We have analyzed leukocyte mono-Ig–like receptor 5 (LMIR5) as an activating receptor among paired LMIRs. Mouse LMIR5 (mLMIR5) is expressed in myeloid cells such as mast cells, granulocytes, macrophages, and dendritic cells. Cross-linking of transduced mLMIR5 in bone marrow–derived mast cells (BMMCs) caused activation events, including cytokine production, cell survival, degranulation, and adhesion to the extracellular matrix. mLMIR5 associated with DAP12 and to a lesser extent with DAP10, and mLMIR5-mediated functions of BMMCs were strongly inhibited by DAP12 deficiency. Importantly, cross-linking of endogenous mLMIR5 induced Syk-dependent activation of fetal liver–derived mast cells. Unlike mLMIR5, cross-linking of human LMIR5 (hLMIR5) induced cytokine production of BMMCs even in the absence of both DAP12 and DAP10, suggesting the existence of unidentified adaptors. Interestingly, hLMIR5 possessed a tyrosine residue (Y188) in the cytoplasmic region. Signaling via Y188 phosphorylation played a predominant role in hLMIR5-mediated cytokine production in DAP12-deficient, but not wild-type BMMCs. In addition, experiments using DAP10/DAP12 double-deficient BMMCs suggested the existence of Y188 phoshorylation-dependent and -independent signals from unidentified adaptors. Collectively, although both mouse and human LMIR5 play activatory roles in innate immunity cells, the functions of LMIR5 were differentially regulated in mouse versus human cells.
Introduction
It is widely accepted that mast cells are major effector cells in allergic inflammation through a high-affinity IgE receptor (FcϵRI). However, recent advances have delineated the significant roles of mast cells in both innate and adaptive immune responses.1,,–4
To find a novel immune receptor expressed on mast cells, we previously performed a signal sequence trap based on retrovirus-mediated expression screening (SST-REX).5 In this screening, we isolated a cDNA for a novel immune receptor, leukocyte mono-Ig–like receptor 1 (LMIR1),6 using cDNA library of bone marrow–derived mast cells (BMMCs). Successively, other members of the LMIR family were cloned by searching for sequences homologous with the Ig-like domain of LMIR1. We and others have demonstrated that LMIR1/CMRF-35–like Ig-like molecule-8 (CLM-8)/myeloid-associated Ig-like receptor-I (MAIR-I)/CD300a and LMIR2/CLM-4/MAIR-II/dendritic cell–derived Ig-like receptor 1 (DIgR1)/CD300d as well as LMIR3/CLM-1 and LMIR4/CLM-5 were a pair of inhibitory and activating, respectively, receptors with high homology in the Ig-like domain.6,,,,,,,–14 LMIR/CLM forms a family of paired receptors mainly expressed in myeloid cells.6,,,,,,,–14 In general, activating receptors do not contain any signaling motifs in the short cytoplasmic tails, but transmit signals by associating with immunoreceptor tyrosine–based activation motif (ITAM) or the related activating motif-bearing molecules via a positively charged residue in the transmembrane domain.15,,,–19 In the present study, we cloned a cDNA for mouse LMIR5 (mLMIR5)/CLM-7 from a BMMC cDNA library. Analysis of DAP10-, DAP12-, and FcRγ-deficient BMMCs demonstrated the predominant role of DAP12 in the activating functions of mLMIR5.
Structural differences in immune receptors in mouse versus human cells sometimes result in differing immunologic responses. For example, human NKG2D associates only with DAP10. On the other hand, mouse NKG2D has 2 splice variants, where the long isoform (NKG2D-L) associates exclusively with DAP10 and the short isoform (NKG2D-S) associates with both DAP10 and DAP12.20,,–23 Interestingly, human LMIR5 (hLMIR5)/CD300b/immune receptor expressed by myeloid cell-3 (IREM-3),24 but not mLMIR5, contained a putative tyrosine phosphorylation motif (YXN) in its short cytoplasmic tail. The present results indicated that DAP12 plays a primary role in functions of mLMIR5, while both DAP12 and DAP10 play roles in functions of its human counterpart hLMIR5. Consistent with a recent report by Martinez-Barriocanal and Sayos,24 our results also implicated an unidentified adaptor in the hLMIR5-mediated signaling pathway, which was activated through phosphorylation of the tyrosine in the absence of DAP12. In addition, the experiment using DAP12-deficient and DAP10/DAP12 double-deficient BMMCs revealed Y188 phosphorylation–dependent and – independent signals downstream of hLMIR5.
Methods
Cells
Murine cell lines used in this study were as follows: FDC-P1, J774-1, RAW264.7, M1, L-G, 32Dcl3, P815, MC/9, L8057, Ba/F3, WEHI231, A20, EL4, BW5147, and DC2.4. L8057 and DC2.4 were a kind gift from Dr Y. Hirabayashi (National Institute of Health Sciences, Tokyo, Japan) and Dr K. L. Rock (University of Massachusetts Medical School, Worcester, MA), respectively. Peripheral blood (PB) cells, bone marrow (BM) cells, splenocytes, thymocytes, and peritoneal cells derived from C57BL/6 mice (or CBA/J mice) were purified as described.14 CBA/J mice or C57BL/6J mice (Charles River Laboratories Japan, Yokohama, Japan) were used at 8 to 10 weeks of age for isolation of tissues and cells. All procedures were approved by an institutional review committee. BMMCs or fetal liver mast cells (FLMCs) were generated and cultured as described.25,–27 BM-derived macrophage (BMMΦ), BM-derived myeloid dendritic cells (BMmDCs), and BM-derived plasmacytoid dendritic cells (BMpDCs) were cultured as described.14 The following mutant mice were used: DAP10−/−,20 DAP12−/−,28 FcRγ −/−,29 and Syk+/−.30
Antibodies and other reagents
Cytokines and anti-mLMIR5 polyclonal antibody (Ab) was obtained from R&D Systems (Minneapolis, MN). Fluorescein isothiocyanate (FITC)–conjugated anti–mouse B220, CD3, CD11b, and Gr-1 mAb were purchased from eBioscience (San Diego, CA). FITC-conjugated anti–mouse IgE, FITC-conjugated anti–mouse Ig polyclonal Ab, R-phycoerythrin (PE)–conjugated anti–mouse c-Kit mAb, and mouse antitrinitrophenyl (TNP) IgE (C38-2) were from BD Pharmingen (San Diego, CA). Anti–Flag mAb (M2), mouse IgG1 mAb (MOPC21), goat IgG polyclonal Ab, and mouse antidinitrophenyl (DNP) IgE mAb (SPE-7) were from Sigma-Aldrich (St Louis, MO). Donkey PE-conjugated F(ab′)2 anti–goat IgG Ab was from Jackson Immunoresearch Laboratories (West Grove, PA). Anti-Myc mAb (9E10) was from Roche Diagnostics (Indianapolis, IN). Rabbit anti–mouse DAP12 polyclonal Ab was a kind gift from Dr N. Aoki (Asahikawa Medical College, Asahikawa, Japan). Mouse antiphosphotyrosine mAb (4G10) was purchased from Upstate Biotechnology (Charlottesville, VA), and other phospho-specific Abs were from Cell Signaling Technology (Beverly, MA). Other Abs were from Santa Cruz Biotechnology (Santa Cruz, CA). Bovine serum fibronectin (FN), human plasma fibrinogen (FB), and N-glycosidase F were purchased from Sigma-Aldrich, Chemicon (Temecula, CA), and New England Biolabs (Beverly, MA), respectively.
Gene expression analysis
Expression of mLMIR5 was analyzed by reverse transcriptase–polymerase chain reaction (RT-PCR) as described.14 Amplification of mLMIR5 as well as β-actin for normalization was performed with the following primers: 5′-TTACCATGGAGATGCTCAGG-3 (base: 266-285) and 5′-TCGCTACAGAGAGTGTGTCTCC-3′ (base: 590-569) for mLMIR5; and 5′-CATCACTATTGGCAACGAGC-3′ and 5′-ACGCAGCTCAGTAACAGTCC-3′ for β-actin. Relative expression levels of DAP10, DAP12, and FcRγ among samples were measured by real-time RT-PCR. cDNA was amplified using a LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics, Mannheim, Germany) under the following conditions: 1 cycle of 95°C for 10 seconds, 40 cycles of 95°C for 5 seconds, and 60°C for 20 seconds. All samples were independently analyzed 3 times. The following primers were used: 5′-CCCCCCAGGCTACCTCC-3′ and 5′-TGACATGACCGCATCTGCA-3′ for DAP10; 5′-CAAGATGCGACTGTTCTTCCG-3′ and 5′-GGTCTCTGACCCTGAAGCTCC-3′ for DAP12; 5′-GCCGTGATCTTGTTCTTGCTC-3′ and 5′-CTGCCTTTCGGACCTGGAT-3′ for FcRγ; and 5′-ATGTGTCCGTCGTGGATCTGA-3′ and 5′-TTGAAGTCGCAGGAGACAACC-3′ for GAPDH. Relative gene expression levels were calculated using standard curves generated by serial dilutions of cDNA and normalized by a GAPDH expression level. Product quality was checked by melting curve analysis via LightCycler software (Roche Diagnostics).
DNA constructs
The GenBank/European Molecular Biology Laboratory (EMBL31 )/DNA Data Bank of Japan (DDBJ32 ) database was searched by using the amino acid sequence of the Ig-like domain of mLMIR1. Based on the sequence data, cDNA of mouse and human LMIR5 were isolated by PCR from a cDNA library of BMMCs (derived from CBA/J or B57BL/6 mice) and a cDNA library of human peripheral mononuclear cells (PMCs), respectively, and confirmed by sequencing as described.6 The cDNA fragment of mLMIR5 or hLMIR5, lacking the signal sequence, was tagged with an Flag or Myc epitope at the N terminus. The resultant Flag or Myc-mLMIR5 or hLMIR5 was subcloned into a pME18s vector containing a SLAM signal sequence (a gift from Hisashi Arase, Osaka University, Osaka, Japan)33 to generate pME-Flag, Myc-mLMIR5, or hLMIR5. The resultant SLAM signal sequence-Flag, Myc-mLMIR5, or hLMIR5 was subcloned into a pMXs-IRES-puror (pMXs-IP)34 retroviral vector to generate pMXs-Flag or Myc-mLMIR5 or hLMIR5-IP. Two-step PCR mutagenesis was performed in the replacement of K158 (lysine with a positive charge) of hLMIR5 with Q (glutamine with a neutral charge) and Y188 of hLMIR5 with F (phenylalanine).
Transfection and infection
Retroviral transfection was as described.6,34 Briefly, retroviruses were generated by transient transfection of PLAT-E packaging cells35 with FuGENE 6 (Roche Diagnostics). BM cells, BMMCs, or Ba/F3 cells were infected with retroviruses in the presence of 10 μg/mL polybrene. After 48 hours, cell selection was started with appropriate antibiotics.34
Flow cytometry
Cells were stained as described.14 Flow cytometric analysis was performed with FACSCalibur (BD Biosciences, Mountain View, CA) equipped with CellQuest software and FlowJo software (Tree Star, Ashland, OR). For mLMIR5 staining, cells were incubated with 20 μg/mL anti-mLMIR5 polyclonal Ab or goat polyclonal IgG Ab as control, before incubation with 10 μg/mL PE-conjugated anti-goat IgG F(ab′)2 Ab.
Immunoprecipitation and Western blotting
Cells were lysed with NP-40 lysis buffer containing protease and phosphatase inhibitor cocktail (Sigma-Aldrich). Cell lysates were assayed using a protein assay kit (Bio-Rad, Hercules, CA). Immunoprecipitation and Western blotting were performed as described.14
Measurement of cytokines and histamines and adhesion assay
BMMCs and FLMCs were stimulated with either 20 μg/mL anti-mLMIR5 Ab, 20 μg/mL control IgG, or 100 nM phorbol-12-myristate13-acetate (PMA). BMMCs transduced with a Flag-tagged hLMIR5 were stimulated with 20 μg/mL anti-Flag mAb or 20 μg/mL control IgG. In some experiments, BMMCs sensitized with 1 μg/mL anti-TNP IgE for 12 hours were stimulated with 100 ng/mL TNP-BSA. TNF-α, IL-6, and MCP-1 concentrations in culture supernatants were measured using enzyme-linked immunosorbent assay (ELISA) kits (BD Pharmingen and R&D Systems). Histamine released during a 50-minute incubation period was measured as described.25 Adhesion assay was described previously.26
Statistical analysis
Data are shown as means plus or minus standard deviation (SD), and statistical significance was determined by the Student t test with P levels less than .05 as statistically significant.
Results
Structure of mLMIR5
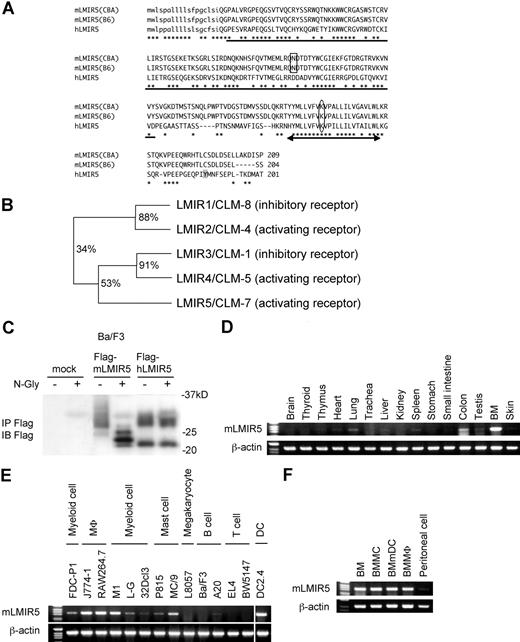
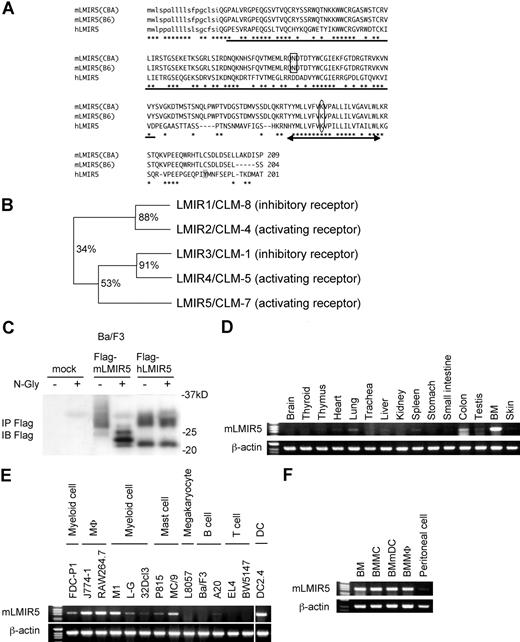
To identify other members of LMIR besides LMIR1/2,6 we searched the database from GenBank/EMBL/DDBJ by using the sequence encoding the Ig-like domain of LMIR1/CLM-8. Based on this, we identified and cloned mLMIR5/CLM-7 in addition to another pair, LMIR3/CLM-1 and LMIR4/CLM-5, from a BMMC cDNA library. An Ig-like domain of mLMIR5 shared 34% and 53% identity at amino acid sequences with that of LMIR1 and LMIR3, respectively (Figure 1A,B).6 We also cloned its human ortholog from a human PMC cDNA library and found that hLMIR5 shared a 55% identity at overall amino acid sequences with mLMIR5 (Figure 1A). The sequence analysis revealed that mLMIR5 and hLMIR5 are identical to CLM-78,9 and IREM-3,24 respectively. Because hLMIR5/IREM-3 has been recently characterized, we analyzed the structure and functions of mLMIR5/CLM-7 to compare them with those of hLMIR5/IREM-3. mLMIR5 was a type I transmembrane protein composed of an N-terminal signal peptide, an extracellular domain containing a single V-type Ig domain, a transmembrane domain with a positively charged amino acid (lysine), and a short cytoplasmic tail without any signaling motif (Figure 1A). mLMIR5 protein derived from CBA/J mice or B57BL/6 mice was 209 aa or 204 aa, respectively, in length, because the latter was devoid of 5 aa in its cytoplasmic tail. We thereafter used CBA/J mouse–derived LMIR5 as mLMIR5 unless otherwise stated. Interestingly, mLMIR5 does not harbor a putative tyrosine phosphorylation site in the cytoplasmic region (Figure 1A), which is found in hLMIR5.24
Molecular characteristics and gene expression of LMIR5. (A) Alignment of amino acid sequences for mLMIR5 and hLMIR5. CBA and B6 indicate CBA/J mice and C57BL/6 mice, respectively. *Identical amino acids in mLMIR5 and hLMIR5. The putative signal sequence is shown in lower case. The variable type Ig-like domain is underlined; the potential N-linked glycosylation site is boxed. The transmembrane domain is marked by an arrow; the positive-charged amino acid residue (lysine) in the transmembrane domain is circled. Y188 in the cytoplasmic tail of hLMIR5 is shaded. (B) The phylogenetic tree of LMIR1/2/3/4/5 is shown based on homology with the Ig-like domain. Percentage of identity in amino acid sequences of the Ig-like domain was indicated. (C) Lysates of Ba/F3 cells transduced with either a Flag-tagged mLMIR5, a Flag-tagged hLMIR5, or mock were immnoprecipitated with anti-Flag mAb. The precipitates treated with or without N-glycosidase F were immunoblotted with anti-Flag mAb. (D-F) RT-PCR analysis on mLMIR5 expression in murine tissues (D), hematopoietic cell lines (E), and primary hematopoietic cells (F). Specific primers were used for mLMIR5 or β-actin as control. Vertical lines have been inserted to indicate a repositioned gel lane.
Molecular characteristics and gene expression of LMIR5. (A) Alignment of amino acid sequences for mLMIR5 and hLMIR5. CBA and B6 indicate CBA/J mice and C57BL/6 mice, respectively. *Identical amino acids in mLMIR5 and hLMIR5. The putative signal sequence is shown in lower case. The variable type Ig-like domain is underlined; the potential N-linked glycosylation site is boxed. The transmembrane domain is marked by an arrow; the positive-charged amino acid residue (lysine) in the transmembrane domain is circled. Y188 in the cytoplasmic tail of hLMIR5 is shaded. (B) The phylogenetic tree of LMIR1/2/3/4/5 is shown based on homology with the Ig-like domain. Percentage of identity in amino acid sequences of the Ig-like domain was indicated. (C) Lysates of Ba/F3 cells transduced with either a Flag-tagged mLMIR5, a Flag-tagged hLMIR5, or mock were immnoprecipitated with anti-Flag mAb. The precipitates treated with or without N-glycosidase F were immunoblotted with anti-Flag mAb. (D-F) RT-PCR analysis on mLMIR5 expression in murine tissues (D), hematopoietic cell lines (E), and primary hematopoietic cells (F). Specific primers were used for mLMIR5 or β-actin as control. Vertical lines have been inserted to indicate a repositioned gel lane.
Next, to examine the characteristics of mLMIR5 at protein levels, we generated Ba/F3 cells expressing either a Flag-tagged mLMIR5 or hLMIR5. Western blot analysis using anti-Flag mAb detected mLMIR5 as a major broad band (26-35 kDa) and a faint band (approximately 24 kDa), while hLMIR5 was detected as a broad band (27-30 kDa) and a discrete band (approximately 22 kDa) (Figure 1C). Treatment with N-glycosidase F diverted the apparent molecular mass of mLMIR5, but not hLMIR5, to several bands ranging from 22 to 26 kDa in size. This was consistent with the presence of a putative N-linked glycosylation site in the extracellular domain of mLMIR5, but not hLMIR5 (Figure 1A,C), suggesting that mLMIR5 was an N-glycoprotein.
mLMIR5 is expressed in myeloid lineage cells
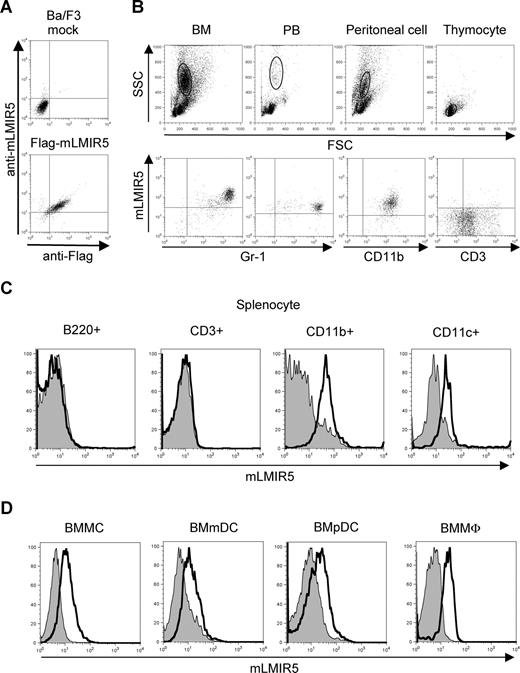
To determine the tissue distribution of mLMIR5, we performed RT-PCR analysis and thereby demonstrated that mLMIR5 was highly expressed in BM and moderately in the lung and colon (Figure 1D). Further investigation of hematopoietic cells revealed that high expression levels of mLMIR5 were observed in myeloid cell lines, including mast, macrophage, and dendritic cell lines, as well as BM-derived cells, but not in T cell lines (Figure 1E,F). Before analyzing surface expression levels of mLMIR5, we confirmed the sensitivity and specificity of polyclonal anti-mLMIR5 Ab, which recognized the extracellular domain of mLMIR5. When Ba/F3 cells were transduced with Flag-tagged mLMIR5, this Ab efficiently detected mLMIR5 on the cell surface, whose expression was confirmed by using anti-Flag mAb (Figure 2A), and anti-mLMIR5 Ab did not detect LMIR1, LMIR2, LMIR3, or LMIR4 transduced into Ba/F3 cells (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition, this polyclonal Ab and anti-Flag Ab gave a similar pattern of several bands in the Western blot of the immunoprecipitates derived from Flag-tagged mLMIR5-transduced Ba/F3 cells. Endogenous mLMIR5 in BMMCs was also detected as a similar pattern. These results confirmed the specificity of this polyclonal Ab raised against mLMIR5 (Figure S3). Surface staining of hematopoietic cell lines with anti-mLMIR5 Ab displayed the results consistent with those from RT-PCR (Figures 1E, S2); A20 cells among the B-cell lines were found to express LMIR5 in both mRNA and protein levels. We then stained a variety of hematopoietic cells using this anti-mLMIR5 Ab. When gated in the population (FSChighSSChigh), immature granulocytes (Gr-1high) in BM and macrophages (CD11bhigh) in peritoneal cells displayed higher expression levels of mLMIR5, while mature granulocytes (Gr-1 high) in PB and dendritic cells (CD11chigh) in spleen showed detectable but lower expression levels as compared with immature granulocytes. However, neither B cells (B220high) in spleen nor T cells (CD3high) in spleen and thymus expressed mLMIR5 on their surfaces (Figure 2B,C). In addition, mLMIR5 was expressed in BM-derived cells such as BMMCs, BMmDCs, BMpDCs, and BMMΦ (Figure 2D). LMIR5 expression was also confirmed in several types of mast cells, including peritoneal mast cells and FLMCs (Figure 2D; data not shown). Collectively, mLMIR5 was mainly expressed in myeloid cells.
Cell-surface expression of mLMIR5. (A) Ba/F3 cells transduced with a Flag-tagged mLMIR5 or mock were stained with FITC-conjugated mouse IgG1 or anti-Flag Ab as well as polyclonal goat IgG or anti-mLMIR5 Ab, followed by PE-conjugated anti-goat IgG F(ab′)2. (B) Analysis of mLMIR5 expression on hematopoietic cells derived from C57BL/6 mice. Single-cell suspensions were prepared from BM, PB, peritoneal cavity, and thymus. Cells were stained with control IgG or anti-mLMIR5 Ab followed by PE-conjugated anti-goat IgG F(ab′)2 and FITC-conjugated mAbs as indicated. In BM, PB, and peritoneal cells, FSChighSSChigh populations representing myeloid lineage were gated and analyzed for mLMIR5 expression. In thymus, the FSClowSSClow populations representing lymphoid lineage were analyzed. (C) Single-cell suspensions were prepared from spleen. After B220+, CD3+, CD11b+, or CD11c+ cells were sorted by using FITC-conjugated Abs, these cells were stained as described in panel B. (D) Analysis of mLMIR5 expression on murine BM-derived cells. BMMCs, BMmDCs, BMpDCs, and BMMΦ were stained with control IgG or anti-mLMIR5 Ab followed by PE-conjugated anti-goat IgG F(ab′)2. The result of control or mLMIR5 staining is shown as a filled or bold-lined histogram, respectively. All the data are representative of 3 independent experiments.
Cell-surface expression of mLMIR5. (A) Ba/F3 cells transduced with a Flag-tagged mLMIR5 or mock were stained with FITC-conjugated mouse IgG1 or anti-Flag Ab as well as polyclonal goat IgG or anti-mLMIR5 Ab, followed by PE-conjugated anti-goat IgG F(ab′)2. (B) Analysis of mLMIR5 expression on hematopoietic cells derived from C57BL/6 mice. Single-cell suspensions were prepared from BM, PB, peritoneal cavity, and thymus. Cells were stained with control IgG or anti-mLMIR5 Ab followed by PE-conjugated anti-goat IgG F(ab′)2 and FITC-conjugated mAbs as indicated. In BM, PB, and peritoneal cells, FSChighSSChigh populations representing myeloid lineage were gated and analyzed for mLMIR5 expression. In thymus, the FSClowSSClow populations representing lymphoid lineage were analyzed. (C) Single-cell suspensions were prepared from spleen. After B220+, CD3+, CD11b+, or CD11c+ cells were sorted by using FITC-conjugated Abs, these cells were stained as described in panel B. (D) Analysis of mLMIR5 expression on murine BM-derived cells. BMMCs, BMmDCs, BMpDCs, and BMMΦ were stained with control IgG or anti-mLMIR5 Ab followed by PE-conjugated anti-goat IgG F(ab′)2. The result of control or mLMIR5 staining is shown as a filled or bold-lined histogram, respectively. All the data are representative of 3 independent experiments.
mLMIR5 associates strongly with DAP12 and to a lesser extent with DAP10 in mLMIR5-transduced cells
The presence of a positively charged residue (lysine) in the transmembrane suggested that LMIR5 associated with adaptor molecules—such as DAP10, DAP12, and FcRγ—containing a negatively charged residue within the transmembrane domain. To test this, we generated Ba/F3 cells cotransfected with retroviruses expressing a Myc-tagged mLMIR5 or hLMIR5 together with either a Flag-tagged DAP10, DAP12, FcRγ, or mock. Transduction of either mLMIR5 or hLMIR5 did not alter the expression levels of DAP10 and DAP12 mRNA when tested by real-time PCR (data not shown). Staining of these transfectants with anti-Myc mAb revealed that surface expression levels of mLMIR5 were significantly lower than those of hLMIR5 when LMIR5 was expressed alone. Surface expression levels of mLMIR5 were weakly or strongly elevated by DAP10 transduction or DAP12 transduction, respectively, while those of hLMIR5 were elevated by DAP10 as efficiently as by DAP12 (Figure 3A). The transduction of FcRγ did not influence the surface expression levels of mLMIR5 and hLMIR5 (Figure 3A). To confirm the physical association of mLMIR5 with either DAP10 or DAP12, we performed coimmunoprecipitation experiments; DAP12, but not DAP10, was coimmunoprecipitated with mLMIR5 probably because mLMIR5 more strongly associated with DAP12 compared with DAP10. On the other hand, total expression levels of mLMIR5 were elevated by the transduction of either DAP10 or DAP12, suggesting that these adaptors stabilized mLMIR5. When similar experiments were conducted on hLMIR5, both DAP10 and DAP12 were coimmunoprecipitated with hLMIR5 (Figure 3B). Furthermore, coimmunoprecipitation of endogenous DAP12 with transduced mLMIR5 was observed in BMMCs (Figure 3C). In conclusion, mLMIR5 was capable of associating strongly with DAP12 and to a lesser extent with DAP10, at least in mLMIR5-transduced cells.
Association of LMIR5 with adaptor molecules such as DAP10, DAP12, and FcRγ. (A,B) Ba/F3 cells were cotransduced with a Myc-tagged mLMIR5 or hLMIR5 and either a Flag-tagged DAP10, DAP12, FcRγ, or mock. (A) Cell-surface expression levels of mLMIR5 (top row) or hLMIR5 (bottom row) were analyzed by flow cytometry by staining cells with control mouse IgG1 or anti-Myc mAb, followed by FITC-conjugated anti-mouse Ig polyclonal Ab. The result of LMIR5 staining in the presence or absence of indicated adaptor molecule was represented by bold- or thin-lined histograms, respectively, while that of control staining was represented by a filled histogram. (B) Lysates of trasduced-Ba/F3 cells were immunoprecipitated with anti-mLMIR5 Ab, anti-Myc Ab, or anti-Flag mAb, and then immunoblotted with anti-Flag mAb or anti-Myc mAb. (C) Lysates of mLMIR5-transduced BMMCs were immnoprecipitated with control IgG or anti-mLMIR5 Ab, and then immunoblotted with anti-DAP12 Ab or anti-Flag mAb.
Association of LMIR5 with adaptor molecules such as DAP10, DAP12, and FcRγ. (A,B) Ba/F3 cells were cotransduced with a Myc-tagged mLMIR5 or hLMIR5 and either a Flag-tagged DAP10, DAP12, FcRγ, or mock. (A) Cell-surface expression levels of mLMIR5 (top row) or hLMIR5 (bottom row) were analyzed by flow cytometry by staining cells with control mouse IgG1 or anti-Myc mAb, followed by FITC-conjugated anti-mouse Ig polyclonal Ab. The result of LMIR5 staining in the presence or absence of indicated adaptor molecule was represented by bold- or thin-lined histograms, respectively, while that of control staining was represented by a filled histogram. (B) Lysates of trasduced-Ba/F3 cells were immunoprecipitated with anti-mLMIR5 Ab, anti-Myc Ab, or anti-Flag mAb, and then immunoblotted with anti-Flag mAb or anti-Myc mAb. (C) Lysates of mLMIR5-transduced BMMCs were immnoprecipitated with control IgG or anti-mLMIR5 Ab, and then immunoblotted with anti-DAP12 Ab or anti-Flag mAb.
Cross-linking of mLMIR5 induces the activation of mast cells
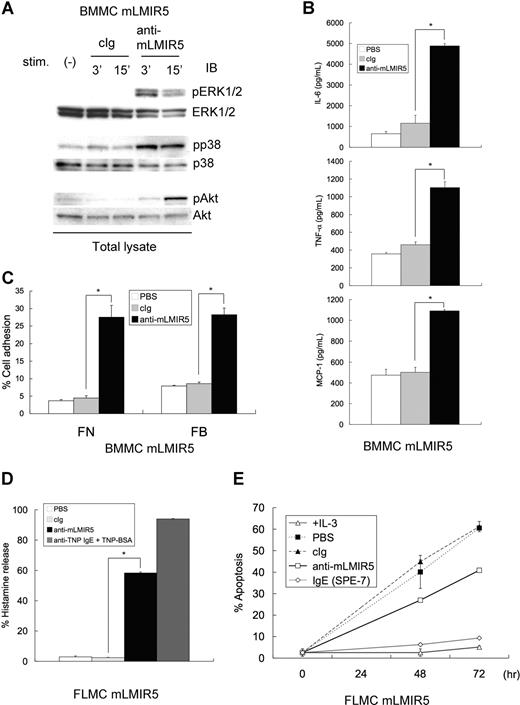
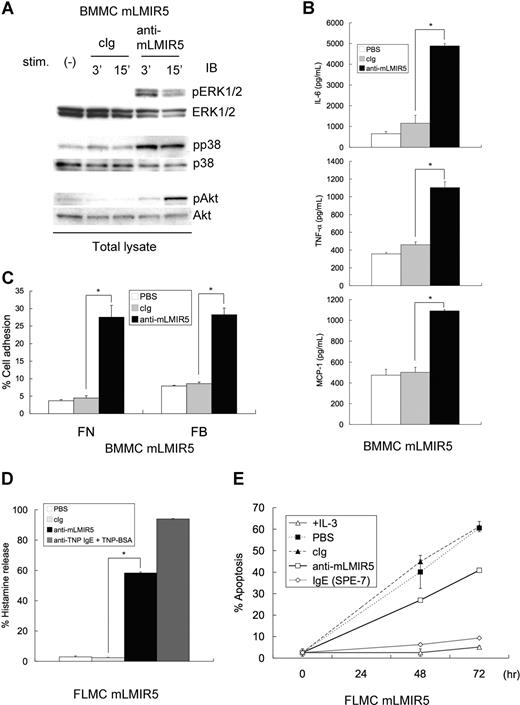
Since mLMIR5 was highly expressed in mast cells, we analyzed the activating functions of mLMIR5 in BMMCs. To obtain strong activation, mLMIR was transduced into BMMCs. When mLMIR5 was engaged by anti-mLMIR5 Ab, but not control Ab, in mLMIR5-transduced BMMCs, strong activation of ERK, p38, and Akt was recognized by using phospho-specific Abs, indicating a cellular activation in mast cells stimulated by LMIR5 cross-linking (Figure 4A). Mast cells, when activated by FcϵRI aggregation, cause a variety of activation events such as cytokine/chemokine production and degranulation characterized by histamine release.1,3,25,36,–38 Therefore, we performed experiments to examine whether similar activation events were induced by mLMIR5 engagement. In line with cellular activation, mLMIR5-transduced BMMCs stimulated by anti-mLMIR5 Ab, but not control Ab, produced IL-6, TNF-α, and MCP-1 (Figure 4B), and adhered efficiently to fibronectin or fibrinogen (Figure 4C). On the other hand, degranulation or cell survival effect was significant but weak in mLMIR5-transduced BMMCs stimulated by anti-mLMIR5 Ab (data not shown). Importantly, mLMIR5-transduced FLMCs in response to anti-mLMIR5 Ab stimulation efficiently released histamine and showed an antiapoptotic effect under IL-3–depleted conditions, thus demonstrating that engagement of endogenous mLMIR5 could also activate the cellular responses (Figure 4D,E).25,39,40 Taken together, mLMIR5 functioned as an activating receptor in mast cells.
Cross-linking of mLMIR5 induced the phosphorylation of several signaling molecules in mLMIR5-transduced mast cells, resulting in cytokine/chemokine production, cell adhesion, histamine release, and cell survival. (A) BMMCs transduced with mLMIR5 were stimulated with either control IgG or anti-mLMIR5 Ab for 3 or 15 minutes. Cell lysates were subjected to immunoblotting with either anti–phospho-p44/42 MAPK (pERK1/2), anti–phospho-p38 MAPK (pp38), or anti–phospho-Akt (pAkt) Ab. Equal loading was evaluated with by reprobing the immunoblots with Abs specific for ERK1/2, p38, or Akt. (B) BMMCs transduced with mLMIR5 were incubated with PBS, control IgG, or anti-mLMIR5 Ab for 12 hours. IL-6, TNF-α, and MCP-1 released into the culture supernatants were measured by ELISA. (C) BMMCs transduced with mLMIR5 in FN- or FB-coated plates were stimulated with PBS, control IgG, or anti-mLMIR5 Ab for 60 minutes. Adherent cells were measured as described in “Measurement of cytokines and histamines and adhesion assay.” (D) FLMCs transduced with mLMIR5 were incubated with PBS, control IgG, or anti-mLMIR5 Ab for 50 minutes. Alternatively, anti-TNP IgE-sensitized cells were incubated with TNP-BSA for 50 minutes. Histamine released in the culture supernatants was measured. (E) FLMCs transduced with mLMIR5 were incubated with either PBS, control IgG, anti-mLMIR5 Ab, or IgE (SPE-7) in the absence of IL-3. At indicated time points, cells were stained with PE-labeled annexin V to monitor apoptosis. Cells incubated in the presence of IL-3 were also analyzed. All data points correspond to the mean and the standard deviation (SD) of 4 independent experiments. Statistically significant differences are shown. *P < .05.
Cross-linking of mLMIR5 induced the phosphorylation of several signaling molecules in mLMIR5-transduced mast cells, resulting in cytokine/chemokine production, cell adhesion, histamine release, and cell survival. (A) BMMCs transduced with mLMIR5 were stimulated with either control IgG or anti-mLMIR5 Ab for 3 or 15 minutes. Cell lysates were subjected to immunoblotting with either anti–phospho-p44/42 MAPK (pERK1/2), anti–phospho-p38 MAPK (pp38), or anti–phospho-Akt (pAkt) Ab. Equal loading was evaluated with by reprobing the immunoblots with Abs specific for ERK1/2, p38, or Akt. (B) BMMCs transduced with mLMIR5 were incubated with PBS, control IgG, or anti-mLMIR5 Ab for 12 hours. IL-6, TNF-α, and MCP-1 released into the culture supernatants were measured by ELISA. (C) BMMCs transduced with mLMIR5 in FN- or FB-coated plates were stimulated with PBS, control IgG, or anti-mLMIR5 Ab for 60 minutes. Adherent cells were measured as described in “Measurement of cytokines and histamines and adhesion assay.” (D) FLMCs transduced with mLMIR5 were incubated with PBS, control IgG, or anti-mLMIR5 Ab for 50 minutes. Alternatively, anti-TNP IgE-sensitized cells were incubated with TNP-BSA for 50 minutes. Histamine released in the culture supernatants was measured. (E) FLMCs transduced with mLMIR5 were incubated with either PBS, control IgG, anti-mLMIR5 Ab, or IgE (SPE-7) in the absence of IL-3. At indicated time points, cells were stained with PE-labeled annexin V to monitor apoptosis. Cells incubated in the presence of IL-3 were also analyzed. All data points correspond to the mean and the standard deviation (SD) of 4 independent experiments. Statistically significant differences are shown. *P < .05.
DAP12 is required for the activation induced by mLMIR5 engagement as well as the sufficient surface expression of mLMIR5 under physiologic conditions
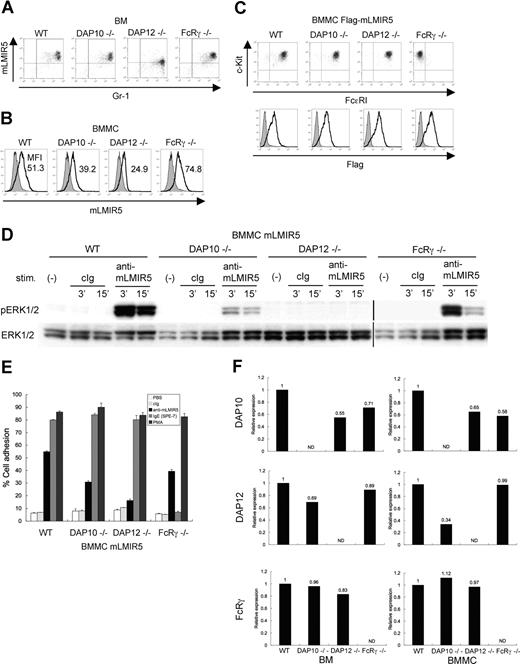
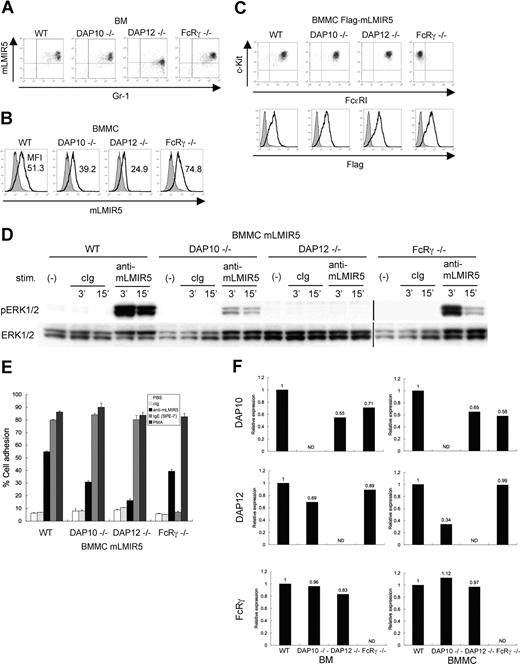
To determine which adaptor protein was a physiologic partner of mLMIR5, we analyzed surface expression levels of mLMIR5 in BM cells or BMMCs derived from wild-type (WT), DAP10−/−, DAP12−/−, or FcRγ−/− mice.20,22,23,28,29,41 As depicted in Figure 5A, surface expression levels of mLMIR5 in BM granulocytes derived only from DAP12-deficient mice were reduced when compared with those from other mice. On the other hand, surface expression levels of mLMIR5 in BMMCs were strongly or moderately reduced by DAP12 or DAP10 deficiency, respectively (Figure 5B). To further address the dependency of activating events caused by mLMIR5 aggregation on each adaptor protein, mLMIR5 was transduced into these BMMCs. The transduced cells exhibited comparable expression levels of FcϵRI and c-kit in addition to transduced mLMIR5, irrespective of the deficiency of respective adaptor molecule (Figure 5C bottom row). FcϵRI expression was not detectable in FcRγ-deficient BMMCs as reported (Figure 5C top row).29,41 Importantly, DAP12- or DAP10-deficient BMMCs in response to mLMIR5 engagement exhibited negligible or weak, respectively, activation of ERK as compared with WT BMMCs (Figure 5D), in proportion to the decreased capacities of DAP12- or DAP10-deficient BMMCs to adhere to fibronectin (Figure 5E). Concurrently, we confirmed that adhesion levels caused by PMA stimulation were comparable among these transfectants, and that FcRγ-deficient BMMCs did not adhere in response to IgE because of the lack of FcϵRI on the cell surface, as expected (Figure 5E). Although the deficiency of adaptor molecules did not show any morphologic difference in mast cells (data not shown), real-time PCR analysis demonstrated significantly low expression levels (approximately 30%) of DAP12 in DAP10-deficient BMMCs and decreased expression levels of DAP10 (approximately 60%) in DAP12- or FcRγ-deficient BMMCs, when compared with those in WT BMMCs (Figure 5F right panel). On the other hand, expression levels of DAP12 in DAP10-deficient BM were approximately 70% of those in WT BM (Figure 5F left panel). Considering the recent report that DAP12 transcript levels were not altered by DAP10 deficiency,20,22 expression levels of DAP12 might have been decreased during the course of differentiation of DAP10-deficient BMMCs in culture. Thus, attenuated activation induced by mLMIR5 cross-linking as well as reduced surface expression levels of mLMIR5 in DAP10-deficient BMMCs can be explained by the decreased expression levels of DAP12 rather than by the deficiency of DAP10. In summary, DAP12 plays a major role in maintaining surface expression levels of mLMIR5 under physiologic conditions and in transmitting activating signals induced by mLMIR5 aggregation.
Analysis of BM cells and BMMCs derived from DAP10−/−, DAP12−/−, and FcRγ−/− mice. (A,B) Surface expression levels of endogenous mLMIR5 on BM cells and BMMCs derived from WT, DAP10−/−, DAP12−/−, or FcRγ−/− mice were analyzed as described in Figure 2. The mean fluorescent intensity (MFI) of mLMIR5 expression was indicated in BMMCs. (C) WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with Flag-tagged mLMIR5 were stained with control IgG or anti-Flag mAb followed by FITC-conjugated anti-mouse Ig Ab to confirm transduced-mLMIR5 expression levels (bottom row). Phenotypical analysis of BMMCs was performed as described in materials and methods (top row). (D) Either WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with mLMIR5 were stimulated with control IgG or anti-mLMIR5 Ab. The amount of phosphorylated ERK1/2 was analyzed as described in Figure 4A. Vertical lines have been inserted to indicate a repositioned gel lane. (E) Either WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with mLMIR5 were stimulated on FN-coated plates. Percentages of adherent cells were estimated. All data points correspond to the mean and the SD of 3 independent experiments as indicated. (F) Relative expression levels of DAP10, DAP12, and FcRγ among WT, DAP10−/−, DAP12−/−, and FcRγ−/− BM or BMMCs were estimated by using real-time PCR as described in “Gene expression analysis.” The amount of expression was indicated relative to that in wild-type BM or BMMCs. Data are representative of 3 independent experiments.
Analysis of BM cells and BMMCs derived from DAP10−/−, DAP12−/−, and FcRγ−/− mice. (A,B) Surface expression levels of endogenous mLMIR5 on BM cells and BMMCs derived from WT, DAP10−/−, DAP12−/−, or FcRγ−/− mice were analyzed as described in Figure 2. The mean fluorescent intensity (MFI) of mLMIR5 expression was indicated in BMMCs. (C) WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with Flag-tagged mLMIR5 were stained with control IgG or anti-Flag mAb followed by FITC-conjugated anti-mouse Ig Ab to confirm transduced-mLMIR5 expression levels (bottom row). Phenotypical analysis of BMMCs was performed as described in materials and methods (top row). (D) Either WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with mLMIR5 were stimulated with control IgG or anti-mLMIR5 Ab. The amount of phosphorylated ERK1/2 was analyzed as described in Figure 4A. Vertical lines have been inserted to indicate a repositioned gel lane. (E) Either WT, DAP10−/−, DAP12−/−, or FcRγ−/− BMMCs transduced with mLMIR5 were stimulated on FN-coated plates. Percentages of adherent cells were estimated. All data points correspond to the mean and the SD of 3 independent experiments as indicated. (F) Relative expression levels of DAP10, DAP12, and FcRγ among WT, DAP10−/−, DAP12−/−, and FcRγ−/− BM or BMMCs were estimated by using real-time PCR as described in “Gene expression analysis.” The amount of expression was indicated relative to that in wild-type BM or BMMCs. Data are representative of 3 independent experiments.
Different signaling pathways between mLMIR5 and hLMIR5
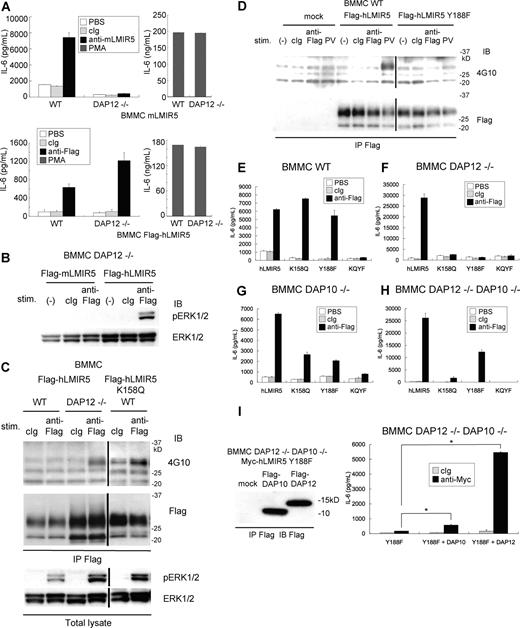
To explore whether the activating functions of hLMIR5 are also regulated by DAP12, Flag-tagged hLMIR5 was transduced into WT or DAP12-deficient BMMCs. Surprisingly, DAP12 deficiency did not inhibit but rather enhanced IL-6 production caused by hLMIR5 cross-linking (Figure 6A bottom row), while it completely abrogated that by mLMIR5 cross-linking (Figure 6A top row), notwithstanding equivalent amounts of cytokine production of WT and DAP12-deficient transfectants stimulated by PMA (Figure 6A). This was consistent with the finding that LMIR5 cross-linking induced strong activation of ERK in DAP12-deficient BMMCs transduced only with hLMIR5, but not mLMIR5 (Figure 6B), and that ERK activation induced by cross-linking of transduced hLMIR5 was rather enhanced in DAP12-deficient BMMCs in comparison with that of WT BMMC (Figure 6C). Since hLMIR5, but not mLMIR5, contained the putative phosphorylation site (Y188) in the cytoplasmic tail, we asked if this is related to DAP12-independent activation of mast cells stimulated by hLMIR5 cross-linking. Intriguingly, the phosphorylation of hLMIR5 was observed only in DAP12-deficient, but not WT, mast cells in response to hLMIR5 aggregation (Figure 6C). To further confirm that Y188 of hLMIR5 was indeed phosphorylated, we transduced either mock, Flag-tagged hLMIR5, or Flag-tagged hLMIR5 (Y188F) into WT BMMCs. As demonstrated in Figure 6D, stimulation of BMMCs with pervanadate induced tyrosine phosphorylation of hLMIR5, but not hLMIR5 (Y188F), suggesting that Y188 was a major phosphorylation site in hLMIR5 (Figure 6D). Interestingly, when hLMIR5 (K158Q)–transduced WT BMMCs were stimulated by hLMIR5 cross-linking, hLMIR5 (K158Q) was strongly phosphorylated (Figure 6C), suggesting that hLMIR5-mediated Y188 phosphorylation could be induced by adaptors that associate with hLMIR5 not through K158. To clarify whether phosphorylation of Y188 is required for cytokine production of mast cells caused by hLMIR5 engagement in the presence or absence of DAP12, we transduced hLMIR5 or hLMIR5 (Y188F) into DAP12-deficient BMMCs as well as WT BMMCs. As depicted in Figure 6E,F, the replacement of Y188 with phenylalanine (F) had no effect on cytokine production of WT BMMCs in response to anti-Flag mAb stimulation (Figure 6E), but abrogated that of DAP12-deficient BMMCs (Figure 6F). Altogether, these data strongly suggest that downstream of hLMIR5 there exists Y188 phosphorylation–independent and – dependent pathways in the presence and absence of DAP12, respectively.
Cross-linking of human LMIR5 induced the activation via phosphorylation of Y188 in its cytoplasmic region in DAP12-deficient BMMCs. (A) WT or DAP12−/− BMMCs transduced with a Flag-tagged mLMIR5 were stimulated with control IgG, anti-mLMIR5 Ab, or PMA (top panels), while WT or DAP12−/− BMMCs transduced with Flag-tagged hLMIR5 were stimulated with control IgG, anti-Flag mAb, or PMA (bottom panels). IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 4 independent experiments. (B) DAP12−/− BMMCs transduced with either a Flag-tagged mLMIR5 or hLMIR5 were stimulated with the indicated Abs for 3 minutes. The amount of phosphorylated ERK1/2 was analyzed as described. (C) WT or DAP12−/− BMMCs transduced with Flag-tagged hLMIR5 or WT BMMCs transduced with Flag-tagged hLMIR5 (K158Q) were incubated with the indicated Abs. Immunoprecipitates of cell lysates with anti-Flag mAb were immunoblotted with anti-phosphotyrosine mAb (4G10) or anti-Flag mAb. Total cell lysates were also analyzed to detect the amount of phosphorylated ERK1/2. Vertical lines have been inserted to indicate a repositioned gel lane. (D) BMMCs transduced with Flag-tagged hLMIR5, hLMIR5 (Y188F), or mock were stimulated with the indicated Ab for 3 minutes or with 100 μm pervanadate (PV) for 10 minutes. Immunoprecipitates of cell lysates with anti-Flag mAb were blotted with antiphosphotyrosine mAb (4G10) or anti-Flag mAb. Vertical lines have been inserted to indicate a repositioned gel lane. (E-H) WT (E) or DAP12−/− (F) DAP10−/− (G), or DAP12−/−DAP10−/− (H) BMMCs transduced with Flag-tagged hLMIR5, hLMIR5 (K158Q), hLMIR5 (Y188F), or hLMIR5 (K158Q) (Y188F) were stimulated with control IgG or anti-Flag mAb. IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 3 independent experiments. K158Q, Y188F, or KQYF indicate hLMIR5 (K158Q), hLMIR5 (Y188F), or hLMIR5 (K158Q) (Y188F), respectively. (I) DAP12−/−DAP10−/− BMMCs transduced with Myc-tagged hLMIR5 (Y188F) were transfected with Flag-tagged DAP10, DAP12, or mock. Immunoprecipitates of cell lysates were immunoblotted with anti-Flag mAb (left panel). These cells were stimulated with control IgG or anti-Myc mAb. IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 3 independent experiments. Statistically significant differences are shown. *P < .05.
Cross-linking of human LMIR5 induced the activation via phosphorylation of Y188 in its cytoplasmic region in DAP12-deficient BMMCs. (A) WT or DAP12−/− BMMCs transduced with a Flag-tagged mLMIR5 were stimulated with control IgG, anti-mLMIR5 Ab, or PMA (top panels), while WT or DAP12−/− BMMCs transduced with Flag-tagged hLMIR5 were stimulated with control IgG, anti-Flag mAb, or PMA (bottom panels). IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 4 independent experiments. (B) DAP12−/− BMMCs transduced with either a Flag-tagged mLMIR5 or hLMIR5 were stimulated with the indicated Abs for 3 minutes. The amount of phosphorylated ERK1/2 was analyzed as described. (C) WT or DAP12−/− BMMCs transduced with Flag-tagged hLMIR5 or WT BMMCs transduced with Flag-tagged hLMIR5 (K158Q) were incubated with the indicated Abs. Immunoprecipitates of cell lysates with anti-Flag mAb were immunoblotted with anti-phosphotyrosine mAb (4G10) or anti-Flag mAb. Total cell lysates were also analyzed to detect the amount of phosphorylated ERK1/2. Vertical lines have been inserted to indicate a repositioned gel lane. (D) BMMCs transduced with Flag-tagged hLMIR5, hLMIR5 (Y188F), or mock were stimulated with the indicated Ab for 3 minutes or with 100 μm pervanadate (PV) for 10 minutes. Immunoprecipitates of cell lysates with anti-Flag mAb were blotted with antiphosphotyrosine mAb (4G10) or anti-Flag mAb. Vertical lines have been inserted to indicate a repositioned gel lane. (E-H) WT (E) or DAP12−/− (F) DAP10−/− (G), or DAP12−/−DAP10−/− (H) BMMCs transduced with Flag-tagged hLMIR5, hLMIR5 (K158Q), hLMIR5 (Y188F), or hLMIR5 (K158Q) (Y188F) were stimulated with control IgG or anti-Flag mAb. IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 3 independent experiments. K158Q, Y188F, or KQYF indicate hLMIR5 (K158Q), hLMIR5 (Y188F), or hLMIR5 (K158Q) (Y188F), respectively. (I) DAP12−/−DAP10−/− BMMCs transduced with Myc-tagged hLMIR5 (Y188F) were transfected with Flag-tagged DAP10, DAP12, or mock. Immunoprecipitates of cell lysates were immunoblotted with anti-Flag mAb (left panel). These cells were stimulated with control IgG or anti-Myc mAb. IL-6 released into the culture supernatants was measured by ELISA. All data points correspond to the mean and the SD of 3 independent experiments. Statistically significant differences are shown. *P < .05.
Since DAP10 also associated with hLMIR5, we next examined the role of DAP10 in hLMIR5-mediated cytokine production of BMMCs. After hLMIR5 or hLMIR5 (Y188F) was transduced into either DAP10- or DAP10/DAP12 double-deficient BMMCs, similar experiments were performed. Surprisingly, cross-linking of hLMIR5 induced cytokine production even in the absence of both DAP12 and DAP10, suggesting the existence of unidentified adaptors of hLMIR5 (Figure 6H). Disruption of Y188 reduced, but did not abrogate, cytokine production of DAP10/DAP12 double-deficient BMMCs, indicating the existence of Y188 phosphorylation–independent and – dependent signaling pathways (Figure 6H). Transduction of DAP10 or DAP12 into hLMIR5 (Y188F)–expressing DAP10/DAP12-double deficient BMMCs enhanced cytokine production weakly or strongly, respectively, suggesting that DAP12 mainly contributed to Y188 phosphorylation–independent signaling (Figure 6I). To further characterize adaptors of hLMIR5, we examined if the replacement of K158 affected hLMIR5-mediated cytokine production. Intriguingly, the disruption of K158 did not affect cytokine production of WT BMMCs, but dampened that of DAP12-deficient BMMCs, while the disruption of both K158 and Y188 completely abrogated cytokine production irrespective of the absence of DAP10 or DAP12 (Figure 6E-H).
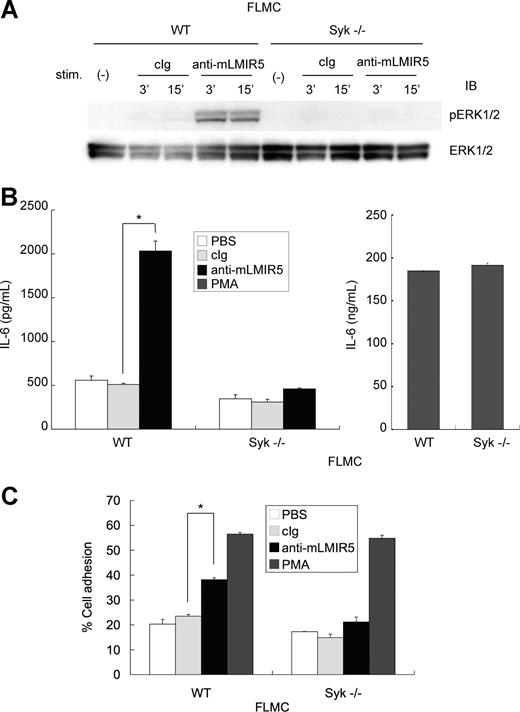
Cross-linking of endogenous mLMIR5 on FLMCs induced a Syk-dependent activation, resulting in cytokine production and adhesion
As mentioned, FLMCs were strongly activated in response to transduced mLMIR5 engagement compared with BMMCs. Next, we tested whether endogenous mLMIR5 could also activate FLMCs. As demonstrated in Figure 7A, activation of ERK was detected in WT FLMCs stimulated by anti-mLMIR5 Ab, but not by control Ab. This ERK activation was severely inhibited in Syk-deficient FLMCs.27 In accordance with this, IL-6 production and adhesive property were observed only in WT but not Syk-deficient FLMCs stimulated by engagement of endogenous mLMIR5, although PMA stimulation induced comparable levels of cytokine production and adhesion between WT and Syk-deficient FLMCs (Figure 7B,C). Altogether, aggregation of endogenous mLMIR5 in FLMCs induced Syk-dependent activation of mast cells, resulting in cytokine production and adhesion.
Cross-linking of endogenous mLMIR5 induced the activation of FLMCs. (A) WT or Syk-deficient FLMCs were stimulated with control IgG or anti-mLMIR5 Ab. The amount of phosphorylated ERK1/2 was measured as described. (B,C) IL-6 production (B) and percentage of adherent cells (C) were analyzed as described. All data points correspond to the mean and the SD of 4 independent experiments. Statistically significant differences are shown. *P < .05.
Cross-linking of endogenous mLMIR5 induced the activation of FLMCs. (A) WT or Syk-deficient FLMCs were stimulated with control IgG or anti-mLMIR5 Ab. The amount of phosphorylated ERK1/2 was measured as described. (B,C) IL-6 production (B) and percentage of adherent cells (C) were analyzed as described. All data points correspond to the mean and the SD of 4 independent experiments. Statistically significant differences are shown. *P < .05.
Discussion
In the present study, we characterized mLMIR5/CLM-7 as a novel member of LMIRs. The structure of mLMIR5 was typical of an activating receptor. mLMIR5 showed a higher homology with the paired receptors LMIR3/CLM-1 and LMIR4/CLM-5 as compared with another pair, LMIR1/CLM-8 and LMIR2/CLM-4, in the sequence of the Ig-like domain. Accordingly, LMIR5 would function as a counterpart of LMIR3 in the absence of LMIR4. Analysis using specific Ab against mLMIR5 revealed that mLMIR5 was mainly expressed in myeloid cells like other activating LMIRs, including LMIR2 and LMIR4, although there existed several differences; for example, LMIR4 or LMIR5 was highly expressed in mature or immature granulocytes, respectively.6,7,14 Thus, these activating receptors of the LMIR family displayed differing expression profiles among different stages of myeloid cells. In addition, high expression levels of mLMIR5 in colon and lung tissues may indicate that mLMIR5 is involved in mucosal immunity, possibly giving a clue to finding the ligands for mLMIR5.
A positively charged residue (lysine) in the transmembrane domain led us to postulate the association of mLMIR5 with ITAM- or the related activating motif–bearing adaptor proteins.15,,,–19,23,42 Our data indicated that mLMIR5 was mainly associated with DAP12 to maintain its surface expression levels under in vivo and in vitro physiologic conditions. However, it was noted that DAP10, when overexpressed, up-regulated mLMIR5 at surface expression levels, indicating that DAP10 could associate with mLMIR5 under conditions where DAP10 is up-regulated. On the other hand, the finding that surface expression levels of exogenously transduced mLMIR5 did not alter between WT and DAP12-deficient BMMCs suggested that mLMIR5, if abundant in cells, did not necessarily require DAP12 for maintaining its surface expression levels. Thus, it is suggested that surface expression levels of mLMIR5 were determined by DAP12 and to a lesser extent DAP10.
We found that mLMIR5 was highly expressed in both mucosal mast cells, including BMMCs and connective tissue–type mast cells, including peritoneal mast cells. Unexpectedly, cross-linking of endogenous mLMIR5 induced a more pronounced activation in FLMCs than in BMMCs despite comparable surface expression levels of mLMIR5. This suggests that mLMIR5 plays an important role in specific types of mast cells, closely related to their differentiation and distribution in tissue. Notably, activation events caused by mLMIR5 cross-linking were almost completely dampened by the deficiency of DAP12 or its ITAM-associated kinase Syk, suggesting that mLMIR5 transmitted an activating signal through the phosphorylation of ITAM in a cytoplasmic region of DAP12.17,–19,27,43 It is noteworthy that Syk-dependent activation was demonstrated by cross-linking of endogenous mLMIR5. Interestingly, the deficiency of DAP10 in BMMCs also inhibited the functions of mast cells, albeit to a lesser degree. This is probably due to decreased expression levels of endogenous DAP12 in the absence of DAP10 (Figure 5F), although we could not completely rule out the involvement of DAP10 in the functions of mLMIR5. Thus, activating functions of mLMIR5 were predominantly regulated by DAP12 among adaptor proteins.
Analysis of differential regulation of immune receptors in mouse versus human cells sometimes sheds light on the evolutionary and physiologic significance of the paired receptor.20,,–23 Indeed, several structural and functional differences were observed in mouse versus human LMIR5. Indeed, several structural differences were observed in mouse versus human LMIR5. For example, mLMIR5, but not hLMIR5, was an N-glycoprotein. In addition, mLMIR5 differed from hLMIR5 by several amino acid residues in the transmembrane domain sequence (Figure 1A). These differences in glycosylation and transmembrane structure may be related to differing surface expression levels or to capacities to associate with DAP10 in mLMIR5 versus hLMIR5 transduced into Ba/F3 cells (Figure 3A). It was noticed that hLMIR5 but not mLMIR5 possessed a putative phosphorylation motif (Y188) in the short cytoplasmic region. From the results using WT and DAP12−/− BMMCs, we concluded that hLMIR5 delivered an activation signal independently of the phosphorylation of Y188 in WT BMMCs, whereas it did so through phosphorylation in DAP12−/− BMMCs. Martinez-Barriocanal and Sayos24 also reported that CD300b/hLMIR5 signal was dependent on Y188 phosphorylation in rat basophilic leukemia (RBL) cells that supposedly do not express DAP12. However, unlike our results, hLMIR5 signal in RBL cells was not completely blocked by Y188 mutation. How Y188 is phosphorylated by cross-linking of hLMIR5 in DAP12-deficient mast cells remains elusive. It is possible that Y188 in a cytoplasmic tail becomes readily accessible in the absence of DAP12 and thereby is phosphorylated by some tyrosine kinases. The finding that cross-linking of hLMIR5 induced cytokine production even in the absence of both DAP12 and DAP10 strongly suggested the existence of unknown adaptors. In addition, hLMIR5-mediated cytokine production was abrogated by the disruption of the charged residue (K158) in DAP12-deficient BMMCs, but not WT BMMCs, whereas the disruption of both K158 and Y188 dampened hLMIR5-mediated cytokine production in WT BMMCs as well as DAP12-deficient BMMCs. In contrast, Martinez-Barriocanal and Sayos24 showed that the disruption of K158 abrogated cytokine production in DAP12-transduced RBL cells. This discrepancy might be explained by the cell-type difference between RBL cells and BMMCs. However, it is also possible that different experimental systems (RBL cells and DAP12 overexpression in RBL cells vs DAP12-knockout cells and the wild-type cells) are responsible for the discrepancy. One strange result is that Y188F mutant but not K158Q mutant of hLMIR5 induced cytokine production in DAP10/DAP12 double-knockout BMMCs but not in DAP12-knockout BMMCs. This difference could be explained by the presence of an additional adaptor that is up-regulated in the absence of both DAP10 and DAP12, but not DAP12 alone.
Recent advances44,,,,,,–51 have demonstrated that DAP12 could mediate an activating or inhibitory signal depending on the associated receptor, and support a model that the avidity of the receptor for its ligand could modulate cellular response. Therefore, identification of the LMIR5 ligand, either endogenous protein or foreign bodies, and analysis of LMIR5 knockout mice will be indispensable for elucidation of in vivo functions of LMIR5.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Takashi Saito and Sho Yamasaki for kindly providing fetal liver cells of syk−/− mice. We also thank Drs Marco Colonna and Hisashi Arase for providing DAP10−/− mice and pME18S expression vector containing a mouse CD150 leader segment, respectively. We are grateful to Dr Dovie Wylie for her excellent language assistance. We thank Mihoko Shibuya and Kaori Ema for technical assistance.
This work was supported by grants from the Ministry of Education, Science, Technology, Sports and Culture and the Ministry of Health and Welfare, Japan.
Authorship
Contribution: Y.Y. did all the experiments and participated in writing the manuscript; J.K. oversaw all the experiments and actively participated in manuscript writing; I.K. assisted with the experiments including retroviral transfection; T.M. assisted with the experiments including retroviral transfection and flow cytometric analysis; T.O., Y.L., F.S., S.Y., H.K., and H.N. did the cloning and made the constructs of LMIRs; M.M.-Y. measured the released histamine; V.L.J.T provided Syk knock-out mice; T.T. provided several knock-out mice; and T.K. conceived of and directed the project, secured funding, and actively participated in manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Toshio Kitamura, Division of Cellular Therapy, Advanced Clinical Research Center, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: kitamura@ims.u-tokyo.ac.jp.