Abstract
Drug-induced immune thrombocytopenia is caused by drug-dependent antibodies (DDAbs) that bind tightly to platelet glycoproteins only when drug is present. How drugs mediate this interaction is not yet resolved. Several studies indicate that sites recognized by DDAbs tend to cluster in specific structural domains, suggesting they may recognize a limited number of distinct epitopes. To address this issue, we characterized the binding sites for 16 quinine-dependent antibodies thought on the basis of preliminary studies to be possibly specific for a single epitope on glycoprotein IIIa (GPIIIa). Fourteen of the antibodies reacted with a 29-kDa GPIIIa fragment comprising only the GPIIIa hybrid and plextrin-semaphorin-integrin homology domains. However, studies with mutant GPIIIa and the blocking monoclonal antibody AP3 showed that the 14 DDAbs recognize at least 6 and possibly more distinct, but overlapping, structures involving GPIIIa residues 50 to 66. The findings suggest that even antibodies specific for restricted domains on a target glycoprotein may each have a slightly different fine specificity; ie, “unique” epitopes recognized by DDAbs may be rare or nonexistent. The observations are consistent with a recently proposed model in which drug reacts noncovalently with both target protein and antibody to promote binding of an otherwise nonreactive immunoglobulin.
Introduction
Drug-induced immune thrombocytopenia is an unusual, but often serious, side effect of drug therapy.1,2 In many examples of drug-induced immune thrombocytopenia, platelet destruction is caused by a remarkable type of antibody that is innocuous in the absence of drug, but binds to specific sites on platelet membrane glycoprotein complexes IIb/IIIa (aphaIIb/beta3 integrin) or Ib/V/IX when drug is present in soluble form.3,4 Although antibodies of this type can also cause hemolytic anemia5 and neutropenia,6 for unknown reasons, platelets are targeted far more often than other cell types. How drug-dependent antibodies (DDAbs) are induced and how, once they are formed, exposure to the immunizing drug causes them to bind tightly to their target(s) and causes platelet destruction is as yet unresolved. It is generally agreed, however, that drug-dependent antibody binding does not require covalent linkage of drug to the target glycoprotein and is therefore not a classic “hapten-dependent” phenomenon.1,4,7 Platelet-specific, drug-dependent antibodies almost invariably recognize epitopes carried on the GPIb/IX and/or the GPIIb/IIIa glycoprotein complexes.8-11 Molecular characterization of the target epitopes recognized by DDAbs on these glycoproteins could provide insights into the mechanism by which soluble drugs promote DDAb binding and cause platelet destruction and could help to explain why platelets are so often targeted by drug-induced antibodies.
In previous studies, we identified a site comprising amino acids 50 to 66 of the “hybrid” domain of glycoprotein IIIa (GPIIIa) that is recognized by a group of 3 quinine-dependent antibodies and showed that certain amino acid residues in this region (Ala50, Arg62, Asp66) are essential for antibody binding.12 Here, we show that these antibodies recognize a recombinant fragment of GPIIIa consisting only of the N-terminal plextrin-semaphorin-integrin (PSI) homology domain and the adjacent hybrid domain and characterize the fine specificity of a total of 16 quinine-induced, GPIIIa-specific antibodies.
Methods
All procedures involving human subjects have been approved by the BloodCenter of Wisconsin's institutional review board. Informed consent was provided in accordance with the Declaration of Helsinki.
Antibodies and reagents
GPIIIa-specific monoclonal antibodies (mAbs) AP3 (specific for GPIIIa residues 50 and 62) and AP5 (specific for GPIIIa residues 1-5) were described previously.12-14 Monoclonal anti-V5 antibody was purchased from Invitrogen (Carlsbad, CA). Quinine-specific DDAbs were from 16 patients who experienced severe thrombocytopenia (platelets < 10 × 109/L) after taking quinine and recovered after drug was discontinued. DDAbs designated 1, 2, and 8 in this report were described previously.12 Alloantibodies specific for HPA-1a (PlA1) were from the Platelet/Neutrophil Immunology Laboratory of BloodCenter of Wisconsin.
Preparation of cDNAs encoding truncated and mutant versions of GPIIIa
Throughout this report, nucleotide (nt) 1 refers to A of the ATG translation start codon of human GPIIIa. All versions of truncated GPIIIa possessed the native signal peptide at the amino terminus and were fused in frame at the carboxyl terminus to a V5 epitope and polyhistidine (6XHis) sequence for detection with an anti-V5 antibody or a nickel-chelating resin. Constructs were generated by polymerase chain reaction with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA) from a full-length human GPIIIa cDNA template12 and were inserted into expression vector pIB/V5-His TOPO (Invitrogen) containing a blasticidin resistance gene. All inserts and ligation sites were validated by direct sequencing.
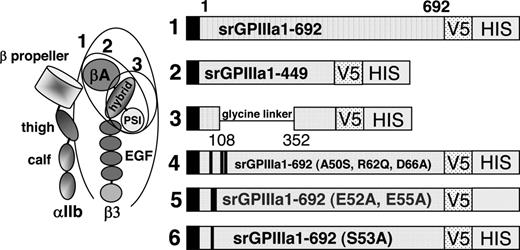
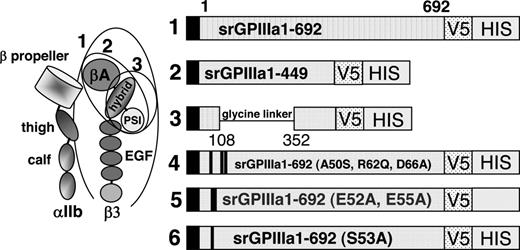
We previously showed that 3 quinine-dependent antibodies (here designated DDAbs 1, 2, and 8) recognize a restricted domain of GPIIIa that includes amino acid residues 50 to 66.12 For the current studies, we produced progressively smaller recombinant fragments of GPIIIa, each of which contains these amino acid residues (Figure 1).15-17 cDNA encoding the entire extracellular domain of GPIIIa amino acid residues 1 to 692 (construct 1) was generated from full-length human GPIIIa cDNA using sense primer 5′GPIIIa (5′ GAG GCG GAC GAG ATG CGA GCG 3′) and antisense primer a3ATM (5′GTC AGG GCC CTT GGG ACA CTC TGG 3′). Construct 2 (srGPIIIa1-449) was generated with sense primer 5′ GPIIIa and antisense primer a3A449 (5′ GTT GCA GCG ATG GCT ATT AGG 3′). Construct 3 is similar to construct 2, except that the βA domain (residues 108 to 352) was deleted and replaced with a 14 amino acid flexible glycine linker.18 Construct 3 was produced by amplifying overlapping polymerase chain reaction products corresponding to GPIIIa residues 1 to 108 and 352 to 449 and splicing a nucleotide sequence encoding a polyglycine linker (SGGGGSGGGGSGGS) between them.12,19 Constructs 4 to 6 were identical to construct 1 except that selected amino acid residues between positions 50 and 66 were mutated as previously described.12 In construct 4, human residues Ala50, Arg62, and Asp66 were changed to the corresponding residues (Ser50, Gln62, Ala66) present in rat GPIIIa.12 Conversion of any of these 3 residues from human to rat was previously shown to abolish the reaction of DDAbs 1, 2, and 8 with GPIIIa.12 In construct 5 (E52A, E55A) and construct 6 (S53A), selected charged and polar residues within the putative binding site for DDAbs 1, 2, and 8 were converted to alanine. The cDNA encoding each of the constructs was validated by sequencing before proteins were expressed. Constructs 1 to 6 are depicted schematically in Figure 1.
Schematic representation of 6 constructs used to investigate the specificity of quinine-dependent antibodies recognizing GPIIIa in Western blot. (Left) Schematic representation of GPIIb/IIIa based on crystallographic studies of alphav/beta315 and alphaIIb/beta316 integrins. Adapted from Humphrey and Mould,17 with permission. Ovals indicate structural domains incorporated into constructs 1 to 3. (Right) Diagram indicating amino acid sequences of constructs 1 to 3 and mutations incorporated into construct 1 to produce constructs 4 to 6. V5 and HIS recognition sites are described in “Preparation of cDNAs encoding truncated mutant versions of GPIIIa.”
Schematic representation of 6 constructs used to investigate the specificity of quinine-dependent antibodies recognizing GPIIIa in Western blot. (Left) Schematic representation of GPIIb/IIIa based on crystallographic studies of alphav/beta315 and alphaIIb/beta316 integrins. Adapted from Humphrey and Mould,17 with permission. Ovals indicate structural domains incorporated into constructs 1 to 3. (Right) Diagram indicating amino acid sequences of constructs 1 to 3 and mutations incorporated into construct 1 to produce constructs 4 to 6. V5 and HIS recognition sites are described in “Preparation of cDNAs encoding truncated mutant versions of GPIIIa.”
Stable expression of truncated and mutant GPIIIa isoforms in insect cells
High five insect cell lines20-22 were transfected using the InsectSelect System (Invitrogen) according to manufacturer's protocol.23,24 Stably transfected cell lines12 were selected in medium supplemented with Blasticidin S HCl 80 μg/mL (Invitrogen) and maintained with Balsticidin S HCl at 20 μg/mL. Secreted proteins were harvested, concentrated 10-fold with a Microcon YM-10 (Amicon), and identified by Western blot.12
Purification of proteins
Soluble recombinant proteins were isolated by immunoaffinity purification. MAb AP3 specific for GPIIIa residues 50 to 6212 was covalently immobilized to cyanogen bromide–activated Sepharose 4B (GE Healthcare, Waukesha, WI) according to manufacturer's protocol. Beads were then blocked with ethanolamine. Harvested culture supernatant was incubated with the AP3- Sepharose beads overnight at 27°C. The beads were then washed (20 mM Tris, 145 mM NaCl, pH 7.6) and bound protein was eluted with Gentle Ab/Ag Elution Buffer (Pierce, Rockford, IL). Eluted fractions were dialyzed in buffer. Constructs 4 to 6 were purified similarly, except that mAb AP5 (anti-GPIIIa residues 1-5) rather than AP3 was used for affinity purification of construct 4.
Antigen capture ELISA
Antigen capture ELISA (ACE) was performed as described previously.12 In brief, mAb AP5 (plated in microtiter wells overnight at 10 μg/mL) was used to capture truncated srGPIIIa and platelet GPIIIa (from platelet lysate) and human antibodies bound to these targets were detected with alkaline-phosphatase labeled goat-antihuman IgG (Fc fragment specific) antibodies (Jackson ImmunoResearch Labs, West Grove, PA).
Western blot analysis was performed as described previously.12 For blots with patient serum equal amounts of pure recombinant protein in 50 mM Tris, pH 6.8, 2% sodium dodecyl sulfate, 4% glycerol were loaded. Patient serum (1:280) was exposed to the blotted proteins in the presence and absence of 400 μM quinine (Sigma-Aldrich, St Louis, MO). Bound human IgG was detected with horseradish peroxidase-conjugated goat antihuman IgG H+L chain specific antibodies (Jackson Immuno-Research Labs).
Results
Characterization of srGPIIIa constructs
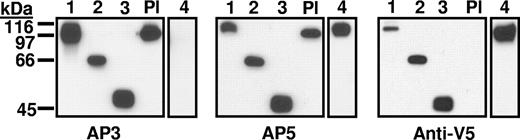
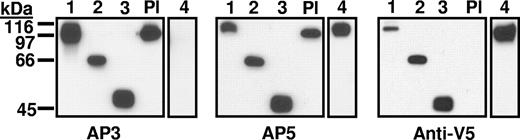
Constructs 1 to 6 (Figure 1) were characterized by Western blotting using mAbs AP3, AP5, and anti-V5 for detection. As shown for constructs 1 to 4 in Figure 2, each construct migrated with the mobility predicted by its molecular weight. As expected,12 construct 4 (Ala50Ser, Arg62Gln, Asp66Ala) was not recognized by mAb AP3 and platelet GPIIIa was not detected with anti-V5. Immunologic integrity of the recombinant proteins was further evaluated with antibodies specific for the HPA-1a (PlA1) alloantigen, determined by a leucine residue at position 33 in GPIIIa.25 Some HPA-1a–specific antibodies (type II) require the PSI homology domain where HPA-1a is located to be in contact with the C-terminal, cysteine-rich domain of GPIIIa for their reactions with the alloantigen, whereas others (type I) do not have this requirement.26 As shown in Table 1, a type I HPA-1a–specific antibody reacted with constructs 1 to 3 as expected. In contrast, a type II antibody recognized only construct 1.
Western blot of constructs 1 to 4 and platelet lysate (Pl) using monoclonals AP3, AP5, and anti-V5 for detection. Constructs 1 to 4 migrated as expected on the basis of their predicted molecular weights and reacted with the appropriate probes. As expected, construct 4 containing mutations A50S, R62Q, and D66A was not recognized by monoclonal AP3, and platelet GPIIIa (Pl) was not recognized by anti-V5.
Western blot of constructs 1 to 4 and platelet lysate (Pl) using monoclonals AP3, AP5, and anti-V5 for detection. Constructs 1 to 4 migrated as expected on the basis of their predicted molecular weights and reacted with the appropriate probes. As expected, construct 4 containing mutations A50S, R62Q, and D66A was not recognized by monoclonal AP3, and platelet GPIIIa (Pl) was not recognized by anti-V5.
Thirteen additional DDAbs were identified that recognize full-length GPIIIa in Western blot
We previously characterized 3 quinine-dependent DDAbs (here designated DDAbs 1, 2, and 8) that react with human, but not rat, GPIIIa in a Western blot assay.12 Specificity of these antibodies for a single epitope comprising GPIIIa residues 50 to 66 was suggested by 2 observations: binding was lost completely when any one of amino acids 50, 62, or 66 was switched to the corresponding amino acid present in rat GPIIIa and the reaction of each DDAb was blocked by mAb AP3, specific for GPIIIa residues 50 to 62.12 To determine the extent to which other quinine-dependent antibodies recognize the same region of GPIIIa, we screened 34 additional DDAbs that reacted strongly and drug-dependently with intact platelets for their ability to react with GPIIIa in Western blot only when quinine was present. Twenty-one of these antibodies did not react with GPIIIa in Western blot either in the presence or absence of quinine. However, 13 (here designated DDAbs 3-7 and 9-16) did have this property (data not shown). Studies were then performed to further characterize the epitopes recognized by all 16 antibodies.
Fourteen DDAbs recognized construct 3 in the presence of quinine
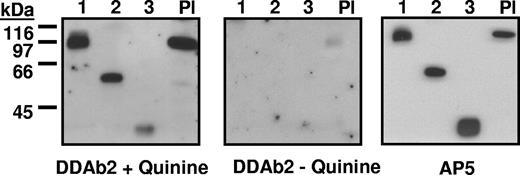
DDAbs 1 to 14 recognized the smallest of the 3 constructs (construct 3) in Western blot only when quinine was present, but DDAbs 15 and 16 did not (Table 2). Representative data are shown in Figure 3. DDAbs 1, 2, and 8 also reacted with constructs 1 to 3 immobilized by capture with mAb AP-5 (Figure 4). The other DDAbs were not tested in this format. Recognition of construct 3 by DDAbs 1 to 14 in Western blot and by DDAbs 1, 2, and 8 in solid phase ELISA indicates that these antibodies bind directly to the smallest (29 kDa) fragment of GPIIIa.
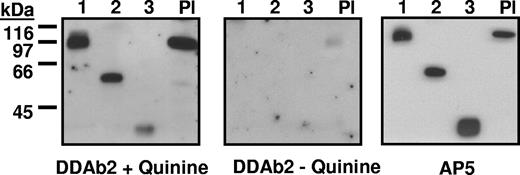
Reactions of DDAb2 with constructs 1 to 3 and platelet GPIIIa (Western blot). (Left) DDAb2 recognizes constructs 1 to 3 and platelet GPIIIa (Pl) in the presence of quinine. (Middle) DDAb2 fails to react with these targets in the absence of drug. (Right) Recognition of the same targets by monoclonal AP5 specific for the 5 N-terminal amino acids of GPIIIa.
Reactions of DDAb2 with constructs 1 to 3 and platelet GPIIIa (Western blot). (Left) DDAb2 recognizes constructs 1 to 3 and platelet GPIIIa (Pl) in the presence of quinine. (Middle) DDAb2 fails to react with these targets in the absence of drug. (Right) Recognition of the same targets by monoclonal AP5 specific for the 5 N-terminal amino acids of GPIIIa.
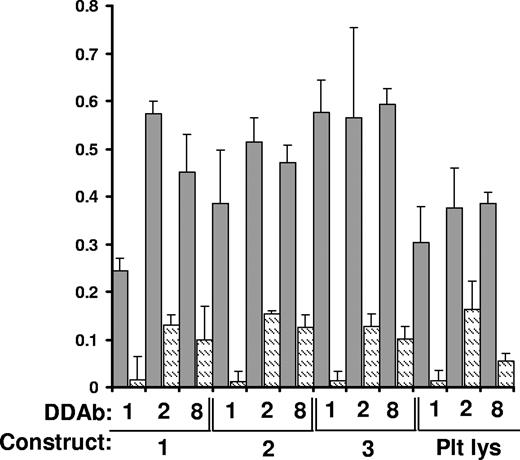
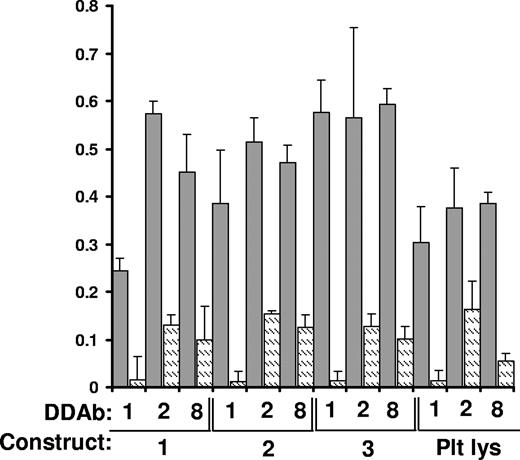
DDAb1, DDAb2, and DDAb8 recognize constructs 1 to 3 captured in a microtiter plate by monoclonal AP5. Bound antibody was detected by ELISA. Dark bars indicate reactions in the presence of 0.4 mM quinine; lighter bars show reactions in the absence of drug. Reactions were performed in triplicate. Error bars represent 1.0 SD. “Plt Lys” indicates reactions with GPIIIa captured from platelet lysate.
DDAb1, DDAb2, and DDAb8 recognize constructs 1 to 3 captured in a microtiter plate by monoclonal AP5. Bound antibody was detected by ELISA. Dark bars indicate reactions in the presence of 0.4 mM quinine; lighter bars show reactions in the absence of drug. Reactions were performed in triplicate. Error bars represent 1.0 SD. “Plt Lys” indicates reactions with GPIIIa captured from platelet lysate.
Seven antibodies behaved identically when tested against the different GPIIIa constructs; 9 others differed in various degrees from this “consensus” reaction pattern
Epitopes recognized by the 16 antibodies were further characterized by determining whether they were blocked by mAb AP3, which requires GPIIIa residues 50 to 62 for binding, and whether they recognized constructs 4 to 6 in which selected amino acids among residues 50 to 66 were mutated. Results are summarized in Table 2. Representative data are shown in Figures 5 and 6. DDAbs 1 to 7 behaved identically in that they recognized constructs 1, 3, 5, and 6 but not construct 4, and their reactions were blocked by AP3. However, deviations from this pattern were seen with DDAbs 8 to 16 (Table 2).
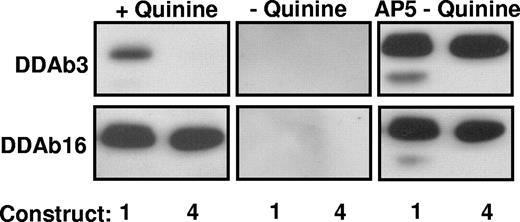
Reactions of DDAb3 and DDAb16 with constructs 1 and 4 (Western blot). (Left) Both antibodies recognize construct 1 (srGPIIIa) in the presence of quinine. However, DDAb3 fails to recognize construct 4 (A50S, R62Q, D66A). (Middle) DDAb3 and DDAb16 fail to recognize either construct in the absence of quinine. (Right) Blot with monoclonal AP5 confirms that adequate quantities of both constructs were present for antibody detection.
Reactions of DDAb3 and DDAb16 with constructs 1 and 4 (Western blot). (Left) Both antibodies recognize construct 1 (srGPIIIa) in the presence of quinine. However, DDAb3 fails to recognize construct 4 (A50S, R62Q, D66A). (Middle) DDAb3 and DDAb16 fail to recognize either construct in the absence of quinine. (Right) Blot with monoclonal AP5 confirms that adequate quantities of both constructs were present for antibody detection.
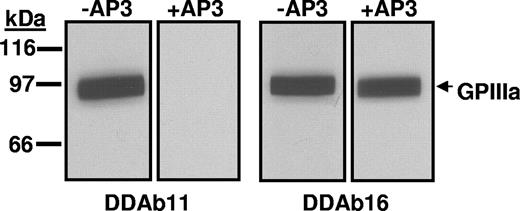
Effect of mAb AP3 on binding of DDAb11 and DDAb16 to construct 1. The quinine-dependent reaction of DDAb11 (left) but not that of DDAb16 (right) with construct 1 (srGPIIIa) is blocked by monoclonal AP3. No reactions occurred in the absence of quinine (not shown). The sharp edges at the lateral margin of each band resulted from cutting the membrane into strips after protein transfer from a preparative gel.
Effect of mAb AP3 on binding of DDAb11 and DDAb16 to construct 1. The quinine-dependent reaction of DDAb11 (left) but not that of DDAb16 (right) with construct 1 (srGPIIIa) is blocked by monoclonal AP3. No reactions occurred in the absence of quinine (not shown). The sharp edges at the lateral margin of each band resulted from cutting the membrane into strips after protein transfer from a preparative gel.
DDAbs 8 and 9 differed only by their failure to react with constructs 5 and 6, but progressively greater differences were seen with DDAbs 10 to 14. The greatest disparity was seen with DDAbs 15 and 16, which failed to recognize construct 3 and were not blocked by AP3. Surprisingly, however, DDAb 15 failed to recognize construct 4 (S50A, R62Q, D66A) and construct 5 (E52A, E55A).
Discussion
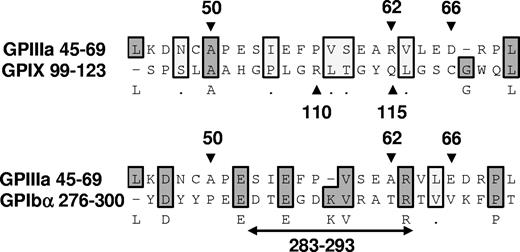
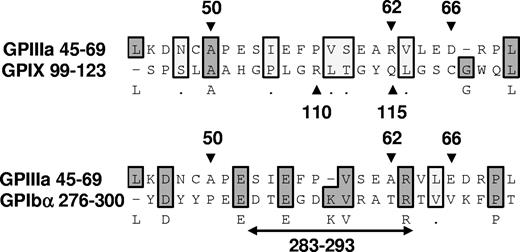
Epitopes recognized by quinine-dependent antibodies have now been identified and partially characterized on 3 platelet membrane glycoproteins: GPIIIa, GPIb alpha, and GPIX. Chong et al10,27 found that reactions of a group of quinine-dependent antibodies with GPIX could be blocked by monoclonal antibody SZ1, specific for a membrane-proximal site on this glycoprotein that is stabilized by its interaction with GPIb beta. Using human/mouse hybrid GPIX constructs and mutated GPIX expressed in CHO cells, they showed that 2 amino acids contained in human, but not mouse, GPIX (Arg110 and Gln115) were critical for binding of 6 quinine-dependent DDAbs.28 Epitopes recognized by quinine-dependent antibodies on GPIb alpha are less well defined, except for an 11 amino acid sequence (residues 283-293) shown to be required for binding of a group of quinine-dependent antibodies to this target.27 Figure 7 shows the alignment of 25 amino acid sequences containing residues demonstrated in these studies to be critical for antibody binding using a program designed to detect homology between sequences.29,30 The “alignment score”29,30 obtained when GPIIIa 45 to 69 is compared with GPIX 99 to 123 is −26 (no homology), but the score obtained when GPIIIa 45 to 69 is compared with GPIb alpha 276 to 300 is 47, 30% of the maximum score possible. This is consistent with the possibility that GPIIIa 45 to 69 and GPIb alpha 276 to 300 share a structural motif recognized by quinine-dependent antibodies, but further comparison is difficult because the crystal structure of GPIb alpha has been resolved only to residue 290.31 It is intriguing that the same region of GPIIIa shown in our studies to be a favored target for quinine-dependent antibodies appears to be a major antigenic determinant for induction of autoantibodies in patients infected with HIV-1.32
Alignment of 25 amino acid sequences known to contain binding sites for quinine-dependent antibodies using the Clustal W algorithm.29,30 GPIIIa residues 50, 62, and 66,12 GPIX residues 110 and 115,28 and GPIb alpha residues 283 to 29327 are known to be critical for binding of selected groups of quinine-dependent antibodies to the respective glycoproteins. Amino acids shaded darkly are identical; those shaded lightly are chemically similar. There is no significant homology between GPIIIa 45 to 69 and GPIX 99 to 123 (alignment score, −26). However, the alignment score between GPIIIa 45 to 69 and GPIb alpha 276 to 300 is 47, approximately 30% of the maximum score possible (2 sequences identical).
Alignment of 25 amino acid sequences known to contain binding sites for quinine-dependent antibodies using the Clustal W algorithm.29,30 GPIIIa residues 50, 62, and 66,12 GPIX residues 110 and 115,28 and GPIb alpha residues 283 to 29327 are known to be critical for binding of selected groups of quinine-dependent antibodies to the respective glycoproteins. Amino acids shaded darkly are identical; those shaded lightly are chemically similar. There is no significant homology between GPIIIa 45 to 69 and GPIX 99 to 123 (alignment score, −26). However, the alignment score between GPIIIa 45 to 69 and GPIb alpha 276 to 300 is 47, approximately 30% of the maximum score possible (2 sequences identical).
Human and rat GPIIIa (with which human DDAbs are nonreactive)12,28 differ in amino acid composition at only 10 positions in the sequence extending from residues 42 to 98. As already noted, the reactions of DDAbs 1, 2, and 8 (Table 2) were previously shown to be abolished when any one of amino acids A50, R62, or D66 were switched to the corresponding rat GPIIIa residue (S50, Q62, or A66) but were unaffected when human to rat substitutions were made upstream at position 42 or downstream at position 67 or 71.12 Reactions of these DDAbs were blocked by mAb AP3, which requires human amino acids at positions 50 and 62 for binding to GPIIIa, but neither the DDAbs nor AP3 recognized a synthetic peptide containing GPIIIa residues 49 to 67.12 The findings indicated that DDAbs 1, 2, and 8 recognize a restricted amino sequence (50-66) in the “hybrid” domain of GPIIIa in its native conformation. Although binding of each of the DDAbs was abolished by disruption of disulfide bonds in GPIIIa, they reacted strongly with the nonreduced protein in a Western blot format, indicating that their target epitope(s) is reconstituted when the protein is partially renatured after being transferred to a supporting membrane after sodium dodecyl sulfate gel electrophoresis.12 Possibly, the stability of this region of GPIIIa is a consequence of the stability provided by 2 intrachain disulfide bonds.33 Drug-dependent binding of DDAbs 1, 2, and 8 under these relatively stringent conditions suggested that they might each recognize the same epitope on GPIIIa.
In the present study, we determined the extent to which other quinine-induced DDAbs mimic the behavior of DDAbs 1, 2, and 8. On screening 34 quinine-dependent DDAbs, we found 13 additional antibodies that reacted with GPIIIa in Western blot, making a total of 16 that were possibly specific for the same epitope. Fourteen of the 16 blotted to a 29-kDa recombinant fragment of GPIIIa (construct 3) containing only the PSI and hybrid domains, and DDAbs 1, 2, and 8 reacted with this recombinant fragment immobilized in a microtiter plate with mAb AP5. Together, the findings indicate that DDAbs 1 to 14 bind directly to construct 3 and are not specific for an epitope created elsewhere in the integrin subunit, eg, in the beta A domain, by a structural change induced by quinine.
Despite these similarities, DDAbs 1 to 14 were found to be heterogeneous when their reactions with GPIIIa mutants and with GPIIIa blocked by mAb AP3 (specific for GPIIIa 50-62) were studied. As shown in Table 2, 7 DDAbs (1 through 7) were each blocked by AP3 and reacted identically with constructs 3 through 6, consistent with the possibility that they recognize the same epitope. However, DDAbs 8 to 14 differed from this pattern to various degrees. It is evident from Table 2 that the epitopes recognized by these DDAbs overlap with the presumptive single epitope seen by DDAbs 1 through 7 but are not identical with it. Because DDAbs 15 and 16 failed to recognize construct 3 and were not blocked by AP3, they appear to be specific for an epitope on GPIIIa spatially separate from the ones recognized by the 7 “consensus” DDAbs 1 through 7 and by DDAbs 8 through 14. The failure of DDAb 15 to recognize constructs 4 and 5 is unexplained, however.
Our studies were initiated to test the hypothesis that the region of GPIIIa defined by amino acid residues 45 to 69 might contain a single epitope for quinine-dependent antibodies which, when more fully characterized, could shed light on the mechanism by which soluble quinine at pharmacologic concentrations promotes tight binding of an otherwise nonreactive antibody to the glycoprotein. However, the data shown in Table 2 indicate that DDAbs 1 to 16 have at least 8 distinct recognition patterns, all but 2 of which (those for DDAbs 15 and 16) involve the region of GPIIIa defined by amino acids 45 to 69. It is possible that mutation of additional amino acids between residues 45 to 69 or in adjacent, noncontiguous parts of the GPIIIa hybrid domain might reveal subtle differences in the fine specificity of DDAbs 1 to 7 as well. Together, the findings suggest that quinine-dependent antibodies, and presumably DDAbs induced by other drugs, inherently recognize a large number of different structures, some of which may cluster in a restricted part of the target protein and may even overlap as appears to be the case for DDAbs 1 to 14. If this is the case, additional studies to characterize epitopes for DDAbs at the molecular level may be unlikely to identify unique structures for which multiple antibodies are specific.
Hypotheses developed to explain drug-dependent antibody binding have stressed several possibilities.1,4,7 One is that drug binds to the target glycoprotein and reconfigures it to create a new epitope elsewhere in the molecule that is recognized by the DDAb. Studies described here and previous reports4,7,34,35 argue against the possibility that this mechanism is operative with quinine-dependent antibodies. A second suggestion is that drug binds noncovalently to the glycoprotein to create a compound epitope consisting of drug and adjacent peptide residues with which the antibody reacts.1,4 A third is that drug binds first to the antibody and modifies it in such a way that it acquires specificity for a site on the glycoprotein.4,35,36 We recently introduced a model that could reconcile the second and third of these hypotheses by proposing that one structural element of the drug reacts noncovalently with antibody and another reacts with the target glycoprotein, increasing the binding energy between the antibody and its target epitope. The resulting “sandwich” leads to trapping of the drug at the antibody-antigen interface and enables an antibody that is only very weakly auto-reactive in the absence of drug to bind tightly and cause platelet destruction.35 It is apparent that whether the drug binds first to antibody or first to the target protein is relatively unimportant and could simply be a function of its relative affinity for one component or the other.
The heterogeneous behavior of the quinine-dependent antibodies described here is consistent with this model. Quinine, like many drugs, contains both charged and hydrophobic structural elements that enable it to bind to multiple sites on albumin and other plasma proteins.37,38 Although these interactions are weak (Kd on the order of 5 × 10−4 M)39 and have not been defined at the molecular level, some are undoubtedly stronger than others. The model would predict that there is one or more preferred binding site(s) for quinine in the region of GPIIIa recognized by DDAbs 1 through 14 (GPIIIa 50-66) and that DDAbs induced by the drug will have been selected for their ability to recognize quinine bound noncovalently to this preferred site. Although each antibody might recognize the same aspect of the quinine molecule, the innate heterogeneity of the immune response is such that antibodies from different patients would be expected to recognize different aspects of the protein component of the target, as was seen with DDAbs 1 through 14. Further studies to test this possibility using human antibodies are made extremely difficult by the limited quantities of antibody available and by the tendency of sensitized patients to produce multiple antibodies recognizing different aspects of the GPIIb/IIIa and GPIb/IX complexes.10,11 We recently succeeded in producing quinine-dependent murine monoclonal antibodies that mimic the behavior of antibodies found in patients with quinine-induced thrombocytopenia40 and hope that these will provide a tool with which to characterize the molecular basis of antibody-drug-protein interaction more fully.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grant HL-13629 from the National Heart, Lung and Blood Institute.
Authorship
Contribution: J.P. designed and performed research, analyzed data, and wrote the manuscript; T.N. helped with experimental design, performed research, and analyzed data; A.K. performed research; and R.A. designed research and wrote most of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Julie A. Peterson, BloodCenter of Wisconsin, Blood Research Institute, PO Box 2178, Milwaukee, WI 53201-2178; e-mail julie.peterson@bcw.edu.