Abstract
Diffuse large B-cell lymphoma (DLBCL) consists of at least 2 phenotypic subtypes; that is, the germinal center B-cell–like (GCB-DLBCL) and the activated B-cell–like (ABC-DLBCL) groups. It has been shown that GCB-DLBCL responds favorably to chemotherapy and expresses high levels of BCL6, a transcription repressor known to play a causative role in lymphomagenesis. In comparison, ABC-DLBCL has lower levels of BCL6, constitutively activated nuclear factor-κB, and tends to be refractory to chemotherapy. Here, we report that the STAT3 gene is a transcriptional target of BCL6. As a result, high-level STAT3 expression and activation are preferentially detected in ABC-DLBCL and BCL6-negative normal germinal center B cells. Most importantly, inactivating STAT3 by either AG490 or small interference RNA in ABC-DLBCL cells inhibits cell proliferation and triggers apoptosis. These phenotypes are accompanied by decreased expression of several known STAT3 target genes, including c-Myc, JunB, and Mcl-1, and increased expression of the cell- cycle inhibitor p27. In addition to identifying STAT3 as a novel BCL6 target gene, our results define a second oncogenic pathway, STAT3 activation, which operates in ABC-DLBCL, suggesting that STAT3 may be a new therapeutic target in these aggressive lymphomas.
Introduction
Diffuse large B-cell lymphoma (DLBCL) accounts for 30% to 40% of newly diagnosed non-Hodgkin lymphoma (NHL) cases in the United States, and yet it accounts for up to 80% of NHL mortality.1 Based upon their gene expression similarities to either normal germinal center (GC) B cells or in vitro–activated peripheral blood B cells, DLBCLs are subdivided into 3 groups: the GC B-cell–like DLBCL (GCB-DLBCL), activated B-cell–like DLBCL (ABC-DLBCL), and an unclassified third type.2 This classification scheme is referred to as the cell of origin (COO) method. In general, the GCB group expresses high levels of the transcription repressor BCL6 and tends to respond better to conventional chemotherapy, whereas the ABC group has lower levels of BCL6 and tends to be refractory to chemotherapeutic treatment.2-5 A second DLBCL molecular classification system has also been devised that includes 3 subgroups, termed BCR, OXPHOS, and host immune response, but the therapeutic implication of this system is not yet clear.6 BCL6 is a transcription repressor that plays important roles in GC formation and lymphoma oncogenesis.7-9 In nearly half of DLBCLs, BCL6 is constitutively expressed due to chromosomal translocations and activating mutations that bypass a negative autoregulation mechanism.10,11 More is understood about GCB-DLBCL, which has high BCL6, than BCL6-low ABC-DLBCL, particularly with regard to important pathogenic/oncogenic pathways. A major function of BCL6 in GC as well as GCB-DLBCL is to suppress terminal B-cell differentiation by inhibiting activation markers as well as PRDM1/Blimp-1, the master regulator of plasma cell program.12-15 BCL6 also contributes to oncogenesis by antagonizing the function of the ARF-p53 axis,16,17 opposing replicative cell senescence18-20 and interacting with cell signaling pathways that are important for normal immune functions and oncogenesis. As an example of the latter, BCL6 can inhibit nuclear factor-κB (NF-κB) function by downregulating NF-κB1 p105/p50.21 In bone marrow–derived macrophages, BCL6 regulates cell morphology and motility via its ability to suppress RhoA activation, and it inhibits the interleukin-6 (IL-6)/STAT3 pathway, preventing autocrine IL-6 production and aberrant proliferation.20,22 Before this study, direct transcriptional repression of STAT3 by BCL6 was not known. In a previous study, forced overexpression of BCL6 in the BCL1 cell line was shown to inhibit STAT3-dependent plasma cell differentiation, but the underlying mechanism was attributed to the ability of BCL6 to compete with STAT3 in binding to dually regulated target genes.23
Compared with the current understanding of GCB-DLBCL, the biology and pathogenic mechanisms of ABC-DLBCL are less understood. It is known that PRDM1 is inactivated by genetic alterations in nearly 24% of ABC-DLBCL cases.24,25 There is also a difference in the ability of GCB and ABC-DLBCL cells to transduce IL-4 signaling, although the relevance of this difference to either oncogenesis or therapy outcome is not yet clear.26 Constitutively activated NF-κB is a prominent feature of ABC-DLBCL; in fact, inactivating NF-κB by drugs or genetic manipulations triggers apoptosis in cultured ABC-DLBCL cells, supporting the notion that NF-κB is a driving force of the chemoresistant behavior of ABC-DLBCL.5,27 Interestingly, primary mediastinal large B-cell lymphoma (PMBL) also has activated NF-κB but responds favorably to chemotherapy,28 suggesting that in ABC-DLBCL, additional tumor-specific factor(s) exist that modify the NF-κB transcription program and render ABC-DLBCL resistant to cytotoxic drugs.
We report in this study that STAT3 is constitutively activated in the ABC group of DLBCL. It is well established that STAT3 activation begins with phosphorylation of Tyr705, which can be carried out by either Jak kinases working downstream of cytokine receptors or several other receptor and nonreceptor tyrosine kinases.29 Tyrosine-phosphorylated STAT3 then homodimerizes and translocates to the nucleus, where its optimal transcriptional activity also depends upon phosphorylation of Ser727, which can be catalyzed by a number of kinases.30 In normal cells, STAT3 activation is usually transient, whereas in many cancers, STAT3 is maintained in a constitutively activated state promoting tumorigenesis by enhancing cell proliferation, survival, and angiogenesis while suppressing the anticancer immune response.29,31,32 In lymphoid malignancies, the role of STAT3 has been best studied in multiple myelomas, where IL-6 autocrine/paracrine action is well known to provide the pivotal survival signal via STAT3 activation.33 Aberrant STAT3 activation has also been documented in Hodgkin disease34 and in CD30+ anaplastic large T-cell lymphomas.35,36 The role of IL-6/STAT3 in the pathogenesis of DLBCL has not been directly investigated.
In the present study, we demonstrate that transcription of the human STAT3 gene is negatively regulated by BCL6 so that high-level STAT3 expression and activation are preferentially associated with DLBCLs of the non-GCB phenotype. Most importantly, sustained STAT3 activity plays an important role in proliferation and survival of ABC-DLBCL cells, suggesting that the specific biologic features and chemoresistance of this DLBCL subgroup are likely to be determined by the simultaneous activation of both NF-κB and STAT3.
Methods
The detailed methods for immunohistochemistry (IHC) analysis of primary DLBDL as well as the segregation of cases into the 3 BCL6 and PY-STAT3 staining groups (negative, moderate, and strong; Figure 1D) are available as Document S1 and Tables S1 to S3 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Also included in supplementary methods are procedures for electrophorectic mobility shift assay (EMSA), chromatin immunoprecipitation (ChIP), Northern and Western blotting, statistical analysis, and sources of lymphoma cell lines.
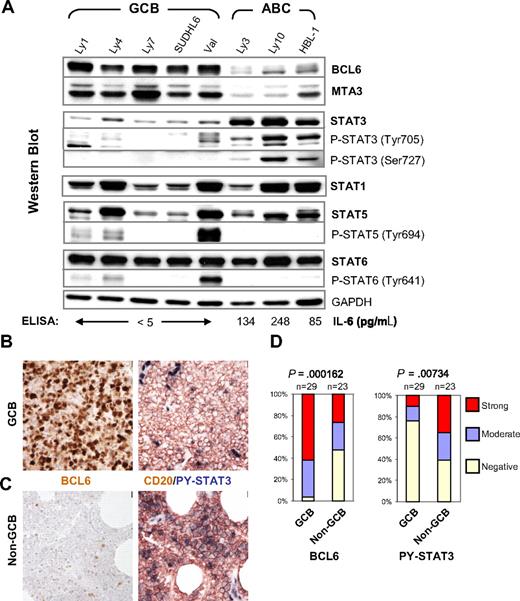
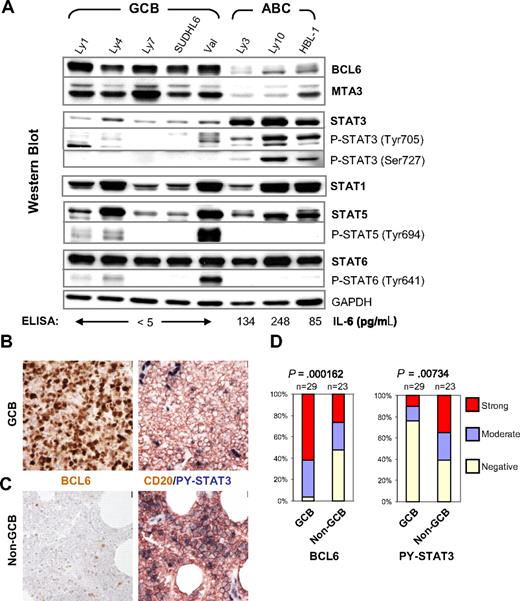
High-level STAT3 expression and activation are preferentially associated with the BCL6-low ABC-DLBCL. (A) IL-6 production, expression, and activation of STAT3 are inversely correlated with BCL6 in DLBCL cell lines. Protein expression of BCL6, MTA3, total and phosphorylated forms of STAT1, STAT3, STAT5, and STAT6 was analyzed by Western blot. GAPDH levels were used as loading control. The COO status of the cell lines are labeled on the top. The titers of secreted IL-6 as measured by ELISA are given at the bottom of the panels. (B,C) Representative IHC staining for BCL6 and CD20/PY-STAT3 in serial sections. (B) Staining of a GCB case that is positive for BCL6 but negative for PY-STAT3. (C) Staining of a non-GCB case that has very few BCL6-positive malignant cells but is strongly positive for PY-STAT3. Note strongly positive endothelial cell nuclei as endogenous positive controls for PY-STAT3 staining. Scale bar = 1 μm (40×). (D) Distribution of BCL6 and PY-STAT3 staining in GCB and non-GCB subgroups. The staining was scored as described in the Document S1. The association between BCL6 expression and DLBCL subgroups or PY-STAT3 and the subgroups was tested by chi-square analysis.
High-level STAT3 expression and activation are preferentially associated with the BCL6-low ABC-DLBCL. (A) IL-6 production, expression, and activation of STAT3 are inversely correlated with BCL6 in DLBCL cell lines. Protein expression of BCL6, MTA3, total and phosphorylated forms of STAT1, STAT3, STAT5, and STAT6 was analyzed by Western blot. GAPDH levels were used as loading control. The COO status of the cell lines are labeled on the top. The titers of secreted IL-6 as measured by ELISA are given at the bottom of the panels. (B,C) Representative IHC staining for BCL6 and CD20/PY-STAT3 in serial sections. (B) Staining of a GCB case that is positive for BCL6 but negative for PY-STAT3. (C) Staining of a non-GCB case that has very few BCL6-positive malignant cells but is strongly positive for PY-STAT3. Note strongly positive endothelial cell nuclei as endogenous positive controls for PY-STAT3 staining. Scale bar = 1 μm (40×). (D) Distribution of BCL6 and PY-STAT3 staining in GCB and non-GCB subgroups. The staining was scored as described in the Document S1. The association between BCL6 expression and DLBCL subgroups or PY-STAT3 and the subgroups was tested by chi-square analysis.
Cell culture and transient transfection
Human lymphoma cell lines Mutu III, Ly1, Ly3, Ly4, Ly7, Ly8, SUDHL6, and Val were cultured in Iscove Modified Dulbecco Medium (IMDM), supplemented with 10% fetal bovine serum (FBS; Gemini Bio-Products, Woodland, CA). Ly10 was cultured in IMDM plus 20% human plasma. HBL-1 was cultured in RPMI medium supplemented with 10% FBS. Multiple myeloma cell line H929 was maintained in RPMI supplemented with 10% FBS, 10 mM HEPES, 1.0 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanal. The 293T cells were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS. Transient transfections of Ly3 and Ly10 cells were performed with the Nucleofector Kit T and program G16 (Amaxa Biosystems, Gaithersburg, MD). Ten-microgram siRNA oligos were used in each transfection with 4 million to 10 million cells.
Antibodies, siRNA, and other reagents
MTA3 antibody was a generous gift from Dr Paul Wade (National Institutes of Health/National Institute of Environmental Health Sciences). Other antibodies used in Western blot were polyclonal anti-BCL6 (N-3), anti–Mcl-1, anti-p27, anti-JunB, monoclonal anti-p53, and anti–c-Myc from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-PARP antibody from R&D Systems (Minneapolis, MN). Antibodies for Erk1/2 and all STAT proteins and their phosphorylated forms were from Cell Signaling Technology (Danvers, MA). The 2 BCL6 siRNA oligos are part of the Stealth Select 3 RNAi Set from Invitrogen (Carlsbad, CA): 5′-TAACGATGTTATTGAGCCGGCTGGC-3′ (no. 1), and 5′-AATCTGAGTACTCAGACTGGGTCTC-3′ (no. 2). The Stealth siRNA oligo for STAT3 (5′-CACATGCCACTTTGGTGTTTCATAA-3′) and its scrambled control (5′-CACACCGTTTCGTGGCTTTATATAA-3′) were also synthesized by Invitrogen. This STAT3 siRNA sequence was identified from the STAT3 SMARTpool (Dharmacon Research, Lafayette, CO); its in vivo targeting specificity has been validated previously.37 Tyrphostin AG490 was purchased from AG Scientific (San Diego, CA) and dissolved in dimethyl sulfoxide. Phorbol 12-myristate 13-acetate (PMA) and ionomycin were obtained from Sigma-Aldrich (St Louis, MO).
Previously published Lymphochip data were downloaded from http://llmpp.nih.gov/DLBCL and imported into GeneSpring 7.1, a microarray analytic software from Agilent Technology (Santa Clara, CA). The signal values of the BCL6, STAT3, and cyclin D2 genes were log2 transformed and compared using a Wilcoxon-Mann-Whitney nonparametric test between ABC-DLBCL and GCB-DLBCL groups. P values less than .05 were considered significant.
Tissue specimens, IHC, and immunofluorescence
Paraffin-embedded tissues were obtained from the files of the Department of Pathology of Columbia University Medical Center after necessary informed consent and/or exemption were obtained. The cases were fully anonymized and coded as per Columbia University institutional review board regulations and federal provisions. All DLBCL diagnoses were made according to the World Health Organization classification system.38 IHC and immunofluorescence (IF) staining with previously validated antibodies (Table S1) were performed as previously published.39 Grayscale and color images were taken with an E600-Nikon Microscope (Nikon, Melville, NY) fitted with Planachromat 4×/0.10/30.0, 20×/0.40/1.3, PlanApo 40×/0.095/0.12-0.16, PlanFluor 40×/0.75/0.72 objectives, a SPOT-2 CCD camera and software (Diagnostic Instruments, Sterling Heights, MI), and edited for optimal color contrast with Adobe Photoshop 7 and Adobe Illustrator 10 (San Jose, CA). Further detailed information, including construction of tissue microarray, antibody dilutions and scoring of IHC staining, is described in Document S1 and Tables S2,S3.
Reporter constructs and reporter assays
A 3-kb genomic DNA fragment corresponding to the 5′ regulatory region of the human STAT3 gene was amplified by polymerase chain reaction (PCR) with Pfx DNA polymerase (Invitrogen) using human placental DNA. Sequences of the 2 PCR primers are: 5′-AGCGAGCTCGGGTCTTGCTCTATCGCCTA-3′ (forward, with a SacI site), 5′-CCGCTCGAGGCTGAATTACAGCCCCTTCA-3′ (reverse, with an XhoI site). The PCR product was digested with SacI and XhoI and cloned into similarly digested pGL3-basic vector (Promega, Madison, WI). Site-directed mutagenesis was performed with the QuikChange II kit (Stratagene, La Jolla, CA) to convert the first 2 consensus Ts to Gs in the core sequence of site B, D, and E. All mutated reporter constructs were sequence verified. For reporter assays, Mutu III cells were transiently transfected with Superfect (Qiagen, Valencia, CA) according to the manufacturer's instructions. Cells in each well of a 12-well plate were transfected with 1.2 μg of either the wild-type or mutant STAT3 reporters, 0.5 μg of a CMV-β-gal plasmid, plus various amounts of pMT2T-BCL6, as indicated in Figure 4C. All transfections were performed in duplicate and harvested after 48 hours. Luciferase activities were measured with the Luciferase Assay System (Promega) and normalized by control readings from the β-gal assays.
Cell proliferation and apoptosis assays
Cell proliferation was measured in triplicate by the MTT[3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay as described previously40 using 2 × 104 cells per 96 wells. For 3H-thymidine incorporation assays, 22 hours after STAT3 siRNA transfection, 2 × 104 Ly3 cells or 3 × 104 Ly10 cells per 96 wells were seeded in triplicate and cultured in normal growth medium. At the indicated time points, cells in each well were pulsed with 1.0 μCi 3H-thymidine for 5 hours before being harvested onto glass fiber filters. The incorporated radioactivity was measured by scintillation counting. To profile the cell-cycle status of AG490-treated cells, Ly3 or Ly10 cells were treated with the indicated doses of AG490 and harvested at different time points, stained with PI, and analyzed by flow cytometry. Ongoing apoptosis was measured 19 hours after AG490 treatment with annexin V and 7-aminoactinomycin D (7-AAD) staining, followed by flow cytometry.
Results
High-level STAT3 expression and activation are detected in ABC-DLBCL but not GCB-DLBCL cell lines
We previously reported that BCL6 negatively regulates IL-6 transcription and STAT3 activation in bone marrow–derived macrophages.20 To determine whether these inhibitory activities of BCL6 can be detected in mature B cells and B-cell lymphomas, we analyzed IL-6 secretion and STAT3 activation status in 8 DLBCL cell lines whose COO status is known. As expected, all 5 GCB-DLBCL cell lines expressed moderate to high levels of BCL6, whereas the 3 ABC-DLBCL lines had very little BCL6 protein (Figure 1A). A similar expression pattern was seen for the GC-specific BCL6 corepressor, MTA3, as reported previously.41 We detected low but measurable amounts of IL-6 in the culture supernatants of all 3 ABC-DLBCL lines but none in the GCB-DLBCL cell lines (Figure 1A bottom). The 3 ABC cell lines also contain high levels of total STAT3 protein and its activated forms (ie, phospho-Tyr705 STAT3 [PY-STAT3] and phospho-Ser727 STAT3). In stark contrast, none of the 5 GCB cell lines had significant amounts of either total or activated STAT3. Interestingly, only STAT3, but not STAT1, STAT5, or STAT6, demonstrates a COO subtype-specific expression/activation pattern. STAT6 is highly expressed in all cell lines tested, whereas STAT1 and STAT5 have a variable expression pattern that does not correlate with COO groups. Activation of STAT5 and STAT6 was detected in 3 of the 5 GCB cell lines, whereas STAT1 was activated at a very low level in only one ABC cell line, HBL-1 (not shown). In summary, ABC-DLCBL but not GCB-DLBCL cell lines contain high levels of STAT3, which is constitutively activated.
STAT3 activation is preferentially associated with primary DLBCLs of the non-GCB phenotype
To extend our study to primary lymphomas, we first determined the COO status of 52 DLBCL cases using a previously published scheme that distinguishes 2 phenotypic subgroups; that is, GCB (Figure 1B) and non-GCB (Figure 1C).42 Based on this method, 29 of the 52 DLBCL cases belonged to the GCB group, whereas the rest had a non-GCB designation. The PY-STAT3 status of these lymphomas was determined by IHC using biopsy samples contained on a tissue microarray (TMA). As expected, we observed a strong association between BCL6 staining and the GCB group (P < .001). Specifically, the majority of BCL6-positive cases (28 of 40; 70%) had a GCB phenotype, and virtually all GCB cases (28 of 29; 97%) stained positive for BCL6 (Figure 1D left panel). Conversely, PY-STAT3 was preferentially associated with the non-GCB classification (P = .007). Among the total of 21 cases that scored positive for PY-STAT3, 70% (14 of 21) had a non-GCB phenotype. Between the 2 COO groups, only 24% (7 of 29) of the GCB cases were positive for PY-STAT3, compared with 61% (14 of 23) in the non-GCB group (Figure 1D). The observation that the non-GCB group contains a sizable fraction of BCL6-positive as well as PY-STAT3–negative cases is in line with the fact that this IHC-defined group also includes type 3 in addition to ABC-DLBCL cases.42
There is an inverse correlation between BCL6 and STAT3 mRNA
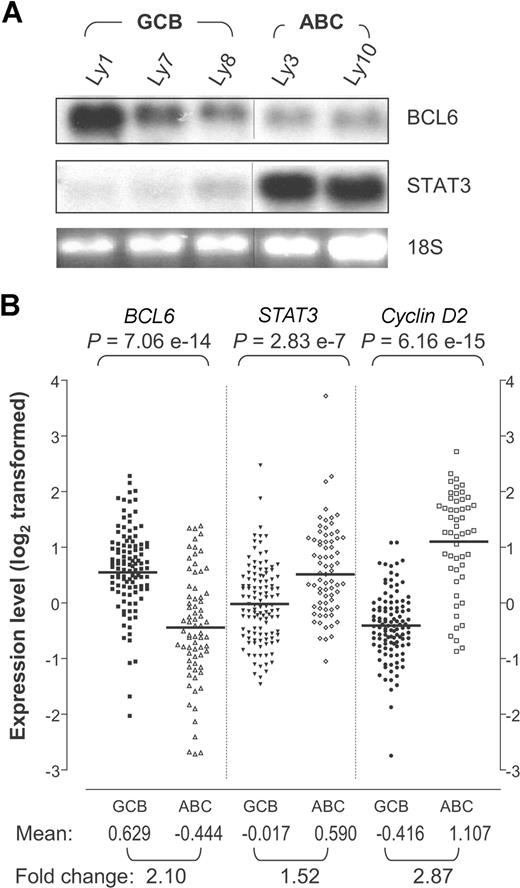
The apparent inverse correlation between BCL6 and total STAT3 proteins in lymphoma cell lines prompted us to examine expression patterns of their mRNAs. Based on Northern blot analysis, abundant STAT3 transcripts were detected only in the ABC-DLBCL cell lines Ly3 and Ly10 but not in the 3 GCB-DLBCL lines tested, whereas a reverse trend was seen for BCL6 (Figure 2A). We also extracted gene-expression patterns of BCL6, STAT3, and Cyclin D2 from the Lymphochip database, which contains 114 primary GCB-DLBCLs and 72 ABC-DLBCLs.3 As shown in Figure 2B, the mRNA levels of all 3 genes vary considerably within each COO group. Nevertheless, based on the group means, BCL6 is expressed significantly higher in GCB-DLBCL compared with ABC-DLBCL; the opposite pattern is seen for STAT3 and cyclin D2, a well-documented BCL6 target gene and a marker of ABC-DLBCL. The gene-expression difference between the 2 COO groups is highly statistically significant for all 3 genes. In terms of the absolute expression values, STAT3 has a smaller intergroup difference than BCL6 and cyclin D2. Possibly, this is because the Lymphochip analysis used whole-biopsy samples, which contain endothelial cells as well as tumor-infiltrating T cells, dendritic cells, and macrophages, all of which can express STAT3, and the relative abundance of these cells in a given biopsy is independent of the tumor's COO subgroup status (Figure 1B and Monti et al6 ).
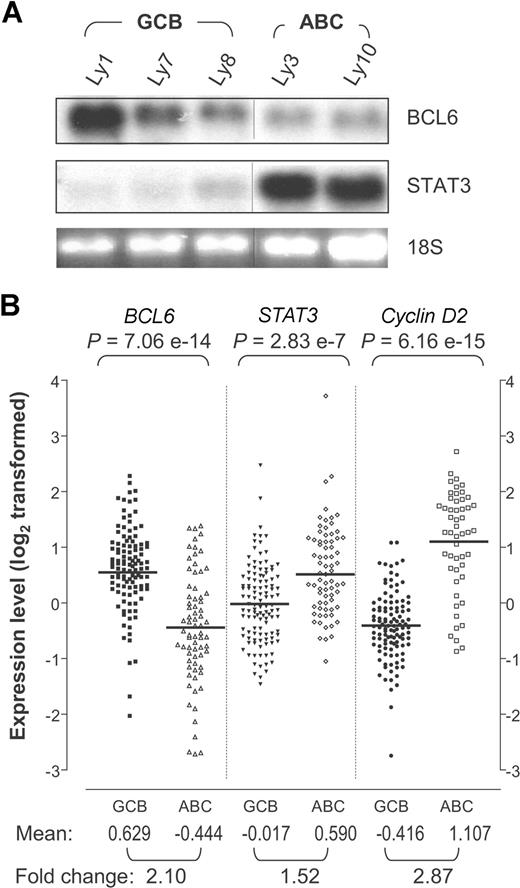
STAT3 mRNA is highly expressed in ABC-DLBCL compared with GCB-DLBCL. (A) Northern blot analysis of BCL6 and STAT3 in DLBCL cell lines. The 28S indicates 28S ribosomal RNA used as loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (B) BCL6, STAT3, and cyclin D2 expression patterns based on previously published DLBCL gene expression data.3 The signal values for BCL6 (probe 24429), STAT3 (probe 31469), and cyclin D2 (probe 16858) were retrieved for 72 ABC-DLBCL and 114 GCB-DLBCL samples. The data had been previously log2 transformed and median centered for each gene across the entire sample set. The P values are based on Wilcoxon-Mann-Whitney nonparametric test performed to compare the values between the 2 subgroups. Bars indicate the group means for each column. Mean, group mean of the log2-transformed values; Fold change, linear fold difference between the group means.
STAT3 mRNA is highly expressed in ABC-DLBCL compared with GCB-DLBCL. (A) Northern blot analysis of BCL6 and STAT3 in DLBCL cell lines. The 28S indicates 28S ribosomal RNA used as loading control. Vertical lines have been inserted to indicate a repositioned gel lane. (B) BCL6, STAT3, and cyclin D2 expression patterns based on previously published DLBCL gene expression data.3 The signal values for BCL6 (probe 24429), STAT3 (probe 31469), and cyclin D2 (probe 16858) were retrieved for 72 ABC-DLBCL and 114 GCB-DLBCL samples. The data had been previously log2 transformed and median centered for each gene across the entire sample set. The P values are based on Wilcoxon-Mann-Whitney nonparametric test performed to compare the values between the 2 subgroups. Bars indicate the group means for each column. Mean, group mean of the log2-transformed values; Fold change, linear fold difference between the group means.
BCL6 and STAT3 have mutually exclusive expression patterns in normal GC
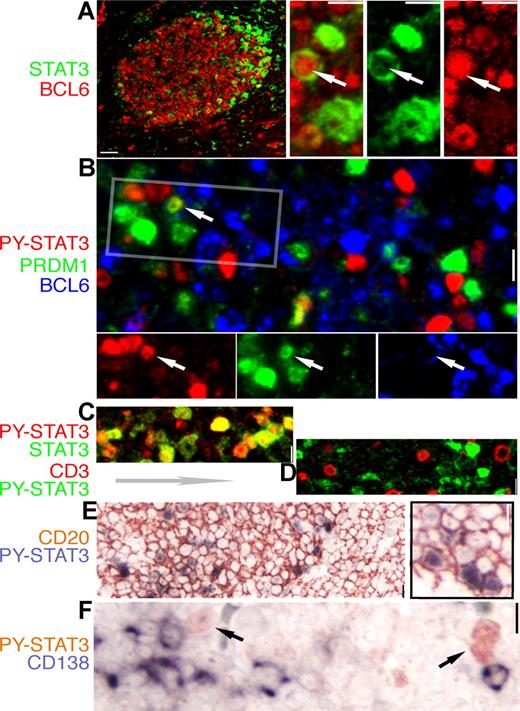
Using IHC and IF staining techniques, we examined the relationship between BCL6 and STAT3 in human tonsillar GC. As shown in Figure 3A,D, STAT3 was not expressed by mantle zone B cells outside of the GC or by T cells within the GC. Consistent with our results from cell-line studies, the great majority of BCL6+ GC B cells were negative for total STAT3 (Figure 3A), and all STAT3+ cells also contained PY-STAT3 (Figure 3C). In the dark zone, a small number of cells that stained weakly positive for total STAT3 were likely to be follicular dendritic/stromal cells based on morphology and negative staining for both CD3 and CD79a (Igα, data not shown). Interestingly, in the light zone, the brightly stained STAT3+ cells were arranged in a crescent pattern and located in the apical light zone right below the GC outer zone (Figure 3A). All these STAT3+ cells were CD20+ and, thus, B cells (Figure 3E). Notably, although cells double positive for BCL6 and PRDM-1/Blimp-1 were never found in the GC (Figure 3B and Cattoretti et al39 ), we detected rare cells with nuclear BCL6 and cytoplasmic STAT3 as well as cells that expressed both PRDM1/Blimp-1 and STAT3 at very low levels (insets of Figure 3A,B). In addition, STAT3+ B cells were uniformly negative for CD138/Syndecan (Figure 3F), a classic marker for mature plasma cells. In summary, results in Figure 3 raise the intriguing possibility that STAT3+ B cells in the apical light zone represent an early stage of plasma cell differentiation (see “Discussion”).
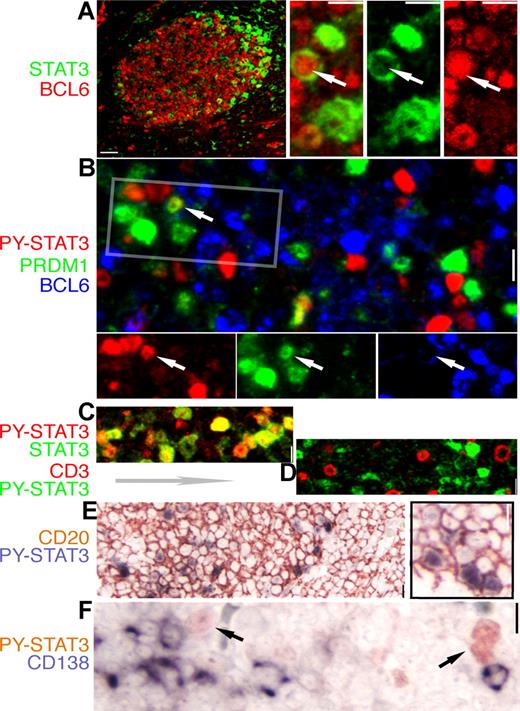
Expression of STAT3 and BCL6 are mutually exclusive in normal GC cells. IHC and IF staining of normal tonsil GC for STAT3 and lineage- and differentiation-associated markers. The color for each marker is depicted by the label beside each panel. (A) A low-power image of BCL6 (red) and STAT3 (green) staining; note the STAT3+ cell localizing to the apical light zone. The high-power insets on the right are from an enlarged apical light zone area showing a rare cell (arrow) with nuclear BCL6 and cytoplasmic STAT3, whereas the other STAT3+ cells have no BCL6. (B) Triple staining for BCL6, PRDM1/Blimp-1, and PY-STAT3 shows mutually exclusive nuclear PY-STAT3 and BCL6, as well as PY-STAT3 and PRDM1. Rare double STAT3+PRDM1+ cells are noted (arrow). The rectangle is selected, and the image is split into a single-color panel below. (C) Double staining for STAT3 and PY-STAT3 shows that these 2 signals are always colocalized (shades of yellow color). (D) Double staining for PY-STAT3 and CD3, a pan-T marker, shows that there is no overlap between these 2 markers. (E) Double staining shows that all STAT3+cells are positive for CD20, a pan-B marker. (F) Double staining for PY-STAT3 and CD138/Syndecan, a late-stage plasma cell marker, shows that these 2 markers are mutually exclusive. Scale bar, 4 μm in the low-power image of panel A, 1 μm in other panels.
Expression of STAT3 and BCL6 are mutually exclusive in normal GC cells. IHC and IF staining of normal tonsil GC for STAT3 and lineage- and differentiation-associated markers. The color for each marker is depicted by the label beside each panel. (A) A low-power image of BCL6 (red) and STAT3 (green) staining; note the STAT3+ cell localizing to the apical light zone. The high-power insets on the right are from an enlarged apical light zone area showing a rare cell (arrow) with nuclear BCL6 and cytoplasmic STAT3, whereas the other STAT3+ cells have no BCL6. (B) Triple staining for BCL6, PRDM1/Blimp-1, and PY-STAT3 shows mutually exclusive nuclear PY-STAT3 and BCL6, as well as PY-STAT3 and PRDM1. Rare double STAT3+PRDM1+ cells are noted (arrow). The rectangle is selected, and the image is split into a single-color panel below. (C) Double staining for STAT3 and PY-STAT3 shows that these 2 signals are always colocalized (shades of yellow color). (D) Double staining for PY-STAT3 and CD3, a pan-T marker, shows that there is no overlap between these 2 markers. (E) Double staining shows that all STAT3+cells are positive for CD20, a pan-B marker. (F) Double staining for PY-STAT3 and CD138/Syndecan, a late-stage plasma cell marker, shows that these 2 markers are mutually exclusive. Scale bar, 4 μm in the low-power image of panel A, 1 μm in other panels.
STAT3 is a direct transcriptional target of BCL6
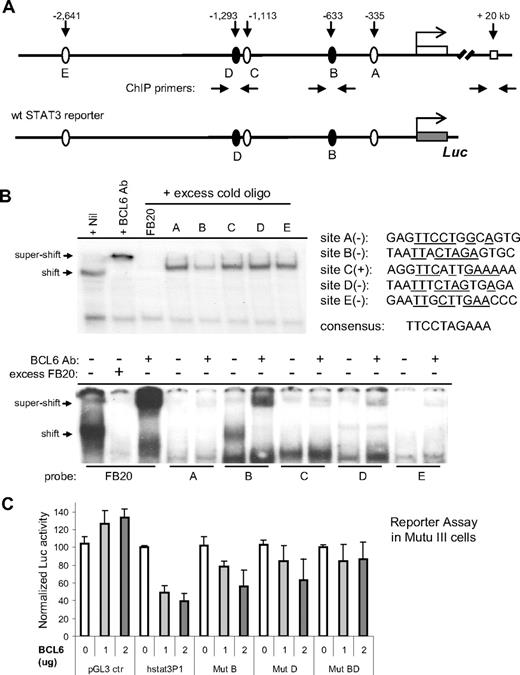
The mutually exclusive expression patterns of STAT3 and BCL6 in both DLBCL and normal GC prompted us to investigate the hypothesis that STAT3 is repressed directly by BCL6. In the 3 kb region upstream of the human STAT3 exon 1, the MatInspector program43 identified 4 candidate BCL6 sites (B-E; Figure 4A). Site A was also included in our analysis, because it mediates STAT3 autoregulation44 and matches the canonical BCL6 binding site at 7 of 10 positions (Figure 4B). In gel-shift assays, oligo probes corresponding to site B but not others could effectively compete with the high-affinity BCL6 probe for binding to the BCL6 protein (Figure 4B top). Furthermore, when all 5 oligo probes were individually labeled and tested in gel-shift assays, only site B formed a specific DNA binding complex with BCL6 (Figure 4B bottom). This indicates that, in vitro, site B has the highest affinity for BCL6 among the 5 candidate motifs.
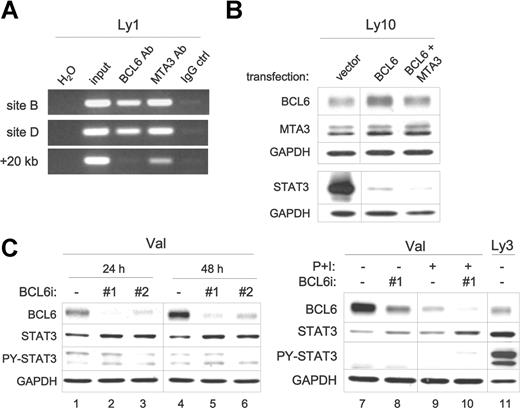
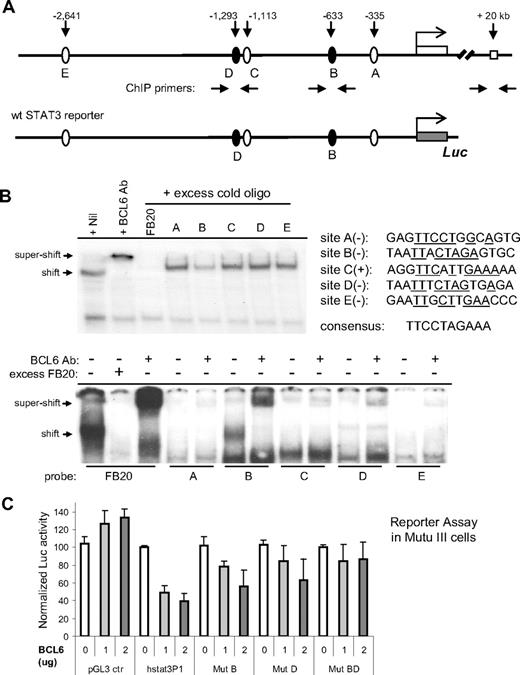
BCL6 inhibits STAT3 transcription via 2 upstream binding sites. (A) Schematic representation of the human STAT3 promoter region showing 5 potential BCL6 binding sites, location of ChIP primers, and the 3-kb fragment contained in the wt STAT3 luciferase reporter construct. (B) EMSA analysis of BCL6 binding to the 5 candidate BCL6 sites. The sequences for these 5 sites are shown together with the 10-bp core of the consensus BCL6-binding site FB20. (+) and (−) indicate the top or bottom strand of the STAT3 promoter region. Underlines indicate sequences identical to those in the consensus. In the top panel, the 20-bp FB20 probe was 32P labeled and used in the assay with nuclear extracts of Ly1 cells. In the bottom panel, all 5 candidate BCL6 sites and FB20 were labeled and subjected to EMSA analysis with or without the use of excess, cold FB20 probe in competition. (C) BCL6 represses transcription from the 3-kb wt STAT3 reporter in BCL6-negative Mutu III cells. Based on site-directed mutagenesis, repression by BCL6 was attributed to both sites B and D. The activity of each reporter in the absence of BCL6 was normalized to 100. Error bars represent SD.
BCL6 inhibits STAT3 transcription via 2 upstream binding sites. (A) Schematic representation of the human STAT3 promoter region showing 5 potential BCL6 binding sites, location of ChIP primers, and the 3-kb fragment contained in the wt STAT3 luciferase reporter construct. (B) EMSA analysis of BCL6 binding to the 5 candidate BCL6 sites. The sequences for these 5 sites are shown together with the 10-bp core of the consensus BCL6-binding site FB20. (+) and (−) indicate the top or bottom strand of the STAT3 promoter region. Underlines indicate sequences identical to those in the consensus. In the top panel, the 20-bp FB20 probe was 32P labeled and used in the assay with nuclear extracts of Ly1 cells. In the bottom panel, all 5 candidate BCL6 sites and FB20 were labeled and subjected to EMSA analysis with or without the use of excess, cold FB20 probe in competition. (C) BCL6 represses transcription from the 3-kb wt STAT3 reporter in BCL6-negative Mutu III cells. Based on site-directed mutagenesis, repression by BCL6 was attributed to both sites B and D. The activity of each reporter in the absence of BCL6 was normalized to 100. Error bars represent SD.
Mindful of a report that a low-affinity BCL6 binding site can synergize with a nearby high-affinity site to facilitate BCL6 repression in vivo,45 we examined the functionality of other BCL6-like motifs in reporter assays. We cloned the 3 kb STAT3 promoter region into a luciferase reporter and generated mutant versions thereof by inactivating sites B, D, and E (Figure 4C). In the BCL6-negative Burkitt lymphoma cell line Mutu III, transcription from the wild-type STAT3 reporter but not the control reporter was inhibited up to 2.5- to 3-fold in a dose-dependent manner by a cotransfected BCL6 expression plasmid (Figure 4C). In 293T cells, the maximum repression by BCL6 was typically 3- to 5-fold (not shown). More importantly, although mutating site B or D individually only led to partial relief from BCL6 repression, mutating both sites led to complete resistance to BCL6 (Figure 4C). Abolishing site E alone had no effect (data not shown). These results indicate that, in vivo, both site B (at −633 bp) and site D (at −1293 bp) contribute to effective repression of STAT3 by BCL6.
Next, we investigated whether BCL6 inhibits expression of the endogenous STAT3 gene. First, we performed ChIP assays to determine whether BCL6 and its GC-specific corepressor MTA3 are associated with the STAT3 promoter region in the GCB-DLBCL cell line Ly1. As shown in Figure 5A, a BCL6 antibody specifically enriched DNA fragments containing either site B or site D but not a randomly chosen downstream site (+20 kb); a strong and specific MTA3 signal was also detected in these 2 regions compared with a weak signal recovered from the +20 kb control site. In the BCL6-negative Mutu III cells, neither the BCL6 antibody nor the MTA3 antibody recovered any specific ChIP signal over IgG background in the STAT3 promoter region (data not shown). This result implies that BCL6 may suppress STAT3 expression with the help of the MTA3/NuRD complex as a corepressor. Next, we evaluated the response of the endogenous STAT3 gene to changes in BCL6 levels. In the ABC-DLBCL cell line Ly10, transient overexpression of BCL6 alone or coexpression of BCL6 and MTA3 dramatically downregulated the endogenous STAT3 protein, which is most obvious 16 to 24 hours after transfection (Figure 5B). The effective downregulation triggered by BCL6 alone may be explained by the fact that Ly10 cells contain substantial amounts of endogenous MTA3 (Figure 1A). We have also knocked down endogenous BCL6 in a GCB-type cell line, Val, by transient small interference RNA (siRNA) transfection. As shown in Figure 5C, both siRNA oligos substantially reduced the levels of BCL6, and this reduction was accompanied by a notable increase in total amount of STAT3. Subjecting Val cells treated with BCL6 siRNA to PMA plus ionomycin (P + I), which activates PKC, enhanced the reduction in BCL6 as well as the increase in total STAT3 (Figure 5C right). That the level of PY-STAT3 remained unchanged in these BCL6 siRNA-treated cells implies that the upstream kinase capable of activating STAT3 is only present in the ABC type of cellular environment. Without PY-STAT3, the positive feedback mechanism of STAT3 transcription44 will not take place. This provides a possible explanation for why the STAT3 increase following BCL6 siRNA did not approach the level in ABC-DLBCL cells. Taken together, the data presented up to Figure 5 provide strong evidence that the human STAT3 gene is a direct transcriptional target of BCL6 in normal GC and GC-derived DLBCL cells.
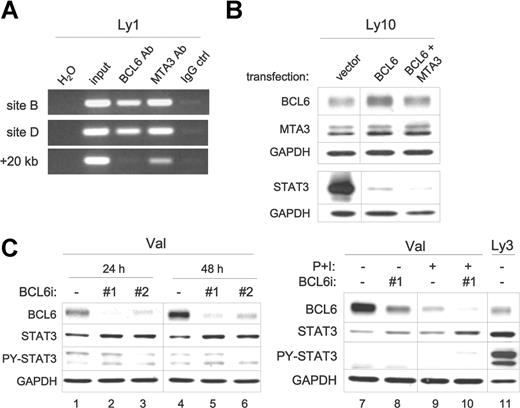
BCL6 inhibits expression of the endogenous STAT3 gene in DLBCL. (A) BCL6 and MTA3 bind to the STAT3 promoter region in vivo. ChIP assays were performed in Ly1 cells. PCR products were amplified from chromatin fragments immunoprecipitated with either anti-BCL6, anti-MTA3, or control rabbit IgG antibodies. The PCR primers flank either site B or the negative control +20-kb site (see arrows in Figure 4A). H2O, negative control PCR without any template; input, purified total genomic DNA before precipitation. (B) Forced expression of BCL6 suppresses endogenous STAT3. Ly10 cells were transiently transfected with the indicated expression plasmids. Western blot was performed to analyze expression of BCL6 and MTA3 12 hours after transfection, and total STAT3 24 hours after transfection. Similar results were obtained from Ly3 cells (not shown). (C) RNAi-mediated BCL6 knockdown increased STAT3 protein expression. Control (ctrl) and 2 different BCL6 siRNA oligos (nos. 1 and 2) were transiently transfected into Val cells, and cell lysates were prepared at either 24 or 48 hours for protein analysis. In lanes 7 to 10, cells that have been transfected with oligos for 45 hours were either left alone (lanes 7 and 8) or exposed to PMA (20 ng/mL) and ionomycin (0.3 μM) (P + I, lanes 9 and 10) for another 24 hours before harvesting. In lane 11, Ly3 lysates were loaded for comparison. BCL6, total STAT3, and PY-STAT3 proteins were examined by Western blot analysis. GAPDH levels were shown as loading control. Vertical lines have been inserted to indicate a repositioned gel lane.
BCL6 inhibits expression of the endogenous STAT3 gene in DLBCL. (A) BCL6 and MTA3 bind to the STAT3 promoter region in vivo. ChIP assays were performed in Ly1 cells. PCR products were amplified from chromatin fragments immunoprecipitated with either anti-BCL6, anti-MTA3, or control rabbit IgG antibodies. The PCR primers flank either site B or the negative control +20-kb site (see arrows in Figure 4A). H2O, negative control PCR without any template; input, purified total genomic DNA before precipitation. (B) Forced expression of BCL6 suppresses endogenous STAT3. Ly10 cells were transiently transfected with the indicated expression plasmids. Western blot was performed to analyze expression of BCL6 and MTA3 12 hours after transfection, and total STAT3 24 hours after transfection. Similar results were obtained from Ly3 cells (not shown). (C) RNAi-mediated BCL6 knockdown increased STAT3 protein expression. Control (ctrl) and 2 different BCL6 siRNA oligos (nos. 1 and 2) were transiently transfected into Val cells, and cell lysates were prepared at either 24 or 48 hours for protein analysis. In lanes 7 to 10, cells that have been transfected with oligos for 45 hours were either left alone (lanes 7 and 8) or exposed to PMA (20 ng/mL) and ionomycin (0.3 μM) (P + I, lanes 9 and 10) for another 24 hours before harvesting. In lane 11, Ly3 lysates were loaded for comparison. BCL6, total STAT3, and PY-STAT3 proteins were examined by Western blot analysis. GAPDH levels were shown as loading control. Vertical lines have been inserted to indicate a repositioned gel lane.
Inhibiting STAT3 activity by AG490 triggers cell-cycle arrest and apoptosis in ABC-DLBCL but not GCB-DLBCL cells
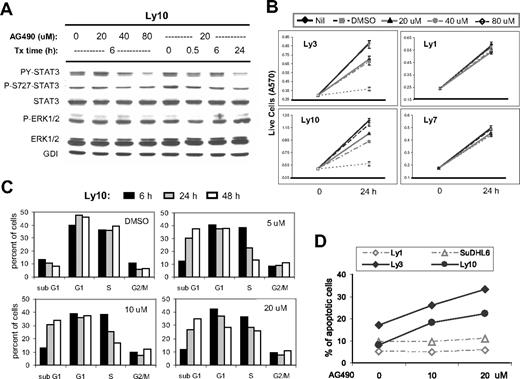
To investigate the cellular function of activated STAT3 in ABC-DLBCL, we used a well-characterized Jak inhibitor, AG490, because the Jak kinases are responsible for STAT3 activation in many cell types. As shown in Figure 6A, in Ly10 cells, AG490 reduced the PY-STAT3 level in a dose- as well as time-dependent manner, while the level of total STAT3 protein remained unchanged. A mild reduction in phospho-Ser727 STAT3 was also detected (especially at later time points when cells were treated with 20 μM AG490). As expected, AG490 specifically inhibited the Jak/STAT3 branch of the signaling cascade without affecting Erk1/2. Similar results were obtained from Ly3 cells (data not shown). Based on MTT assays, proliferation of the 2 ABC-type cell lines, Ly3 and Ly10, was inhibited by 20 to 80 μM of AG490, and yet similar treatment had negligible effect on the 2 GCB-type cell lines, Ly1 and Ly7 (Figure 6B). Thus, within the 20- to 80-μM range, AG490 appears to have a very specific effect on the STAT3-active ABC-DLBCL cells. These data also indicate that in DLBCL cells, constitutive STAT3 activation requires at least one upstream Jak kinase, as has been described in other cell types.
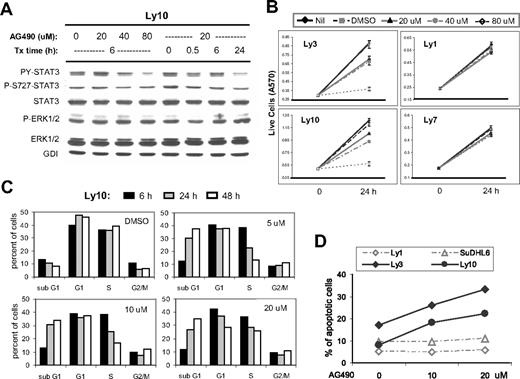
AG490 treatment reduced STAT3 activation, triggered cell-cycle arrest and apoptosis in ABC-DLBCL cells. (A) Western blot analysis showed that AG490 treatment inhibited STAT3 activation but not Erk1/2 in Ly10 cells. Cells were treated with the indicated drug concentrations and sampled at the indicated time points. Similar results were obtained from Ly3 cells (not shown). (B) MTT assays indicated that AG490 selectively reduced cell growth of ABC-type DLBCL cells (Ly3 and Ly10) without affecting GCB-DLBCL cells (Ly1 and Ly7). (C) Cell-cycle profiles of AG490-treated Ly10 cells were analyzed by PI staining followed by flow cytometry. Cells were treated with the indicated drug concentrations and sampled at the indicated time points. Result shown is representative of 2 independent experiments. (D) Annexin V and 7-AAD staining was used to monitor ongoing apoptosis in Ly1, SUDHL6, Ly3, and Ly10 cells treated for 24 hours with 10 or 20 μM AG490. The percentage of annexin V+7-AAD− cells is plotted in the graph.
AG490 treatment reduced STAT3 activation, triggered cell-cycle arrest and apoptosis in ABC-DLBCL cells. (A) Western blot analysis showed that AG490 treatment inhibited STAT3 activation but not Erk1/2 in Ly10 cells. Cells were treated with the indicated drug concentrations and sampled at the indicated time points. Similar results were obtained from Ly3 cells (not shown). (B) MTT assays indicated that AG490 selectively reduced cell growth of ABC-type DLBCL cells (Ly3 and Ly10) without affecting GCB-DLBCL cells (Ly1 and Ly7). (C) Cell-cycle profiles of AG490-treated Ly10 cells were analyzed by PI staining followed by flow cytometry. Cells were treated with the indicated drug concentrations and sampled at the indicated time points. Result shown is representative of 2 independent experiments. (D) Annexin V and 7-AAD staining was used to monitor ongoing apoptosis in Ly1, SUDHL6, Ly3, and Ly10 cells treated for 24 hours with 10 or 20 μM AG490. The percentage of annexin V+7-AAD− cells is plotted in the graph.
To further characterize the growth-inhibitory effect of STAT3 inactivation, we performed experiments to study cell-cycle behavior and apoptosis. Ly10 cells treated with different concentrations of AG490 were sampled after 6, 24, and 48 hours and stained with propidium iodide (PI) followed by flow cytometric analysis. All drug concentrations revealed a time-dependent decrease in the S-phase fraction. This is likely due to reduced S-phase entry, because 3H-thymidine incorporation was also reduced in these cells (data not shown). AG490 treatment also caused a steady increase in the sub-G1 (apoptosis/necrosis) fraction (Figure 6C), which was primarily derived from non–S-phase cells, as revealed by BrdU labeling experiments (Figure S1). Meanwhile, the total number of G1-phase cells remained unchanged in the presence of 5 and 10 μm AG490. These results are most compatible with the interpretation that AG490 caused a partial cell-cycle arrest at the G1/S boundary, which increased the G1 fraction, whereas a concomitant apoptotic response removed G1 cells from the cell cycle. However, at 20 μM, the total G1 fraction was reduced, suggesting that apoptosis became the dominant effect. Annexin V and 7-AAD double staining confirmed that AG490 triggered dose-dependent apoptosis in both Ly3 and Ly10 cells but not in the 2 GCB-type cell lines Ly1 and SUDHL6 (Figure 6D). The apoptotic nature of the cell death was further demonstrated by a PARP cleavage assay, which indicated that caspases 3 and/or 7 were activated during AG490 treatment in a time- as well as dose-dependent fashion (data not shown).
RNAi-mediated STAT3 knockdown inhibits cell proliferation and increases apoptosis in ABC-DLBCL cells
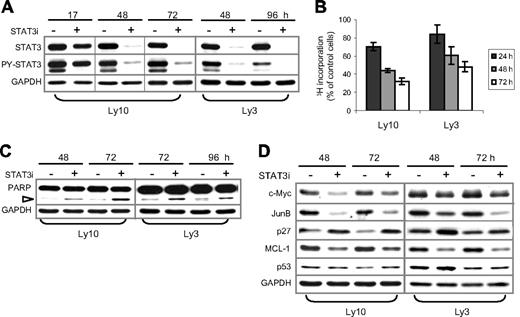
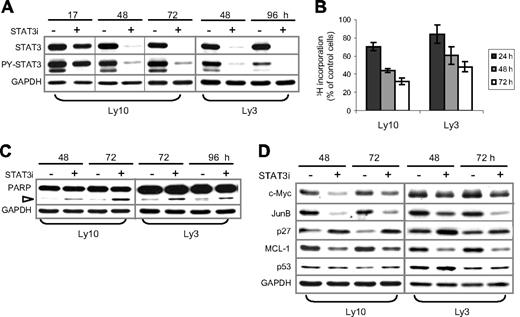
Two lines of evidence support STAT3 as the main mediator of AG490 action in ABC-DLBCL cells. First, it is unlikely that non-Jak tyrosine kinases are involved in our experiments, because Lck, Lyn, Btk, Syk, and Src are not inhibited by up to 50 μM AG490.46 Second, among the STAT proteins, STATs 1, 5, and 6 are not activated in Ly3 or Ly10 cells (Figure 1A); in addition, STAT4 is barely expressed in these cell lines, according to the Lymphochip database.3 Nevertheless, to conclusively establish the role of STAT3 in ABC-DLBCL, we carried out STAT3 knockdown experiments using siRNA. As shown in Figure 7A, transient transfection of Ly3 and Ly10 cells with STAT3 siRNA oligos resulted in a near-complete knockdown of endogenous STAT3 beginning on day 2 and lasting for 3 days. PY-STAT3 was also reduced proportionally. Compared with control siRNA oligos, STAT3 siRNA substantially diminished 3H-thymidine incorporation of both Ly3 and Ly10 (up to 52% and 68%, respectively, at 72 hours). Pronounced cell-cycle inhibition was also triggered by STAT3 siRNA in HBL-1, another ABC-DLBCL cell line (data not shown). An increase in apoptosis was also observed, as evidenced by elevated PARP cleavage (Figure 7C) and an increased number of annexin V+/7AAD− cells (not shown). We also investigated molecular changes associated with STAT3 inactivation by Western blot analysis. Among the cell proliferation and survival regulators previously shown to be controlled by STAT3,47-50 expression of c-Myc, JunB, and Mcl-1 was reduced, whereas the levels of CDK inhibitor p27 (CDKN1B) were increased by STAT3 knockdown (Figure 7D). Expression of p53 was not notably affected, suggesting that transcriptional inhibition of p53 by STAT3, which was observed in nonlymphoid cell types,51 did not occur in these lymphoma B cells. Collectively, our experiments described in Figures 6 and 7 demonstrate that constitutive STAT3 activation plays a very important role in cell proliferation and survival of ABC-DLBCL cells.
RNAi-mediated STAT3 knockdown in ABC-DLBCL cells also triggered cell-cycle arrest and apoptosis. (A) STAT3 siRNA (STAT3i, +) and control (−) oligos were transiently transfected into Ly3 and Ly10 cells. At the indicated time points after transfection, total STAT3 and PY-STAT3 expression was examined by Western blot analysis. (B) Proliferation of STAT3i-transfected Ly3 and Ly10 cells was measured by 3H-thymdine incorporation assays. The amount of radioactivity recovered from STAT3i-transfected cells was normalized to that from the control cells defined as 100%. Represented in the graph are the mean plus or minus standard derivation of 3 independent STAT3 siRNA experiments. (C) Western blot showing increased PARP cleavage (▷) and, thus, activation of caspase 3/7 in STAT3i-transfected cells. (D) Western blot analysis demonstrating alterations in selected cell cycle and survival regulators following STAT3 knockdown. Vertical lines have been inserted to indicate a repositioned gel lane.
RNAi-mediated STAT3 knockdown in ABC-DLBCL cells also triggered cell-cycle arrest and apoptosis. (A) STAT3 siRNA (STAT3i, +) and control (−) oligos were transiently transfected into Ly3 and Ly10 cells. At the indicated time points after transfection, total STAT3 and PY-STAT3 expression was examined by Western blot analysis. (B) Proliferation of STAT3i-transfected Ly3 and Ly10 cells was measured by 3H-thymdine incorporation assays. The amount of radioactivity recovered from STAT3i-transfected cells was normalized to that from the control cells defined as 100%. Represented in the graph are the mean plus or minus standard derivation of 3 independent STAT3 siRNA experiments. (C) Western blot showing increased PARP cleavage (▷) and, thus, activation of caspase 3/7 in STAT3i-transfected cells. (D) Western blot analysis demonstrating alterations in selected cell cycle and survival regulators following STAT3 knockdown. Vertical lines have been inserted to indicate a repositioned gel lane.
Discussion
In this study, we demonstrate that STAT3 is a novel BCL6 target gene in GC B cells and DLBCL. As a result, STAT3 is predominantly expressed in BCL6− GC B cells, whereas high-level STAT3 expression and activation are preferentially associated with BCL6-low ABC-DLBCLs. Most importantly, constitutively activated STAT3 promotes proliferation and survival in ABC-DLBCL cells, suggesting STAT3 as a new drug target for this group of lymphomas that are refractory to current chemotherapy.
Before this study, negative regulation of Jak/STAT signaling by BCL6 had been reported in bone marrow–derived macrophages; however, in macrophages, BCL6 suppresses STAT3 activation indirectly by inhibiting basal level IL-6 production, whereas its influence on total STAT3 protein is very mild.20 In IL-2– and IL-5–treated BCL1 cells, the inhibitory effect of overexpressed BCL6 on STAT3-dependent plasma cell differentiation was attributed to its ability to compete with STAT3 in binding to dually regulated target genes.23 PRDM1/Blimp-1 was thought to be one such target at the time, but later studies revealed that BCL6 represses PRDM1/Blimp-1 independently of any STAT3 sites.52 Results from our current work demonstrate that STAT3 is a direct BCL6 target gene, raising the possibility that BCL6 prevents premature plasma cell differentiation by simultaneously inhibiting both Blimp-1 and STAT3. We also observed an interesting phenomenon of coupled STAT3 expression and activation in both ABC-DLBCL cell lines and a subset of GC B cells (Figures 1A,3F). This suggests that in GC, Jak/STAT3 signaling is limited by STAT3 protein expression, which is a departure from the conventional model in which cytokine-triggered STAT activation occurs rapidly via posttranslational modifications. STAT3-activating cytokines, such as IL-6, IL-10, and IL-21, which are capable of acting at both short and long range, are unlikely to be limiting in the GC. Therefore, B cells within the GC seem to operate under a licensed-to-respond mechanism, in which STAT3 expression, not cytokine triggering, is the rate-limiting step controlling STAT3 activity. In GC B cells, an on/off switch for STAT3 expression may represent one strategy for regulating stepwise reprogramming toward post-GC plasma cell fate.
A recent report indicates that during development of conventional B cells and the course of GC response, STAT3 is only required within a narrow window after immunoglobulin class switching and before full plasma cell differentiation.53 This conclusion is consistent with our observation that within GC, STAT3 expression and activation appears to follow the downregulation of BCL6 and precede the upregulation of Blimp-1 (Figure 3B). Thus, it is possible that the STAT3+ B cell in the apical light zone may be at a transitional stage between a BCL6-governed GC program and commitment to a Blimp-1–driven plasma cell fate. As a group, ABC-DLBCLs highly express a number of plasma cell markers, including IRF4 and XBP1,4 and yet they do not display recognizable plasmablast features, possibly due to genetic inactivation of the PRDM1/Blimp-1 gene.24,25 These studies, together with our observation that STAT3 is highly expressed and constitutively activated in BCL6-low ABC-DLBCLs, prompt us to speculate that ABC-DLBCLs are phenotypically frozen at a late stage of GC reaction with only the earliest signs of plasma cell differentiation/commitment. We cannot formally exclude the possibility that ABC-DLBCL may also derive from activated post-GC cells bearing a fixed load of Ig mutation and sharing a similar transcription profile with in vitro–activated naive B cells.
Before our study, it was known that ABC-DLBCL is associated with constitutively activated NF-κB, which is required for survival of these tumor cells, at least in culture.5,27 Interestingly, PMBL also has activated NF-κB and yet responds much more favorably to chemotherapy than ABC-DLBCL; and only ABC-DLBCL but not PMBL carries high loads of aberrant Ig class switch recombinations.28,54 In light of these clinical and immunologic differences between these 2 lymphoma groups, it is not surprising that the NF-κB signaling pathway regulates very different sets of target genes in PMBL and ABC-DLBCL.55 Clearly, the net output of the NF-κB transcriptional program is modulated by additional, tumor-specific oncogenic pathway(s). In this regard, it was previously reported that STAT6 is constitutively activated in PMBL,56 whereas our results in this study showed that STAT3 but not STAT6 is constitutively activated in ABC-DLBCL. Finally, a survival-promoting role of STAT3 in ABC-DLBCL also has several implications for lymphoma prognosis and therapy. On one hand, it implies that chemosensitivity of GCB-DLBCL is at least partially attributable to the ability of BCL6 to suppress 2 prominent antiapoptosis pathways, NF-κB and STAT3; on the other, it also suggests that the chemoresistant property of ABC-DLBCL is the result of simultaneous activation of both NF-κB and STAT3. In summary, our findings demonstrate that constitutive STAT3 activation represents a second oncogenic pathway in ABC-DLBCL, potentially providing an additional therapeutic target for treatment of these lymphomas.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Masafumi Abe for his generous gift of the HBL-1 cell line; Dr Paul Wade for sharing the MTA3 antibody; Drs Richard Kitsis, Liang Zhu, Arthur Skoultchi, and Bridget Shafit-Zagardo for assistance with other antibodies; Dr Wayne Tam for the initial validation of STAT3 antibodies on tissue sections; and Drs Barbara Birshtein and Amit Verma and Mr J. Tsoni Peled for critically reading the manuscript.
This work was supported by grants from the National Institute of Health (RO1 CA85573) and the G & P Foundation for Cancer Research to B.H.Y., and a grant from the Lauri Strauss Leukemia Foundation to B.B.D. R.Y.-L.Y was supported during this study by the Harry Eagle Postdoctoral Scholarship from the Department of Cell Biology, Albert Einstein College of Medicine. G.C. was a recipient of a Ms and Mr Aboodi Professorship.
Authorship
Contribution: B.B.D., J.J.Y., R.Y.-L.Y, and L.M.M. performed research and analyzed data. L.M.M. also edited the paper. R.S. constructed the DLBCL MTA. G.G. performed IHC and IF staining. Y.Z. analyzed DLBCL gene expression patterns. B.H.Y designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: B. Hilda Ye, Department of Cell Biology, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461; e-mail: hye@aecom.yu.edu.