The Epstein-Barr virus (EBV) contributes to the growth and survival of Hodgkin lymphoma (HL) cells. Here we report that down-regulation of the transforming growth factor-beta (TGF-beta) target gene, protein tyrosine phosphatase receptor kappa (PTPRK), followed EBV infection of HL cells and was also more frequently observed in the Hodgkin and Reed-Sternberg (HRS) cells of EBV-positive compared with EBV-negative primary HL. The viability and proliferation of EBV-positive HL cells was decreased by overexpression of PTPRK, but increased following the knockdown of PTPRK expression in EBV-negative HL cells, demonstrating that PTPRK is a functional tumor suppressor in HL. EBV suppressed the TGF-beta–mediated activation of PTPRK expression, suggesting disruption of TGF-beta signaling upstream of PTPRK. This was confirmed when we showed that the Epstein-Barr nuclear antigen-1 (EBNA1) decreased Smad2 protein levels and that this was responsible for PTPRK down-regulation. EBNA1 decreased the half-life of Smad2 but did not interact with Smad2. By down-regulating Smad2 protein expression, EBNA1 apparently disables TGF-beta signaling, which subsequently decreases transcription of the PTPRK tumor suppressor. We speculate that loss of the phosphatase function of PTPRK may activate as-yet-unidentified growth-promoting protein tyrosine kinases, which in turn contribute to the pathogenesis of EBV-positive HL.
Introduction
In approximately half of patients with Hodgkin lymphoma (HL), the Epstein-Barr virus (EBV) can be localized to the malignant Hodgkin and Reed-Sternberg (HRS) cells where it expresses a restricted subset of virus genes, including the latent membrane protein 1 (LMP-1) and LMP-2 and the Epstein-Barr nuclear antigen-1 (EBNA1). Although LMP1 and LMP2 are believed to contribute to the pathogenesis of HL,1 it is not known if EBNA1 is also involved. Although EBNA1 enables efficient episome replication and segregation and is therefore essential for the maintenance of latent EBV infection,2,3 several studies suggest it might also contribute more directly to tumorigenesis. For example, EBNA1 alone can inhibit apoptosis4 and can enhance the tumorigenicity of HL cells in nonobese severe combined immunodeficient (SCID) mice.5 Transgenic mice in which EBNA1 expression was targeted to B cells also show an increased incidence of lymphomas,6 although another study recently failed to replicate these findings.7
We previously generated KM-H2 HL cells stably infected with a recombinant EBV and also produced EBV-negative clones of the parental EBV-positive L591 cell line; in both cell lines the presence of EBV was associated with significantly increased growth and survival.8 Here we show that expression of the protein tyrosine phosphatase receptor K (PTPRK) gene is down-regulated by EBV infection in these cell lines and also in the primary tumor cells of EBV-positive HL. Furthermore, we show that PTPRK is a transforming growth factor-beta (TGF-beta) responsive gene in HL cells and that its down-regulation contributes to increased cellular growth and survival and is a consequence of EBNA1-mediated decreases in Smad2 protein. Our data point to an important role for EBNA1 in conferring resistance to the growth-suppressive effects of TGF-beta in EBV-positive HL.
Methods
This study received ethical approval from the South Birmingham Research Ethics Committee (LREC no. 0844).
Cell lines
KM-H2 was originally established from the pleural effusion of a patient with mixed cellularity HL.9 L591 is a cell line that originated from the pleural effusion of a female patient with histologically confirmed nodular sclerosis HL.10 KM-H2 cells infected with Akata-derived recombinant EBV11 were established as previously described8 and subsequently maintained under drug selection (G418 sulfate, 1 mg/mL). Control KM-H2 cells were generated by electroporation with vector only. EBV-positive L591 cells were serially diluted for up to 6 weeks to generate EBV-negative clones as previously described.8 All cell lines were cultured at 37°C in 5% CO2 in normal growth media: RPMI1640 supplemented with 10% B-cell serum, 2 mM glutamine (Invitrogen, Paisley, United Kingdom), and 0.5% penicillin-streptomycin solution (Sigma-Aldrich, Gillingham, United Kingdom), which was exchanged twice weekly.
Cell viability and proliferation assays
Viability of cell lines was determined using trypan blue reagent (Sigma-Aldrich). Cell proliferation was determined using WST-1 reagent (Roche Diagnostics, Burgess Hill, United Kingdom). In this assay, tetrazolium salts are cleaved to formazan by cellular enzymes. An expansion in the number of metabolically active cells results in an increase in the amount of formazan dye formed, which can be quantified by a scanning multiwell spectrophotometer (enzyme-linked immunosorbent assay [ELISA] reader) by measuring the absorbance of the dye solution at 450 nm. Results from all assays were taken from triplicate wells, averaged out, plotted as a graph showing standard error of mean, and subjected to statistical analysis in Microsoft Excel (Reading, United Kingdom) using a 2-tailed Student t test assuming the 2 samples displayed unequal variance. The generated probability (P) values were taken to indicate a significant difference between the datasets when the P value was less than .05.
TGF-beta stimulation of HL cells
KM-H2 and KM-H2-EBV cells were washed in PBS and resuspended in OptiMEM serum-free media (Invitrogen), supplemented with 0.5% penicillin-streptomycin solution, with or without recombinant human TGF-beta1 (PeproTech EC, London, United Kingdom) at a concentration of 20 ng/mL. Cells were incubated at 37°C for 24 hours before protein or RNA was extracted.
Transfection of individual genes into HL cells
HL cells were transfected with either pcDNA/HisMaxA-LMP2A, pSG5-EBNA1, pCDNA3-A73, pCDNA3-RPMS1, or pCMV-Tag2A-(Flag)-PTPRK (a kind gift from Shizhen Emily Wang and Carlos L. Arteaga, Department of Cancer Biology, Vanderbilt University School of Medicine, Nashville, TN12 ) expression plasmids or appropriate empty vector. Transfection was performed by the process of Nucleofection using the Cell Line Nucleofector Kit T (Amaxa, Cologne, Germany) for the Nucleofector Device (Amaxa), according to the manufacturer's protocol. Cells were incubated at 37°C for 24 hours before protein or RNA was extracted.
Reverse-transcriptase–polymerase chain reaction
RNA was extracted using the NucleoSpin RNA II kit (AB Gene, Epsom, United Kingdom) according to the protocol of the manufacturer. cDNA was generated in a reaction consisting of 1.5 μg RNA, 4 μL avian myeloblastosis virus (AMV) buffer, 5 units reverse transcriptase AMV (Roche Diagnostics), 20 pmol oligo (dT) primers (Alta Bioscience, Birmingham, United Kingdom), 2 μL deoxyribonucleotide triphosphates (dNTPs), 1 unit RNase inhibitor (Roche Diagnostics), and diethylpyrocarbonate (DEPC)–treated water to make the final reaction volume up to 20 μL. Tubes were incubated for 1 hour at 42°C. Of each reaction product, 2 μL was then added to a second tube reaction containing 5 μL 10× polymerase chain reaction (PCR) buffer, 5 μL dNTPs, 5 μL 3′ primer, 5 mL 5′ primer, 3 μL 25 mM magnesium chloride, 2.5 units Platinum Taq DNA polymerase (Invitrogen), and DEPC-treated water to make the final reaction product up to 50 μL. PCR amplification using an Eppendorf Thermal Cycler (Cambridge, United Kingdom) involved an initial 2-minute denaturation at 94°C, followed by 30 cycles consisting of a denaturing step for 30 seconds at 94°C, an annealing step for 1 minute at a temperature specific for primers, and an extension for 1 minute at 72°C. Primers used were as follows: PTPRK forward 5′-CAGAGGAAGGGATGCTACGAT-3′ and reverse 5′-CAGTTCGCCCGCCACCATTT-3′, annealing at 59°C; Smad2 forward 5′-CGAAATGCCACGGTAGAAAT-3′ and reverse 5′-CGGCTTCAAAACCCTGATTA-3′, annealing at 57°C; EBNA1 forward 5′-CCGCAGATGACCCAGGAGAA-3′ and reverse 5′-TGGAAACCAGGGAGGCAAAT-3′, annealing at 55°C; and GAPDH forward 5′-CCACCCATGGCAATTCCATGGCA-3′ and reverse 5′-TCTAGACGGCAGGTCAGGTCCACC-3′, annealing at 60°C.
Quantitative PCR
Each 25-μL quantitative PCR (Q-PCR) reaction consisted of the following: 12.5 μL 2× TaqMan Universal PCR Mastermix; 1.25 μL of each TaqMan Gene Expression Assay primer and probe mix for either the target gene of interest (SMAD2 Hs00183425, PTPRK Hs00267788) or β-2-microglobulin (B2M) used as the “housekeeping” gene; 50 ng cDNA together with RNase-free water in a volume of 11.25 μL. Each sample was analyzed in triplicate. Q-PCR was performed on an ABI 7500 Fast Real Time PCR System (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's instructions. Data were analyzed using 7500 Fast System SDS Software 1.3.1 (Applied Biosystems). This uses the 2-DeltaDelta CT method for quantifying expression relative to the B2M housekeeping control. The lowest 2-DeltaDelta CT value was set to a relative quantity (RQ) value of 1, and all other samples were expressed as a ratio of this.
Gene expression analysis
Affymetrix HG Focus arrays (http://www.affymetrix.com/products/arrays/specific/focus.affx) were used for all experiments. Total RNA from mycoplasma-free cell lines was used to prepare biotinylated RNA.13 3′/5′ ratios for GAPDH and beta-actin were within acceptable limits (GAPDH, 0.72-0.81; beta-actin, 0.89-1.14), and BioB, BioC, BioD, and CreX spike controls were also present in increasing intensity. When scaled to a target intensity of 100 (Affymetrix MAS 5.0), scaling factors for all arrays were within acceptable limits (HG Focus chips 0.54-0.85), as were background, Q values, and mean intensities.
Images of GeneChips were analyzed by Affymetrix Microarray Suite 5.0. Probe level quantile normalization14 and robust multiarray analysis15 on the raw.CEL files were performed using the Affymetrix package of the Bioconductor (http://www.bioconductor.org) project. Differentially expressed probe sets were identified using significance analysis of microarrays (SAM)16,17 ; only those with fold change equal to or greater than 1.5 and false discovery rate less than or equal to 10% were included. Hierarchic clustering was performed using dChip (http://www.dchip.org, Harvard School of Public Health).
Immunoblot analysis
Cells were lysed in buffer (50 mM tris(hydroxymethyl)aminomethane [pH 7.5], 9 M urea, 0.15 M beta-mercaptoethanol) and protein was quantified by BioRad DC Protein Assay Kit. Gel sample buffer was added to samples prior to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transfer to BioTrace NT membrane (VWR International, Lutterwath, United Kingdom), and incubation at 4°C overnight with relevant primary antibodies diluted in 2.5% milk. Antibodies used were as follows: PTPRK rabbit polyclonal antibody (ab13225; Abcam, Cambridge, United Kingdom) at 1:500 dilution; USP 7 rabbit polyclonal antibody (Bethyl Laboratories, Montgomery, TX) at 1:1000 dilution; Smad2 rabbit polyclonal antibody (51-1300; Zymed Laboratories, South San Francisco, CA) at 1:500 dilution; and phospho-Smad2 (Ser465/467) (138D4) rabbit monoclonal antibody (3108S; Cell Signaling Technology, Danvers, MA) at 1:1000 dilution. Actin (AC-38; Sigma-Aldrich) at a 1:20 000 dilution and minichromosome maintenance (MCM)-7 (M7931; Sigma-Aldrich) at a 1:2000 dilution were used as loading controls. Following TTBS rinsing, blots were incubated for 2 hours with HRP-conjugated goat anti–rabbit or goat anti–mouse secondary IgG (DakoCytomation, Ely, United Kingdom). Detection was with ECL (Amersham-Pharmacia Biotech, Little Chalfont, United Kingdom).
Immunohistochemistry
PTPRK immunohistochemistry was carried out on a panel of HL biopsies obtained from the Queen Elizabeth Hospital (Birmingham, United Kingdom). All cases were reviewed by a single pathologist. Sections of paraffin-embedded tissues were cut at 4-μm thicknesses onto microscope slides treated with Vectabond Reagent (Vector Laboratories, Peterborough, United Kingdom). Sections were dewaxed and rehydrated, and endogenous peroxidase activity was blocked. Antigen retrieval was achieved by microwaving slides in citric acid buffer (pH 5.8) for 15 minutes. Sections were incubated in goat polyclonal PTPRK antibody (M-20; Autogen Bioclear, Wiltshire, United Kingdom) at a 1:20 dilution. Secondary detection was achieved using the VECTASTAIN Universal Quick Kit (Vector Laboratories) as described in the manufacturers' protocol. Visualization was carried out using the Sigma Fast 3,3′-diaminobenzidine (DAB) substrate system (Sigma-Aldrich) for 10 minutes, and then sections were counterstained with hematoxylin, dehydrated, and mounted for microscopic analysis. In parallel, LMP1 immunohistochemistry was performed to determine the EBV status of each biopsy, using a mouse monoclonal LMP1 antibody (CS1-4; DakoCytomation) at 1:50 dilution.
RNA interference
Cells suspended in OptiMEM serum-free media (Invitrogen) were incubated in the presence of 10 nM specific small inhibitory RNA (siRNA) in RiboJuice transfection reagent (Merck Biosciences, Nottingham, United Kingdom) according to the protocol for 6-well plates. PTPRK siRNAs (5′-CCACCAGGAUCUGUAUGAUUU-3′) were designed and produced by Eurogentec (Southampton, United Kingdom); HP Validated Smad2 siRNAs (1027400) were supplied by QIAGEN (Crawley, United Kingdom). Cells were incubated at 37°C for 4 hours after which an equivalent volume of OptiMEM media containing 0.5% penicillin-streptomycin solution and 20% B-cell serum was added, followed by further incubation at 37°C. RT-PCR, immunoblotting, and proliferation and viability assays were performed as described earlier. RiboJuice only, OptiMEM only, and irrelevant siRNAs (scrambled LMP1 siRNA 5′-GGGUAGAUAGACUCUCGCU-3′) acted as negative controls.
Immunoprecipitation
L428 HL cells that stably expressed a GFP-EBNA1 fusion protein were pelleted and washed 3 times in 10 mL cold PBS. After the last wash, all supernatant was aspirated on ice and replaced with 500 μL prechilled CHAP lysis buffer. Cells were incubated on ice for 30 minutes, vigorously vortexed, and centrifuged at 25 160g for 20 minutes at 4°C, and the supernatants were removed for protein concentration determination. Protein (1 mg from each sample) was equalized to 800 μL with cold lysis buffer containing protease inhibitors. Samples were precleared for 1 hour by addition of 100 μL of a 1:1 slurry of prewashed protein G beads (CRUK) and lysis buffer, while rotating at 4°C. Samples were then centrifuged at 500g at 4°C to pellet the beads. The supernatants were transferred to another tube and 2 μg GFP antibody was added to the precleared lysates and the samples were rotated at 4°C for 16 hours. Control immunoprecipitation (IP) samples incubated with 2 μg of the relevant isotype control antibody (Sigma-Aldrich) were run alongside tests. Anti–rabbit IgG agarose beads (30 μL; Sigma-Aldrich) were added to the samples and left to rotate for 2 hours at 4°C. The agarose and any bound immunocomplexes were recovered by centrifuging at 500g, 4°C for 5 minutes. The supernatants were then discarded and the beads washed 5 times in 500 μL cold lysis buffer with protease inhibitors added. After the final wash, supernatants were aspirated and the immunocomplexes were eluted from the agarose by addition of 30 μL preheated 4× Laemmli buffer and analyzed by subsequent SDS-PAGE and immunoblotting for USP7 and Smad2.
Determination of Smad2 half-life
EBV-positive and -negative KM-H2 cells were incubated with 200 μg/mL of the protein synthesis inhibitor cycloheximide (Sigma-Aldrich) for 6, 12, and 24 hours before protein was extracted. Lysates were then subjected to immunoblotting for Smad2 and actin protein expression. Densitometric scanning was performed on the immunoblots and the values for Smad2 were normalized against actin. The results were plotted on a scatter plot, from which the half-life of Smad2 could be determined.
Results
PTPRK expression is down-regulated by EBV infection or expression of EBNA1 in HL cells
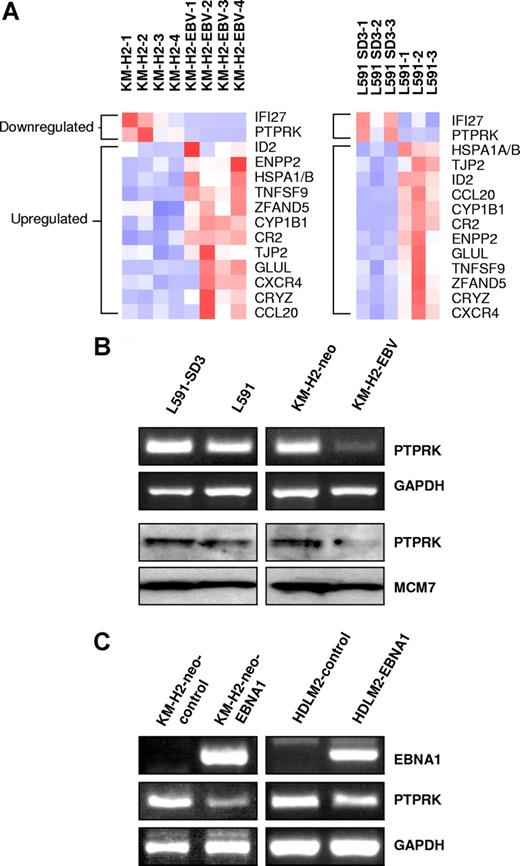
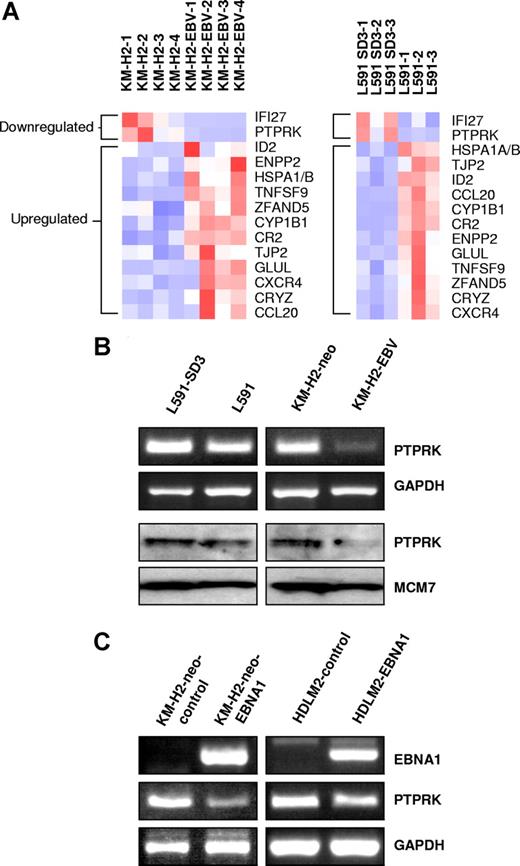
We initially compared cellular gene expression in paired EBV-positive and -negative HL cell lines by microarray analysis. We compared our previously published KM-H2 microarray data8 with the L591 microarray data presented here. A total of 12 genes were up-regulated and 2 genes down-regulated in the presence of EBV in both cell lines (Figure 1A). Genes differentially expressed in KM-H2-EBV cells and L591 cells are given in Tables S1 and S2 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Genes differentially expressed in both cell lines are given in Table 1 and include autotaxin, which we have previously reported to be up-regulated by EBV infection of HL cells.8 Of the 12 genes up-regulated by EBV (Table 1), 4 (CYP1B1, ID2, autotaxin, and Glutamate-ammonia ligase) have previously been reported to be suppressed by TGF-beta.18,,–21 Furthermore, both genes down-regulated by EBV (PTPRK and IFI27) are known to be induced by TGF-beta.12,22
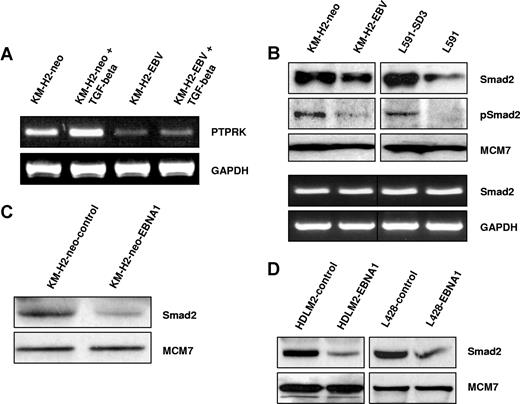
PTPRK expression is down-regulated by EBV infection or expression of EBNA1 in Hodgkin lymphoma cells. (A) Heat map showing genes differentially expressed in the presence of EBV in both L591 and KM-H2 cells. Twelve genes were up-regulated and 2 genes down-regulated in both HL cell lines in the presence of EBV. Down-regulated genes included PTPRK, a putative tumor-suppressor gene. L591-SD3 is an EBV-negative variant of the EBV-positive parental L591 line. KM-H2-EBV is an EBV-positive HL cell line generated by infection of EBV-negative KM-H2 cells with recombinant Akata-derived EBV. (B) Semiquantitative RT-PCR for PTPRK shows lower levels of PTPRK mRNA in EBV-positive L591 and KM-H2 cells compared with their EBV-negative counterparts (L591-SD3 and KM-H2-neo, respectively). Immunoblotting demonstrates the down-regulation of PTPRK protein in EBV-positive HL cells compared with their EBV-negative counterparts. KM-H2 neo is a control cell line generated by the transfection of EBV-negative KM-H2 cells with a plasmid containing a neomycin resistance gene. (C left panel) RT-PCR analysis of EBV-negative KM-H2 cells transfected with EBNA1 reveals that expression of EBNA1 is sufficient to reduce PTPRK mRNA expression. No other EBV latent gene affected PTPRK expression (data not shown). (C right panel) RT-PCR analysis demonstrates that transient transfection of the EBV-negative HL cell line, HDLM-2, with EBNA1 also results in the down-regulation of PTPRK expression. Similar results were observed following ectopic EBNA1 expression in the EBV-negative HL cell line L428 (data not shown).
PTPRK expression is down-regulated by EBV infection or expression of EBNA1 in Hodgkin lymphoma cells. (A) Heat map showing genes differentially expressed in the presence of EBV in both L591 and KM-H2 cells. Twelve genes were up-regulated and 2 genes down-regulated in both HL cell lines in the presence of EBV. Down-regulated genes included PTPRK, a putative tumor-suppressor gene. L591-SD3 is an EBV-negative variant of the EBV-positive parental L591 line. KM-H2-EBV is an EBV-positive HL cell line generated by infection of EBV-negative KM-H2 cells with recombinant Akata-derived EBV. (B) Semiquantitative RT-PCR for PTPRK shows lower levels of PTPRK mRNA in EBV-positive L591 and KM-H2 cells compared with their EBV-negative counterparts (L591-SD3 and KM-H2-neo, respectively). Immunoblotting demonstrates the down-regulation of PTPRK protein in EBV-positive HL cells compared with their EBV-negative counterparts. KM-H2 neo is a control cell line generated by the transfection of EBV-negative KM-H2 cells with a plasmid containing a neomycin resistance gene. (C left panel) RT-PCR analysis of EBV-negative KM-H2 cells transfected with EBNA1 reveals that expression of EBNA1 is sufficient to reduce PTPRK mRNA expression. No other EBV latent gene affected PTPRK expression (data not shown). (C right panel) RT-PCR analysis demonstrates that transient transfection of the EBV-negative HL cell line, HDLM-2, with EBNA1 also results in the down-regulation of PTPRK expression. Similar results were observed following ectopic EBNA1 expression in the EBV-negative HL cell line L428 (data not shown).
Semiquantitative RT-PCR confirmed the down-regulation of PTPRK mRNA in EBV-positive cells and immunoblotting showed a corresponding down-regulation of PTPRK protein (Figure 1B). mRNA expression changes were confirmed by quantitative PCR (Q-PCR) (Figure S1A,B). To determine which of the EBV genes was responsible for this effect, EBV-negative KM-H2 cells were transfected with expression vectors encoding individual latent genes that are known to be expressed in EBV-positive KM-H2 cells (LMP2, EBNA1, A73, and RPMS1 ORFs). EBNA1 induced the down-regulation of PTPRK expression in KM-H2 cells (Figures 1C and S1C), whereas no other EBV gene affected PTPRK expression (data not shown). Transient EBNA1 expression in 2 other EBV-negative HL cell lines, HDLM2 (Figure 1C right panel) and L428 (data not shown), also resulted in the down-regulation of PTPRK transcription. Taken together these data show that EBV infection of HL cells results in the down-regulation of PTPRK expression and that this effect is mediated by EBNA1. Given that LMP1 is expressed in primary HL, we also studied whether this viral oncogene influenced the expression of PTPRK. Expression of LMP1 in EBV-negative HL cells had no effect on the levels of PTPRK mRNA (data not shown). We also performed Q-PCR for PTPRK on EBV-positive and EBV-negative paired BL cell lines (Akata, Awia, and Mutu). However, we could not detect PTPRK expression in any of these cell lines (data not shown).
EBV infection or EBNA1 expression decreases Smad2 protein levels in HL cells
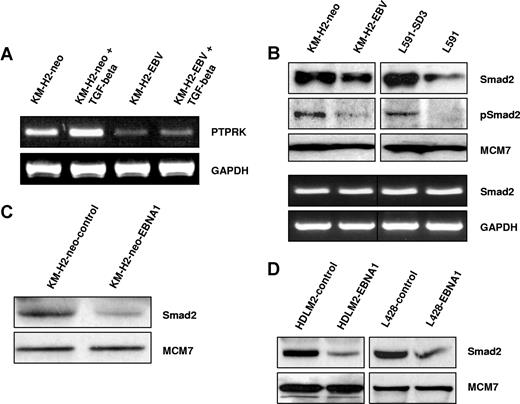
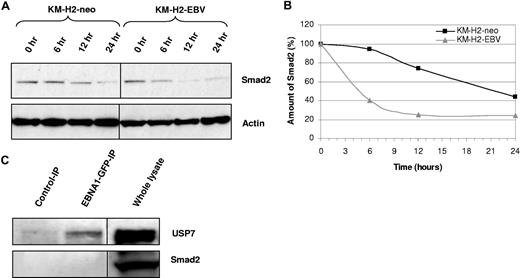
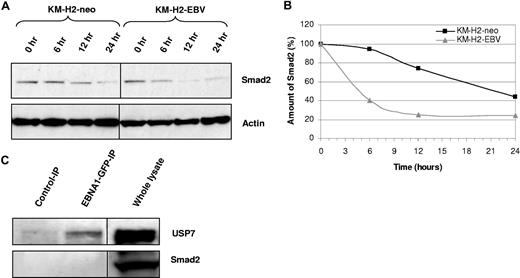
We first wished to confirm that PTPRK is a TGF-beta target gene in HL cells. Figure 2A shows that treatment of EBV-negative HL cells with recombinant human TGF-beta1 resulted in the robust up-regulation of PTPRK transcription. However, the same treatment in EBV-positive HL cells led to only a slight increase in PTPRK transcription. These data confirm that in HL cells PTPRK expression can be up-regulated by TGF-beta treatment but that this effect is markedly inhibited by the presence of EBV. Next, we investigated whether EBV infection or EBNA1 expression in HL cells affected the levels of Smad proteins. Figure 2B shows that levels of total and phosphorylated Smad2 were decreased in both EBV-positive KM-H2 and L591 cells compared with their EBV-negative counterparts. In contrast, the levels of Smad3, Smad4, Smad6, and Smad7 proteins did not vary with EBV status (data not shown). The decrease in Smad2 protein levels was not a consequence of altered SMAD2 transcription since Smad2 mRNA levels were unaffected by the presence of EBV in these cell lines (Figure 2B). Q-PCR confirmed no change in Smad2 mRNA in these cells (Figure S2). Furthermore, we could show that this effect was due to EBNA1 since transient expression of EBNA1 in EBV-negative KM-H2 cells reduced Smad2 protein levels (Figure 2C). Expression of EBNA1 in other EBV-negative HL cell lines also decreased the levels of total Smad2 protein (Figure 2D). Treatment of cells with the protein synthesis inhibitor cycloheximide for different times followed by immunoblotting revealed that the presence of EBV in HL cells was associated with a marked reduction in the half-life of Smad2 (Figure 3A,B). To determine whether EBNA1 physically interacted with Smad2, we studied L428 HL cells that stably expressed a GFP-EBNA1 fusion protein (Figure 3C). Pull-down with an anti-GFP antibody followed by immunoblotting revealed that, whereas EBNA1 was bound to its known interactor, USP7, we could not detect any interaction between Smad2 and EBNA1. We conclude that EBNA1 alters the half-life of Smad2 but does not physically interact with Smad2.
EBV infection or EBNA1 expression inhibits the TGF-beta–mediated activation of PTPRK expression and decreases total and phosphorylated Smad2 protein levels in HL cells. (A) TGF-beta stimulation of KM-H2 HL cells results in the robust up-regulation of PTPRK transcription. However, responsiveness to TGF-beta is reduced by the presence of EBV as demonstrated by a less marked increase in PTPRK transcription in EBV-positive HL cells compared with their EBV-negative counterparts. (B top panel) Immunoblotting shows a reduction in the levels of both total and phosphorylated Smad2 protein in EBV-positive HL cells. (B bottom panel) Semiquantitative RT-PCR demonstrates that SMAD2 transcription is unaffected by the presence of EBV in HL cells. (C) Immunoblotting demonstrates that expression of EBNA1 in EBV-negative KM-H2 cells is sufficient to reduce levels of total Smad2 protein. (D) Immunoblotting shows that expression of EBNA1 in 2 other EBV-negative HL cell lines, HDLM2 and L428, also results in the reduction of total Smad2 protein.
EBV infection or EBNA1 expression inhibits the TGF-beta–mediated activation of PTPRK expression and decreases total and phosphorylated Smad2 protein levels in HL cells. (A) TGF-beta stimulation of KM-H2 HL cells results in the robust up-regulation of PTPRK transcription. However, responsiveness to TGF-beta is reduced by the presence of EBV as demonstrated by a less marked increase in PTPRK transcription in EBV-positive HL cells compared with their EBV-negative counterparts. (B top panel) Immunoblotting shows a reduction in the levels of both total and phosphorylated Smad2 protein in EBV-positive HL cells. (B bottom panel) Semiquantitative RT-PCR demonstrates that SMAD2 transcription is unaffected by the presence of EBV in HL cells. (C) Immunoblotting demonstrates that expression of EBNA1 in EBV-negative KM-H2 cells is sufficient to reduce levels of total Smad2 protein. (D) Immunoblotting shows that expression of EBNA1 in 2 other EBV-negative HL cell lines, HDLM2 and L428, also results in the reduction of total Smad2 protein.
Smad2 half-life is decreased in the presence of EBV. (A) Treatment of KM-H2 cells with cycloheximide reveals that the turnover of Smad2 is increased in EBV-positive cells relative to EBV-negative controls (KM-H2-neo). (B) Scatter plot generated from normalized densitometry values from Smad2 and actin immunoblots from cycloheximide-treated KM-H2 cells. The half-life of Smad2 in EBV-positive cells is decreased in comparison with EBV-negative controls (KM-H2-neo). (C) Immunoprecipitation from L428 HL cells stably expressing an EBNA1-GFP fusion protein using anti-GFP antibody (which pulls down EBNA1-GFP) followed by immunoblotting for either USP7 (top panel) or Smad2 (bottom panel). A positive band is observed for USP7, a known EBNA1 interactor, in the EBNA1-GFP-IP, but not in the IP with isotype control antibody. An interaction between USP7 and EBNA1 was also confirmed by IP with USP7 and subsequent immunoblotting for EBNA1 (data not shown). No bands are observed in the Smad2 immunoblot, suggesting that EBNA1 and Smad2 do not physically associate. Immunoblotting on whole lysates confirms that USP7 and Smad2 are expressed in these cells. The black dividing lines signify where images from different parts of the same immunoblot have been moved together to bring samples next to each other for direct comparison.
Smad2 half-life is decreased in the presence of EBV. (A) Treatment of KM-H2 cells with cycloheximide reveals that the turnover of Smad2 is increased in EBV-positive cells relative to EBV-negative controls (KM-H2-neo). (B) Scatter plot generated from normalized densitometry values from Smad2 and actin immunoblots from cycloheximide-treated KM-H2 cells. The half-life of Smad2 in EBV-positive cells is decreased in comparison with EBV-negative controls (KM-H2-neo). (C) Immunoprecipitation from L428 HL cells stably expressing an EBNA1-GFP fusion protein using anti-GFP antibody (which pulls down EBNA1-GFP) followed by immunoblotting for either USP7 (top panel) or Smad2 (bottom panel). A positive band is observed for USP7, a known EBNA1 interactor, in the EBNA1-GFP-IP, but not in the IP with isotype control antibody. An interaction between USP7 and EBNA1 was also confirmed by IP with USP7 and subsequent immunoblotting for EBNA1 (data not shown). No bands are observed in the Smad2 immunoblot, suggesting that EBNA1 and Smad2 do not physically associate. Immunoblotting on whole lysates confirms that USP7 and Smad2 are expressed in these cells. The black dividing lines signify where images from different parts of the same immunoblot have been moved together to bring samples next to each other for direct comparison.
Down-regulation of PTPRK expression is a consequence of decreased Smad2 protein levels
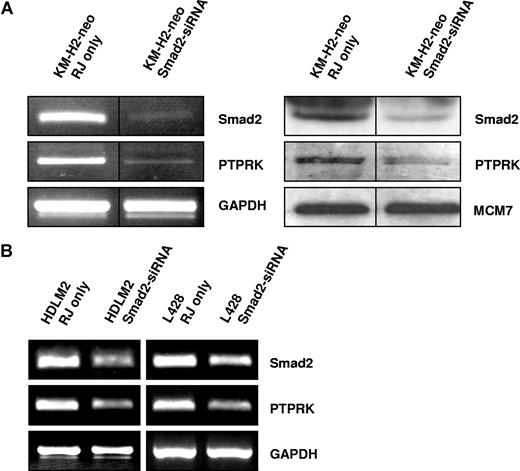
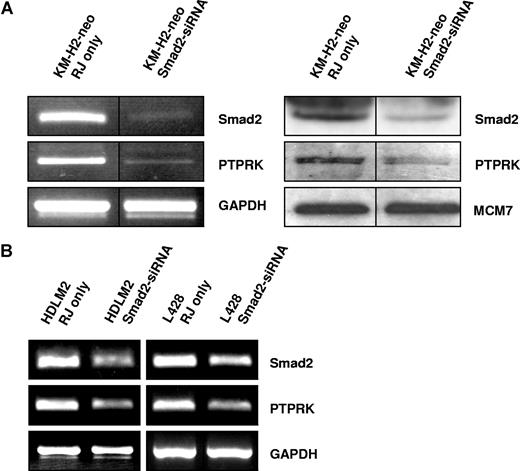
We next wished to establish whether PTPRK down-regulation in EBV-positive HL cells is a direct result of the decreased levels of Smad2 protein. To do this, we treated the EBV-negative HL cell lines with Smad2-specific siRNA. Semiquantitative PCR (Figure 4) and quantitative PCR (Figure S3) demonstrated that knockdown of Smad2 expression in these cells led to a decrease in PTPRK expression. We conclude that loss of PTPRK expression in EBV-positive HL cells is a consequence of the decrease in Smad2 protein.
Knockdown of Smad2 expression down-regulates PTPRK expression in EBV-negative HL cells. (A) Semiquantitative RT-PCR for Smad2 and PTPRK mRNA (left panel) and immunoblotting for Smad2 and PTPRK (right panel) in KM-H2 cells show that knockdown of Smad2 expression decreases PTPRK expression. The black dividing lines signify where images from different parts of the same gel have been moved together to bring samples next to each other for direct comparison. (B) Semiquantitative RT-PCR analysis of 2 other EBV-negative HL cell lines (HDLM-2 and L428) shows that PTPRK transcription is also reduced following the knockdown of Smad2 expression in these cells. In all cell lines, transfection with irrelevant siRNA oligonucleotides had no effect on Smad2 or PTPRK expression (data not shown).
Knockdown of Smad2 expression down-regulates PTPRK expression in EBV-negative HL cells. (A) Semiquantitative RT-PCR for Smad2 and PTPRK mRNA (left panel) and immunoblotting for Smad2 and PTPRK (right panel) in KM-H2 cells show that knockdown of Smad2 expression decreases PTPRK expression. The black dividing lines signify where images from different parts of the same gel have been moved together to bring samples next to each other for direct comparison. (B) Semiquantitative RT-PCR analysis of 2 other EBV-negative HL cell lines (HDLM-2 and L428) shows that PTPRK transcription is also reduced following the knockdown of Smad2 expression in these cells. In all cell lines, transfection with irrelevant siRNA oligonucleotides had no effect on Smad2 or PTPRK expression (data not shown).
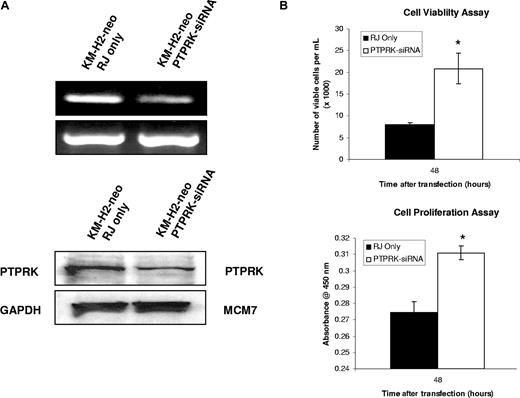
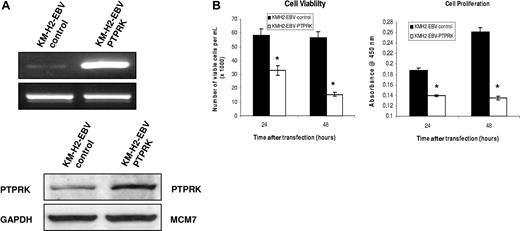
PTPRK is a functional tumor suppressor in HL cells
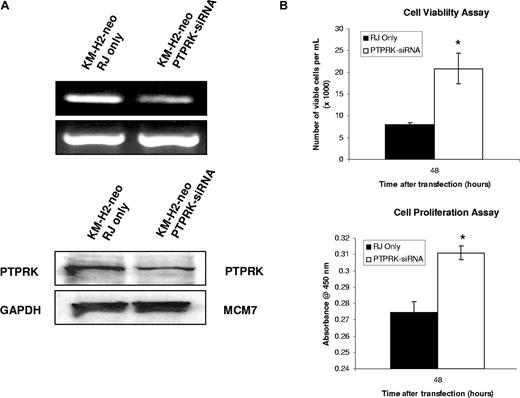
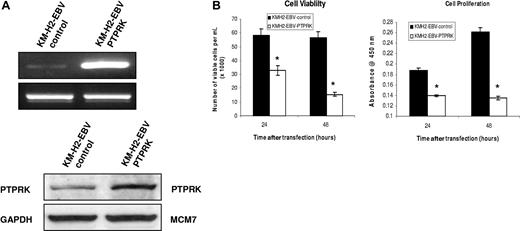
To investigate the contribution of the down-regulation of PTPRK to the growth and survival of HL cells, we first knocked down its expression in EBV-negative KM-H2 cells using specific siRNAs. Figure 5A shows that this treatment resulted in the down-regulation of PTPRK mRNA and protein and that this was accompanied by a significant increase in cell viability and cell proliferation as assessed by trypan blue and WST-1 assays, respectively (Figure 5B). These differences were statistically significant (P values are .02 for cell viability and .003 for cell proliferation). To confirm these effects, PTPRK was overexpressed in EBV-positive HL cells, where PTPRK expression is normally low (Figure 6A). Figure 6B shows that the re-expression of PTPRK resulted in a significant decrease in both the viability and proliferation of EBV-positive KM-H2 cells (P values are .003 and .001, respectively, for viability and P values are both less than .001 for cell proliferation). Taken together, these data show that the down-regulation of PTPRK expression by EBV contributes to the growth and survival of HL cells.
Knockdown of PTPRK expression in EBV-negative KM-H2 cells increases the growth and survival of HL cells. (A) Semiquantitative RT-PCR for PTPRK mRNA (top) and immunoblotting for PTPRK protein (bottom) following siRNA-mediated knockdown of PTPRK expression in EBV-negative KM-H2 cells. (B) Cell viability (trypan blue; left) and cell proliferation (WST-1; right) assays on KM-H2 cells following treatment with PTPRK-specific siRNA. The down-regulation of PTPRK expression in these cells results in a significant increase in cell viability and proliferation (* denotes significant difference; P values are .02 for viability and .003 for proliferation). Transfection with irrelevant siRNA oligonucleotides had no effect on PTPRK expression, cell viability, or proliferation (data not shown).
Knockdown of PTPRK expression in EBV-negative KM-H2 cells increases the growth and survival of HL cells. (A) Semiquantitative RT-PCR for PTPRK mRNA (top) and immunoblotting for PTPRK protein (bottom) following siRNA-mediated knockdown of PTPRK expression in EBV-negative KM-H2 cells. (B) Cell viability (trypan blue; left) and cell proliferation (WST-1; right) assays on KM-H2 cells following treatment with PTPRK-specific siRNA. The down-regulation of PTPRK expression in these cells results in a significant increase in cell viability and proliferation (* denotes significant difference; P values are .02 for viability and .003 for proliferation). Transfection with irrelevant siRNA oligonucleotides had no effect on PTPRK expression, cell viability, or proliferation (data not shown).
Transient overexpression of PTPRK in EBV-positive KM-H2 cells significantly decreases their growth and survival. (A) Semiquantitative RT-PCR for PTPRK mRNA (top) and immunoblotting for PTPRK (bottom) following overexpression of PTPRK in EBV-positive KM-H2 cells. (B) Cell viability (trypan blue; left) and cell proliferation (WST-1; right) assays on KM-H2-EBV cells following PTPRK overexpression. The transient expression of PTPRK in these cells results in a significant decrease in cell viability and cell proliferation at both 24 and 48 hours (* denotes significant difference; P values are .003 at 24 hours and .001 at 48 hours for viability and P values are less than .001 at both 24 and 48 hours for cell proliferation).
Transient overexpression of PTPRK in EBV-positive KM-H2 cells significantly decreases their growth and survival. (A) Semiquantitative RT-PCR for PTPRK mRNA (top) and immunoblotting for PTPRK (bottom) following overexpression of PTPRK in EBV-positive KM-H2 cells. (B) Cell viability (trypan blue; left) and cell proliferation (WST-1; right) assays on KM-H2-EBV cells following PTPRK overexpression. The transient expression of PTPRK in these cells results in a significant decrease in cell viability and cell proliferation at both 24 and 48 hours (* denotes significant difference; P values are .003 at 24 hours and .001 at 48 hours for viability and P values are less than .001 at both 24 and 48 hours for cell proliferation).
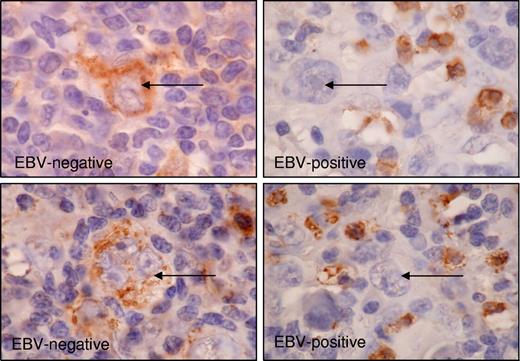
Frequent down-regulation of PTPRK expression in EBV-positive HRS cells of primary HL
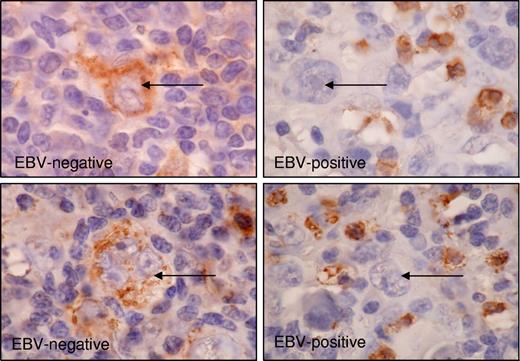
Finally, we studied whether PTPRK expression was associated with EBV status in primary HL. Fifty-three classic HL tumors were investigated by immunohistochemistry for the PTPRK protein. We observed cytoplasmic PTPRK staining in malignant HRS cells in the majority of cases and in a subset of nonmalignant lymphocytes in all cases (Figure 7). Staining in tumor cells was assessed as either negative or lower than surrounding lymphocytes (PTPRKlow) or equivalent to or higher than surrounding lymphocytes (PTPRKhi). PTPRKlow cases were significantly more frequent in the EBV-positive group (P value of Fisher exact test is .003) (Table 2). We conclude that the down-regulation of PTPRK is more frequent in the primary tumor cells of EBV-positive HL compared with EBV-negative HL.
Expression of PTPRK in primary HL biopsies. Immunohistochemistry significantly demonstrates the frequent down-regulation of PTPRK expression in HRS cells (→) of EBV-positive cases of HL. Shown is representative staining for 2 EBV-negative cases (left panel) and 2 EBV-positive cases (right panel). Subsets of surrounding lymphoid cells were PTPRK positive and provided an internal positive control. Images were acquired using Nikon TE2000 with 60×/1.4 NA oil-immersion lens (Nikon, Kingston-upon-Thames, United Kingdom). Cells were stained with DAB immunoperoxidase and counterstained with hematoxylin. Images were captured with a Nikon Coolpix 2100, and Paint Shop Pro 8.0 (Jase Software, Maidenhead, United Kingdom).
Expression of PTPRK in primary HL biopsies. Immunohistochemistry significantly demonstrates the frequent down-regulation of PTPRK expression in HRS cells (→) of EBV-positive cases of HL. Shown is representative staining for 2 EBV-negative cases (left panel) and 2 EBV-positive cases (right panel). Subsets of surrounding lymphoid cells were PTPRK positive and provided an internal positive control. Images were acquired using Nikon TE2000 with 60×/1.4 NA oil-immersion lens (Nikon, Kingston-upon-Thames, United Kingdom). Cells were stained with DAB immunoperoxidase and counterstained with hematoxylin. Images were captured with a Nikon Coolpix 2100, and Paint Shop Pro 8.0 (Jase Software, Maidenhead, United Kingdom).
Discussion
PTPRK is a putative tumor-suppressor gene located within a region on chromosome 6 (6q22.2-q22.3) that is frequently deleted in a variety of tumors, including breast cancer, ovarian carcinoma, melanoma, leukemia, and non-Hodgkin lymphoma.23,24 It has also been suggested in a recent study that cleavage of PTPRK protein is implicated in colon cancer metastasis.25 Here we show that EBV infection of HL cells down-regulates PTPRK expression and that this leads to their increased growth and survival. We observed these effects not only in KM-H2 cells that were artificially infected with a recombinant EBV but also following the loss of the EBV genome from L591 cells that are presumed to derive from EBV-positive primary HRS cells. However, given the limitations of these cell lines it was important to investigate PTPRK expression in relation to EBV status in primary HRS cells. We observed that in primary tumors, PTPRK was more frequently down-regulated in EBV-positive HRS cells compared with EBV-negative HRS cells, suggesting that EBV has similar effects on PTPRK expression in vivo. The demonstration that EBNA1 was responsible for this effect suggests a potential mechanism through which this virus protein may contribute more directly to oncogenesis, aside from its role in episome maintenance.
PTPRK expression can be activated by TGF-beta12 and has been shown to mediate the TGF-beta–dependent inhibition of cell proliferation in human keratinocytes.26 Knockdown of PTPRK accelerates cell cycle progression, amplifies the response to epidermal growth factor (EGF), and overturns TGF-beta–mediated antimitogenesis.12 Our findings that EBV suppresses the TGF-beta–mediated activation of PTPRK expression suggest that the virus might disrupt TGF-beta signaling upstream of PTPRK. This was supported by the observation that several genes previously reported to be suppressed by TGF-beta were up-regulated by EBV infection18,,–21 and that both genes down-regulated by EBV are known to be induced by TGF-beta.12,22
Binding of TGF-beta to its receptors results in the phosphorylation of Smad2 and Smad3, which assemble with Smad4. Smad2/3/4 complexes then translocate to the nucleus, where they interact with other transcription factors to transactivate TGF-beta target genes.27 Loss of function mutations in both TGF-beta receptor and Smad genes have been detected in many tumor types; SMAD2 and SMAD4 are known tumor-suppressor genes.28 We observed that either EBV infection or the presence of EBNA1 alone in HL cells decreased Smad2 protein levels; this was not due to alterations in SMAD2 transcription, since Smad2 RNA levels were unaffected by the presence of EBV. Furthermore, we showed that the loss of Smad2 expression was directly responsible for the down-regulation of PTPRK. We were able to demonstrate that the half-life of Smad2 was decreased in the presence of EBV. Smad protein turnover is regulated by 2 HECT-domain–containing E3 ubiquitin ligases known as Smad ubiquitination regulatory factor-1 (Smurf1) and Smurf2. Smurf1 causes the ubiquitination and proteosomal degradation of Smad1 and Smad5,29 whereas Smurf2 targets Smad1 and Smad2.30,31 Other E3 ligases reported to target Smad2 include Wwp1 (or TGIF interacting ubiquitin ligase 1; Tiul1)32 and NEDD4-L (or NEDD4-2).33 However, we did not observe significant changes in the mRNA expression of any of these E3 ligases following EBV infection or EBNA1 expression (data not shown). Furthermore, the increased Smad2 degradation was not the consequence of its direct binding to EBNA1, since we were unable to demonstrate a physical interaction between Smad2 and EBNA1 in HL cells, although we could show that EBNA1 bound to USP7, a known interactor of EBNA1, in these cells. Thus, at present, we are unable to identify the precise mechanism by which EBNA1 down-regulates Smad2 protein. It should be noted that similar effects of EBNA1 on Smad2 protein expression have been recently described in epithelial cells.34
HL cells have been shown to produce TGF-beta, which contributes to the shift away from a Th1-biased toward a Th2-biased T-cell infiltrate characteristic of HL. Thus, EBNA1 might confer EBV-infected HRS cells with resistance to the growth-suppressive effects of TGF-beta, while at the same time allowing these cells to secrete the high levels of TGF-beta required to inhibit cell-mediated immune responses to virus-infected cells.
PTPRK is a type R2B receptor protein tyrosine phosphatase PTP). PTPs dephosphorylate and thereby antagonize receptor tyrosine kinase (RTK) signaling; by virtue of these effects, many are established or putative tumor suppressors. The structural features of PTPRK and its ability to mediate homophilic interactions between cells indicate that it could serve as a direct connection between physical cell contact and downstream cellular signaling events incorporating tyrosine phosphorylation.35 It has been shown that PTPRK colocalizes with β- and γ-catenin in breast cancer cells at the point where adjacent cells are touching.36 This observation suggests that PTPRK could negatively regulate the action of tyrosine kinase events at cell junctions. In vitro phosphatase activity of PTPRK toward β-catenin as a substrate has been demonstrated, thus implying that this negative regulation might occur through the dephosphorylation of β-catenin.37 PTPRK has also recently been shown to be a regulator of both basal- and ligand-activated epidermal growth factor receptor (EGFR) phosphorylation and function in keratinocytes.37 HL is characterized by the aberrant expression of a number of RTKs.38 Studies using phosphotyrosine-specific antibodies have revealed the specific activation of PDGFRA and TRKA/B and a general elevation of cellular phosphotyrosine levels in HL.39 Thus, although the precise targets of PTPRK have yet to be defined in HL cells, it is likely they will be tyrosine kinases that function to promote cell growth and survival.
LMP1 has previously been shown to repress TGF-beta signaling through NF-κB–mediated depletion of transcriptional coactivators required for Smad-dependent transcription.40,41 Our data show that the essential EBV-encoded EBNA1 can also suppress TGF-beta signaling through a mechanism that involves down-regulation of Smad2 protein. This results in the altered transcription of TGF-beta target genes, among them PTPRK. Our data show that loss of PTPRK expression in HL cells increases their survival and proliferation. Repression of TGF-beta responses induced by EBNA1 and LMP1 may contribute to oncogenesis in HL by providing HRS cells with resistance to the growth-suppressive effects of TGF-beta, while at the same time allowing these cells to secrete the high levels of TGF-beta required to inhibit cell-mediated immune responses to virus-infected cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Leukemia Research Fund and Cancer Research UK.
V.H.J.W. was funded by the European Commission's FP6 Life-Sciences-Health Program (INCA Project: LSHC-CT-2005-018704).
Authorship
Contribution: J.R.F. designed and performed research and prepared the paper; K.R.N.B. performed microarray analysis; V.H.J.W. and G.L.D. performed immunoprecipitation experiments; W.W. provided statistical and bioinformatics support; G.M.R. performed immunocytochemistry experiments; S.M. performed the pathology studies; A.B. and G.L.K. analyzed Burkitt lymphoma cells; P.G.M. and L.S.Y. designed research and reviewed paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul G. Murray, CRUK Institute for Cancer Studies, The Medical School, University of Birmingham, Vincent Drive, Edgbaston, Birmingham, B15 2TT, United Kingdom; e-mail: p.g.murray@bham.ac.uk.