifferentiated cells from 6 and 5 independent cultures, respectively, were studied.
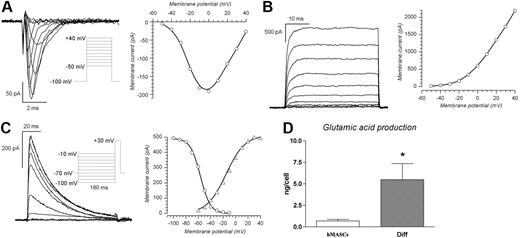
Undifferentiated cells shared similar characteristics, independently from their tissue of origin. Specifically, the membrane capacitance was 11.6 (± 0.7) pF (n = 19), whereas the resting membrane potential (RMP) was somewhat variable, ranging from −3 to −20 mV (−12.2 ± 1.7 mV, n = 19). The cells were tested for the presence of voltage-dependent currents under voltage-clamp conditions by applying depolarizing steps from −50 to + 40 mV after a 180-ms preconditioning to −100 mV, and no inward currents could be detected in any of them. A small delayed rectifier potassium current (6.2 ± 1.1 pA at + 40 mV) was recorded in 8 of 19 tested cells, whereas the other cells showed a purely ohmic behavior. The presence of an inward rectifying current was tested in 13 cells by applying hyperpolarizing steps from − 20 to − 80 mV, with no results.
Differentiated cells presented a rather different setting. Their membrane capacity ranged from 14 to 152 pF (30.8 ± 4.6 pF, n = 29), and their RMP ranged from −5 to −60 mV (−22.5 ± 3.8 mV, n = 29). Concerning the presence of voltage-dependent currents, 17 of 29 differentiated cells presented a distinct TTX-sensitive transient inward current whose maximal amplitude, at 0 mV, ranged from 50 to 1000 pA (237 ± 60.9 pA, n = 17). Usually the Na+ current was isolated by subtraction of the TTX-sensitive component, and occasionally by equimolar substitution of intracellular K+ ions with Cs+. The Na+ current, evoked by depolarizing steps ranging from − 50 to + 40 mV from a holding potential of −100 mV, showed a typical current/voltage (I/V) relationship, peaking at 0 mV and reverting close to the theoretic sodium equilibrium potential (Figure 6A right). Twenty-five of 29 differentiated cells presented large outward currents, 21 of 25 of the conventional delayed rectifier type (Figure 6B), and 4 of 25 of the transient A-type (Figure 6C). The delayed rectifier K+ current averaged 489 (± 113) pA (n = 21) at + 40 mV, and the A-type potassium current showed a peak amplitude of 324 (± 81) pA.
Neuronal differentiation of human single-cell–derived clones. (A) Na+ currents; the tracings were elicited by 80-ms pulses from a holding potential of − 100 mV to test potentials ranging from − 50 to + 40 mV, in 10-mV increments, applied every 10 seconds (protocol in the inset); to the right is represented the corresponding I/V relationship. The potassium currents were blocked by equimolar substitution of intracellular K+ with Cs+. (B) Delayed rectifier potassium currents obtained using the same activation protocol described in panel A; to the right is the corresponding I/V relationship. Here, as for the recordings shown in panel C, the sodium current was blocked by TTX 1 μM. (C) A-type K+ currents; traces obtained with the inactivation protocol shown in the inset: a test potential to − 30 mV was preceded by a 180-ms conditioning step to potentials ranging from − 100 to − 10 mV, and the resulting peak amplitudes are represented to the right as a function of the conditioning potential (○). The peak currents obtained in the same cell with the activation protocol (from − 60 to + 40 mV in 10-mV increments, not shown) are represented to the right (▵). (D) Glutamic acid production. Fluorometric determination of glutamic acid production (expressed in pg and normalized for the seeded cell number) produced between day 10 and day 12 from 10 single-cell–derived clones, either in undifferentiated (▭) or differentiated state ( ). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.
). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.
Neuronal differentiation of human single-cell–derived clones. (A) Na+ currents; the tracings were elicited by 80-ms pulses from a holding potential of − 100 mV to test potentials ranging from − 50 to + 40 mV, in 10-mV increments, applied every 10 seconds (protocol in the inset); to the right is represented the corresponding I/V relationship. The potassium currents were blocked by equimolar substitution of intracellular K+ with Cs+. (B) Delayed rectifier potassium currents obtained using the same activation protocol described in panel A; to the right is the corresponding I/V relationship. Here, as for the recordings shown in panel C, the sodium current was blocked by TTX 1 μM. (C) A-type K+ currents; traces obtained with the inactivation protocol shown in the inset: a test potential to − 30 mV was preceded by a 180-ms conditioning step to potentials ranging from − 100 to − 10 mV, and the resulting peak amplitudes are represented to the right as a function of the conditioning potential (○). The peak currents obtained in the same cell with the activation protocol (from − 60 to + 40 mV in 10-mV increments, not shown) are represented to the right (▵). (D) Glutamic acid production. Fluorometric determination of glutamic acid production (expressed in pg and normalized for the seeded cell number) produced between day 10 and day 12 from 10 single-cell–derived clones, either in undifferentiated (▭) or differentiated state ( ). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.
). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.
In conclusion, cells in neurogenic medium acquired electrophysiological properties (ie, increased resting membrane potential and the appearance of voltage-dependent sodium and potassium currents) indicative of functional differentiation toward neuronal phenotypes. Furthermore, differentiated cells released the excitatory neurotransmitter l-glutamate (Figure 6D).
Mesodermic differentiation of single-cell–derived clones.
When exposed to an osteogenic medium, the clones became positive to (1) osteocalcin immunostaining (Figure S9A), (2) alkaline phosphatase reaction (Figure 5F), and (3) von Kossa staining (Figure 5G). As an additional proof of the acquisition of a functional competence, we cultured hMASCs in the presence of tetracycline, a calcium chelator often used to label the mineralizing front of forming bone. In cultures induced to differentiate toward an osteogenic fate, a gradual deposition of tetracycline-labeled material was demonstrated, indicating that the newly formed matrix was also mineralized (Figures 5H,S9B).
The ability of single-cell–derived clones to differentiate into muscle cells was tested in a medium containing VEGF, bFGF, and IGF-1. After the differentiation period, cells showed organized filaments of α-actinin and α-sarcomeric actin (Figures 5D,S10A). Gap-junctions were demonstrated by the presence of connexin-43 in proximity to cell-to-cell contact sites (Figure S10A). Ryanodine receptors were organized in tubular structures interdigitating α-actinin filaments (Figure 5D), suggesting their localization in the developing sarcoplasmic reticulum.28 L-Type calcium channels were also identified in differentiated cells (Figure S10B).
The presence of functional competent receptors involved in calcium handling was corroborated by the demonstration of spontaneous intracellular calcium transients, as displayed by Fluo4 assays (Video S1). Spontaneous calcium transients were indeed detectable in 36.3% (± 13.5%), 16.7% (± 9.8%), and 36.7% (± 8.8%) of bone marrow–derived, heart-derived, and liver-derived differentiated cells, respectively.
Moreover, cells maintained in differentiation medium for at least 3 months showed, although at a very low frequency (2-5 cells/104 seeded cells), a spontaneous contractile activity, as documented by time-lapse microscopy (Video S2). Cells continued to beat for up to 4 weeks in culture.
Importantly, undifferentiated cells did not show spontaneous calcium oscillations or contractile activity (data not shown).
Next, we evaluated the ability of hMASCs to differentiate into endothelial cells. In fact, after a 14-day treatment with VEGF, the majority of cells expressed von Willebrand factor (VWF) and actively uptook acetylated LDL (Figure 5O).
Endodermic differentiation.
In order to verify if hMASCs were able to generate endodermal derivatives we induced hepatic differentiation.
Differentiated cells assumed a globular shape with an eccentric nucleus, progressively decreasing their anchorage to the substrate. These cells stained positive for cytokeratins 18 and 19 and for the transcription factor GATA-4 (Figure 5E,M). Finally, they acquired some hepatocytic functions such as the ability to store glycogen, as demonstrated by PAS staining (Figure 5I,K), and to produce albumin, as tested by dosing its concentration in culture supernatants (Figure S11B). Moreover, an inducible cytochrome P450 activity was revealed by PROD assay (Figures 5L,S11A).
Multipotency of single-cell–derived clones.
In order to verify the fraction of clones that displayed multipotency, we decided to score as multipotent a clone that was able to generate at least one derivative of each germ layer. Of the tested clones, 65% (± 9%) possessed a wide multilineage differentiation potential (Table S3).
Altogether the accumulated evidence shows that clonal cell populations, derived from the hMASC lines, maintain the differentiation potential described for parental lines. Therefore, the reported clonal assay excludes the possibility of contamination with other cell types that could be instrumental to support the stem- cell phenotype.
Transcript expression profiling of heart-, liver- and bone marrow–derived hMASCs by cDNA microarrays
Using the cDNA microarray technology, we characterized the gene expression profiles of BM-derived (n = 4), liver-derived (n = 5), and cardiac-derived (n = 7) hMASC lines together with 8 different somatic cell lines (IOSE-h-TERT [n = 20], U2-OS [n = 10], HCT-116 [n = 6], NT2/D1 [n = 3], U-118 MG [n = 3], HT-29 [n = 3], OVCAR-3 [n = 3], CAOV-3 [n = 1]) and 2 adult human tissues, heart (n = 2) and liver (n = 2). The 8 different cell lines and the 2 adult tissues were integrated into the hMASCs transcriptional profiling study to provide a framework for interpretation of the hMASC-specific expression patterns.
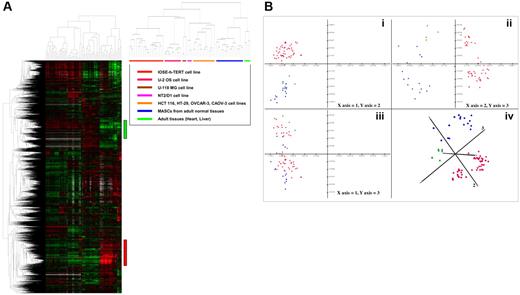
In order to identify gene expression signatures differentially expressed among the different groups of the 69 profiled samples, we performed unsupervised hierarchical cluster analysis and principal component analysis (PCA) on FEM1 matrix (“Materials and methods”). The hierarchical clustering results for the 14 902 features of the filtered expression matrix (FEM1) are displayed in Figure 7A. Based solely on their gene expression patterns, hMASCs derived from the 3 different organs are characterized by the expression of a unique set of genes or transcriptional module (red bar in Figure 7A, ∼ 2000 array elements), characterized by genes up-regulated in hMASCs with respect to the profiled somatic cell lines and adult tissues, and able to unequivocally distinguish the hMASCs from all the other samples. The most up-regulated transcripts, with respect to their functional roles, can be classified as follows: cell adhesion molecules (CDH11, ITGA2, ITGB1); extracellular matrix–related elements (FN1, BGN, DCN, FBLN2, COL3A1, COL6A1, MMP2, TFPI2, THBS1, SPARC); immune response proteins (CD59, B2M); markers of the undifferentiated state (VIM, LMO4, LAMC1, LAMA4, THY1, SNAI2, ENG, CD200); cytokines, growth factors, and receptors (CXCL12, CXCL1, CXCL2, IL6, IL6R, LIF, FGF2, FGFR4, VEGFC). This transcriptional signature also contains elements of the Wnt (WNT2, FZD1, DKK3, TCF4), TGF-β (TGFB3, TGFBR2), Hedgehog (SMO, GLI3), and Notch (NOTCH2, NOTCH4, HES1) pathways that are known to regulate the self-renewal/maintenance of different stem cells. Cluster analysis highlighted also the existence of an opposite gene module (green bar in Figure 7A, ∼ 1500 array elements) characterized by genes down-regulated in hMASCs with respect to the profiled somatic cell lines. Among the most down-regulated transcripts there were CD24, cytokeratin genes (KRT7, KRT8, KRT19), and diverse Histone 1 genes. This gene module was also highly enriched for cell cycle/mitotic surveillance elements (CDC2, CDC6, CDC20, CCNA2, AURKB, FOXM1, BUB1, BIRC5, TOP2A, PCNA, RAD21, PLK1, MCM6, MCM7, TUBA1, TUBB2). Some of the genes more highly up-regulated and down-regulated in hMASCs with respect to the profiled somatic cell lines and adult tissues are reported in Table S4A and Table S4B, respectively.
hMASC gene expression profile. (A) Left panel shows the unsupervised hierarchical clustering analysis for the 14 902 features of the filtered expression matrix (FEM1). hMASCs derived from the 3 different organs are characterized by the expression of a unique set of genes or transcriptional module (red vertical bar), characterized by genes up-regulated in hMASCs with respect to the profiled somatic cell lines and adult tissues. Green vertical bar highlights the existence of an opposite gene module characterized by genes down-regulated in hMASCs with respect to the profiled somatic cell lines. Right panel schematizes the sample arrangement with respect to the dendrogram. (B) Principal component analysis (PCA) of log-transformed data starting from filtered FEM1 matrix (14 902 features). In panels Bi-iii, the first 3 principal components are plotted in pairs, while in panel Biv the first 3 principal components are plotted in 3D view. The emerging sample groups confirmed that the 3 hMASC types (blue dots) constitute a homogeneous cell population, irrespective of their tissue of origin and distinct from that of somatic cell lines (red dots) and adult tissues (green dots).
hMASC gene expression profile. (A) Left panel shows the unsupervised hierarchical clustering analysis for the 14 902 features of the filtered expression matrix (FEM1). hMASCs derived from the 3 different organs are characterized by the expression of a unique set of genes or transcriptional module (red vertical bar), characterized by genes up-regulated in hMASCs with respect to the profiled somatic cell lines and adult tissues. Green vertical bar highlights the existence of an opposite gene module characterized by genes down-regulated in hMASCs with respect to the profiled somatic cell lines. Right panel schematizes the sample arrangement with respect to the dendrogram. (B) Principal component analysis (PCA) of log-transformed data starting from filtered FEM1 matrix (14 902 features). In panels Bi-iii, the first 3 principal components are plotted in pairs, while in panel Biv the first 3 principal components are plotted in 3D view. The emerging sample groups confirmed that the 3 hMASC types (blue dots) constitute a homogeneous cell population, irrespective of their tissue of origin and distinct from that of somatic cell lines (red dots) and adult tissues (green dots).
Principal component analysis, performed on the 69 expression profiled samples, confirmed that the 3 hMASC types exhibited a very similar gene expression profile, distinct from that of somatic cell lines and adult tissues (Figure 7B).
Both cluster analysis and PCA plots demonstrate that, in terms of their overall transcript expression profiles, hMASCs constitute a homogeneous cell population, irrespective of their tissue of origin.
In order to identify similarities/differences among the 3 hMASC types, we performed unsupervised hierarchical cluster analysis on the FEM2 matrix (“Materials and methods”). This revealed, once again, a strong similarity among cells of different origin, with no significant gene clusters associated to each class (data not shown). Expression data were then analyzed using Student t test (P < .01) and analysis of variance test (ANOVA) (5% level of significance): in both cases only very few genes were differentially expressed in a statistically significant manner (data not shown).
On the whole, the analyses revealed that, among the 3 hMASC types, the overall gene expression profiles were markedly similar, indicative of their intrinsic transcriptional uniqueness.
Discussion
In vivo and in vitro studies performed in animal models have suggested that stem cells obtained from different tissues can differentiate into unrelated ones, even crossing lineage boundaries.29,30 This stem-cell property, named stem-cell plasticity, has not reached a consensus either in terms of its basic properties or of the responsible molecular mechanisms.24,31,32 A potential explanation for this phenomenon is the presence in tissues of adult organisms of a single, rare, multipotent stem cell that copurifies in protocols designed to enrich for resident stem cells.24 Another possibility is that adult stem-cell multipotency is an in vitro artifact, a claim that has been extended, by some authors, even to embryonic stem cells.33,34 Although this distinction is fundamental from a biologic standpoint, the potential clinical utility of obtaining and expanding plastic cells in vitro remains unquestionable.33,34
In humans, the most convincing evidence, although very debated,7 for the in vitro generation of a population of multipotent cells has been obtained only for BM.10
We therefore investigated the hypotheses that populations of widely multipotent cells similar to those identified in young mice9 could be obtained in culture from adult human organs, other than BM, and that these cells shared common features, possibly responsible for their multipotent state.
Adopting systematically a well-standardized methodological approach9 we grew in vitro, with a high reproducibility, 99 cell lines starting from 107 different human samples. We found that, similarly to adult BM used as parallel reference, an analogous population could be obtained in vitro from adult human liver and heart. Specifically, it was possible to produce in culture a population of multipotent, clonogenic, telomerase-positive cells, expressing pluripotent state–specific embryonic genes. Expanded single-cell–derived clones retained the expression of oct-4 and nanog, and the ability to differentiate into cell types morphologically and functionally corresponding to derivatives of the 3 germ layers.
Interestingly, cell lines generated in culture from different tissues and expanded in vitro displayed high levels of similarity, as confirmed by their global transcriptional profiles. Specifically, we observed a common gene signature indicative of (1) maintenance of an undifferentiated state, (2) active expression of cytokines, growth factors, and related receptors, (3) high expression of extracellular matrix–related elements, and (4) cell adhesion molecules. In addition, this transcriptional signature contains elements of various pathways such as Wnt, Hedgehog, and Notch that are known to regulate the self-renewal/maintenance in different stem cells.35
Although the resulting similarities found within the derived cells do not necessarily imply their identity, it can be speculated that hMASCs obtained and expanded in culture from the most accessible tissues (eg, bone marrow or adipose tissue) could be used for the regeneration of tissues, even unrelated to the one of origin.
The results obtained, although exciting, raise some crucial issues.
First, the demonstration of cells that possess in vitro multipotency does not imply the existence of an in vivo counterpart. In fact, a possible explanation for the observed multipotency is that cells, extensively expanded under low-serum conditions, could be reprogrammed to a more embryonic phenotype (dedifferentiation).36
Second, it would be valuable from both a biologic and a clinical viewpoint to recognize which tissue conditions could interfere with the ability to produce hMASCs in vitro. From our data, we have seen that peripheral blood, in steady-state conditions, could not be used as a source, suggesting that the artificial microenvironment per se is not a sufficient condition to reprogram these cells. On the other hand, it is important to underline that hMASC lines can be obtained from both pathological and nonpathological samples. In fact, considering the heart model, we isolated similar cells both from explanted and transplanted organs.37
In conclusion, (1) we have optimized a method that allows to obtain, from different human tissues, with high efficiency, proliferating cell lines characterized by the expression of pluripotent state–specific transcription factors, multipotency, and telomerase activity (hMASCs); (2) proliferating cell lines can be cloned and single-cell–derived clones retain the ability to differentiate toward several mature cell types; and (3) hMASCs share strong similarities, independently from the tissue of origin.
The isolated hMASCs could represent a valid system to dissect the molecular mechanisms regulating cell proliferation and differentiation toward specific lineages, thus providing new molecular targets to be used for regenerative approaches.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the following grants: MURST-PRIN pr.2004061130 (Udine University/CAB), Italian Ministry of Health Research Project pr.10/04 (Udine University/CAB), AIRC-Regional Grant 2005 pr.1023 (Udine University/CAB; LNCIB/CS), AIRC-Onco Genomic Center Grant (LNCIB/CS), and FIRB projects n.RBNE01ZYMR (LNCIB/CS) and RBNE01MAWA (LNCIB/CS).
We thank Drs Maura Pandolfi and Moira Bottecchia for their important technical help, and Dr Vanna Pecile (Burlo Garofolo Hospital, Trieste, Italy) for the karyotype analyses.
Authorship
Contribution: A.P.B. designed and performed research, analyzed data, and wrote the paper; D.C. designed and performed research, analyzed data, and wrote the paper; N.B. designed and performed research, analyzed data, and wrote the paper; P.M. designed and performed research; S.R. designed and performed research; F.D. designed and performed research; E.P. designed and performed research; R.V. designed and performed research, and analyzed data; S.P. designed and performed research, and analyzed data; A.P. performed research; U.B. performed research; D.D. performed research; R.F. performed research; L.M. performed research; N.F. performed research; P.M. performed research; S.B. performed research; O.B. designed and performed research; C.S. designed research and wrote the paper; and C.A.B. designed research and wrote the paper. A.P.B. and D.C. contributed equally to this work
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo Alberto Beltrami, Istituto di Anatomia Patologica, Università degli Studi di Udine, Piazzale S. Maria della Misericordia, 33100 Udine, Italy; e-mail: beltrami@uniud.it.










 ). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.
). Results are expressed as mean (± SEM). *P < .01 versus single-cell–derived clones.