Abstract
Atypical hemolytic uremic syndrome (aHUS) is a severe renal disease that is associated with defective complement regulation caused by multiple factors. We previously described the deficiency of factor H–related proteins CFHR1 and CFHR3 as predisposing factor for aHUS. Here we identify in an extended cohort of 147 aHUS patients that 16 juvenile individuals (ie, 11%) who either lacked the CFHR1/CFHR3 completely (n = 14) or showed extremely low CFHR1/CFHR3 plasma levels (n = 2) are positive for factor H (CFH) autoantibodies. The binding epitopes of all 16 analyzed autoantibodies were localized to the C-terminal recognition region of factor H, which represents a hot spot for aHUS mutations. Thus we define a novel subgroup of aHUS, termed DEAP HUS (deficiency of CFHR proteins and CFH autoantibody positive) that is characterized by a combination of genetic and acquired factors. Screening for both factors is obviously relevant for HUS patients as reduction of CFH autoantibody levels represents a therapeutic option.
Introduction
The atypical form of hemolytic uremic syndrome (aHUS) is characterized by microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure.1 aHUS is associated with defective complement regulation. The complement system represents an innate immune defense system that eliminates invading microbes. Mutations in genes coding for complement regulators factor H (CFH), membrane cofactor protein (MCP), the components factor B (CFB), C3, and factor I (CFI)2-9 cause impaired regulation of the alternative pathway convertase C3bBb. This results in defective local complement control on host cell surfaces.10,11 In addition, CFH gene conversion, deletion of the complement factor H–related genes CFHR1 and CFHR3 by nonallelic homologous recombination, and the presence of CFH autoantibodies have been reported in aHUS patients.12-16
These diverse scenarios are responsible for approximately 50% of the reported cases, indicating that additional factors contribute to aHUS. On the surface of human cells, multiple regulators control complement activation. Under physiological conditions, defective function of one mutated protein is compensated by the additional regulators, which display redundant activities. This situation might explain the incomplete penetrance of the genetic mutations. We have recently shown that CFH autoantibodies of 5 patients bind to the C-terminus of CFH and reduce CFH-C3b interaction.16
To extend the understanding of the molecular basis of aHUS, we determined the frequency of CFH autoantibodies in the Jena aHUS cohort and correlated the presence of CFH autoantibodies with CFHR1 and CFHR3 expression.
Methods
This study was approved by the Research Ethics Committee of the Friedrich Schiller University, Jena, Germany; the University of Cologne, Cologne, Germany; and the Hospital for Sick Children, Toronto, Canada. Informed consent was obtained in accordance with the Declaration of Helsinki.
Patients
The cohort analyzed here represents an extended cohort of 147 patients with atypical HUS of whom 121 patients were recently reported.14 Information regarding the patients is summarized in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Western blot analysis
Plasma samples of all patients (not shown) and of members of 3 selected families were investigated by Western blotting.14 CFHR1 was detected using monoclonal antibody C18 and CFHR3 was detected with CFHR3 antiserum.
Identification and domain mapping of CFH autoantibodies
The binding domains of the CFH autoantibodies in CFH were determined by enzyme-linked immunosorbent assay (ELISA) as described.16 Briefly, microtiter plates (Nunc, Wiesbaden, Germany) were coated with CFH fragments17 and incubated with plasma of the patients, and CFH autoantibodies were detected with HRP-conjugated anti–human IgG antibodies (Sigma-Aldrich, Taufkirchen, Germany).
Results and discussion
Frequency of CFH autoantibodies
In a cohort of 147 aHUS patients, we identify 16 children (ie, 11%) as positive for CFH autoantibodies by ELISA (Table 1). CFH autoantibodies were completely absent in a control group of 100 healthy individuals, thus indicating that CFH autoantibodies are associated with aHUS. Similar to the young age of the patients of the Jena cohort, the 8 previously identified CFH autoantibody–positive HUS patients (5-17 years)15,16 were also juvenile, suggesting related mechanisms for autoantibody induction.
Further analyses of the CFH autoantibody–positive group revealed by Western blotting that the patients either showed the complete absence of CFHR1 and CFHR3 in plasma (14 patients) or displayed low, barely detectable levels of CFHR1 and CFHR3 (Table 1 and data not shown). The strong correlation between the occurrence of CFH autoantibodies and absence or reduction of CFHR1/CFHR3 in plasma suggests that this deficiency represents a risk factor for CFH autoantibody formation. The mechanism involved in how a deficiency of these plasma proteins leads to the generation of CFH autoantibodies is currently unknown and requires further investigations. The 22 CFHR1/CFHR3-deficient patients of the Jena cohort include 16 CFH autoantibody–positive and 6 patients who have no autoantibodies to CFH. The frequency of the deficient group without CFH autoantibodies is 4% in this cohort and thus slightly higher than in the Jena and Newcastle control groups (2% each)14 or in the Iowa, Columbia, and Finnish AMD study cohorts (2.7%, 3.0%, and 2.5%, respectively).18 Concurrence of 2 risk factors in development of aHUS has been reported for combined mutations in either the CFI and the MCP genes19 or for various CFH haplotypes.20 Here we report a new combination of 2 disease-associated conditions in predominantly juvenile aHUS patients, namely the presence of CFH autoantibodies and absence of CFHR1/CFHR3 in plasma.
Family studies
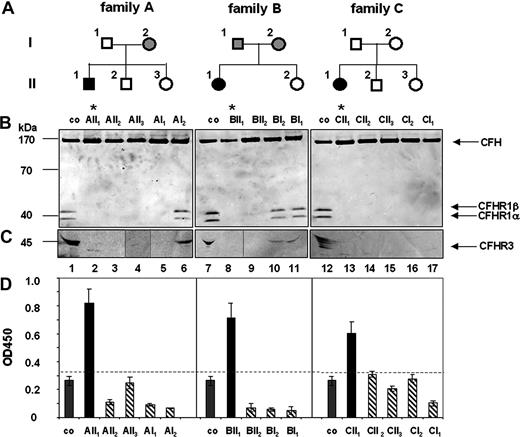
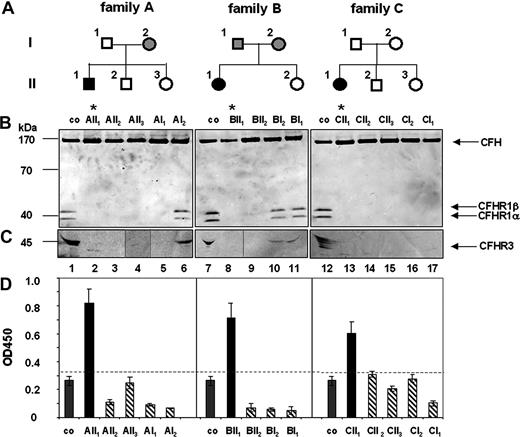
Family studies were performed to analyze how autoantibodies to CFH or CFHR1/CFHR3 deficiency influences or predisposes to the disease. Three CFH autoantibody–positive, CFHR1- and CFHR3-deficient patients and their family members were assayed for both parameters (Figure 1). In family A, the patient (AII1) (Figure 1A) was positive for CFH autoantibodies (Figure 1D) and CFHR1 and CFHR3 proteins were absent in his plasma (Figure 1B lane 2). The mother (AI2, Figure 1B lane 6) showed lower plasma levels of CFHR1 and CFHR3 proteins, indicating heterozygous deficiency. The other family members lacked CFHR1 and CFHR3 proteins, which corresponds to homozygous deficiency. Genetic analyses confirmed homozygous CFHR1 and CFHR3 deficiency for the patient (AII1), the healthy brother (AII2), the healthy sister (AII3), and the healthy father (AI1). The CFH gene was intact in all family members (data not shown). A similar scenario was observed for families B and C. In family B the patient, but no other relative, was positive for CFH autoantibodies (Figure 1D). CFHR1 and CFHR3 proteins were absent in the plasma of the patient (BII1) and the unaffected healthy sister (BII2; Figure 1B lanes 8,9), but were detected in sera of the healthy mother and the father (Figure 1B lanes 10,11). Genetic analyses confirmed that the patient and his sister were homozygous for the CFHR1/CFHR3 gene deletion. Similarly, in family C the aHUS patient was positive for CFH autoantibodies (Figure 1D) and CFHR1/CFHR3 proteins were absent. The remaining 4 healthy family members lacked CFH autoantibodies and also CFHR1/CFHR3 proteins in plasma (Figure 1B lanes 13-17). Genetic analyses confirmed a homozygous deletion of CFHR1/CFHR3 genes and nonrearranged CFH genes for all members of this family (data not shown). Thus, in each family the HUS patient was positive for CFH autoantibodies and deficient for CFHR1 and CFHR3. The chromosomal breakpoints in each case were located in the same chromosomal repeat region as recently described.14 All 11 members of the 3 families who lacked CFH autoantibodies and showed either homozygous or heterozygous CFHR1/CFHR3 deficiency were healthy. Thus, these family studies demonstrate that CFH autoantibodies develop on a background of CFHR1 and CFHR3 deficiency.
Family analysis: deficiency of CFHR1 and CFHR3 in aHUS patients and their family members. (A) A pedigree is shown for each family. Black boxes indicate patients; open symbols, family members with homozygous CFHR1 and CFHR3 deletion; and gray symbols, individuals with heterozygous CFHR1 and CFHR3 deficiency. (B) Plasma of the patients or their healthy family members were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a membrane, and analyzed by Western blotting using a mAB that identifies the conserved C-termini of CFH (150 kDa) and the 2 differently glycosylated forms CFHR1α and CFHR1β) (37 and 42 kDa). For detection of CFHR3, antiserum reacting with different glycosylated forms of CFHR3 (45 kDa, multiple bands) was used. Western blot analysis of plasma derived from individual family members demonstrated deficiency of CFHR1 in the aHUS patients (* lanes 2, 8, 13) and also in healthy relatives (lanes 3-5,9,14-17). CFHR1α and CFHR1β are detected in plasma of a healthy control (lanes 1,7,12). CFH is detected in all plasma samples. (C) Complete deficiency of CFHR3 is detected in the 3 aHUS patients (lanes 2,8,13) and several relatives (lanes 3-5,9,14-17), but CFHR3 is observed in the plasma of a healthy volunteer (lanes 1,7,12) and of heterozygous relatives (lanes 6,10,11). The band at 30 kDa in lane 2 is unspecific. (D) CFH autoantibody levels were detected by ELISA. CFH autoantibodies (black bars) are present in serum of the patients (AII1, BII1, and CII1) but not of their relatives (dashed bars) and in plasma derived from controls (co, gray bars). The dotted line represents the background level (OD450 − 0.35), that is, the highest absorbancy of plasma samples derived from 100 control individuals (Document S1).
Family analysis: deficiency of CFHR1 and CFHR3 in aHUS patients and their family members. (A) A pedigree is shown for each family. Black boxes indicate patients; open symbols, family members with homozygous CFHR1 and CFHR3 deletion; and gray symbols, individuals with heterozygous CFHR1 and CFHR3 deficiency. (B) Plasma of the patients or their healthy family members were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a membrane, and analyzed by Western blotting using a mAB that identifies the conserved C-termini of CFH (150 kDa) and the 2 differently glycosylated forms CFHR1α and CFHR1β) (37 and 42 kDa). For detection of CFHR3, antiserum reacting with different glycosylated forms of CFHR3 (45 kDa, multiple bands) was used. Western blot analysis of plasma derived from individual family members demonstrated deficiency of CFHR1 in the aHUS patients (* lanes 2, 8, 13) and also in healthy relatives (lanes 3-5,9,14-17). CFHR1α and CFHR1β are detected in plasma of a healthy control (lanes 1,7,12). CFH is detected in all plasma samples. (C) Complete deficiency of CFHR3 is detected in the 3 aHUS patients (lanes 2,8,13) and several relatives (lanes 3-5,9,14-17), but CFHR3 is observed in the plasma of a healthy volunteer (lanes 1,7,12) and of heterozygous relatives (lanes 6,10,11). The band at 30 kDa in lane 2 is unspecific. (D) CFH autoantibody levels were detected by ELISA. CFH autoantibodies (black bars) are present in serum of the patients (AII1, BII1, and CII1) but not of their relatives (dashed bars) and in plasma derived from controls (co, gray bars). The dotted line represents the background level (OD450 − 0.35), that is, the highest absorbancy of plasma samples derived from 100 control individuals (Document S1).
We have previously localized the binding epitope of 5 CFH autoantibodies, derived from aHUS patients, 2 of whom are also part of the Jena aHUS cohort, to the C-terminus of CFH. In addition, these CFH autoantibodies inhibit the regulatory function of CFH at the cell surface.16 To define if this phenomenon holds true for the newly identified CFH autoantibodies, their binding epitopes were also identified.
CFH autoantibodies from each of the 16 patients bound to the C-terminal fragments of CFH (ie, SCRs 15-20 and SCRs 19,20) but neither to SCRs 1-7, SCRs 11-15, SCRs 15-18, nor to SCRs 15-19 (Table S1). Four CFH autoantibodies also showed weak binding to a fragment representing SCRs 8-11 of CFH. This profile reveals that all 16 analyzed CFH autoantibodies bind preferentially within the C-terminal recognition region of CFH,21-25 which represents also a hot spot for aHUS-associated mutations.9 This overlap suggests similar functional consequences for the CFH autoantibodies and for the genetic mutations, namely reduced cell recognition functions of CFH.
In summary, we identify a new subgroup of aHUS patients who are deficient for CFHR1 and CFHR3 in plasma and positive for CFH autoantibodies. This deficiency may favor development of specific autoantibodies that bind to the recognition region of CFH and likely block cell binding. It remains to be shown if disease progression of this new subgroup differs from that of other HUS patients (eg, patients with CFHR1/CFHR3 deficiency and the absence of CFH autoantibodies or patients with CFH mutations).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Gerlinde Heckrodt and Ina Löschmann for expert technical assistance. We thank the patients and physicians for cooperation. Five probes in the Jena cohort were derived from the HUS registry collected by the Arbeitsgemeinschaft Pädiatrische Nephrologie (APN) in Germany.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, Zi 432) and KIDNEEDS.
Authorship
Contribution: M.J., C.L., P.F.Z., and C.S. designed the research and wrote the paper; S.S., S.L.H.Z., H.R., and S.H. performed experiments and analyzed data; C.L. provided patient specimen.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter F. Zipfel, Department of Infection Biology, Leibniz Institute for Natural Product Research and Infection Biology–Hans Knöll Institute, Beutenbergstr. 11a, D-07745 Jena, Germany; e-mail peter.zipfel@hki-jena.de.
References
Author notes
M.J. and C.L. contributed equally to this work.