The relationship among neuroinflammation, blood-brain barrier (BBB) dysfunction, and progressive HIV-1 infection as they affect the onset and development of neuroAIDS is incompletely understood. One possible link is signal transducers and activators of transcription (STATs) pathways. These respond to proinflammatory and regulatory factors and could affect neuroinflammatory responses induced from infected cells and disease-affected brain tissue. Our previous works demonstrated that HIV-1 activates pro-inflammatory and interferon-alpha–inducible genes in human brain microvascular endothelial cells (HBMECs) and that these genes are linked to the Janus kinase (JAK)/STAT pathway. We now demonstrate that HIV-1 activates STAT1, induces IL-6 expression, and diminishes expression of claudin-5, ZO-1, and ZO-2 in HBMECs. The STAT1 inhibitor, fludarabine, blocked HIV-1–induced IL-6, diminished HIV-1–induced claudin-5 and ZO-1 down-regulation, and blocked HIV-1– and IL-6–induced monocyte migration across a BBB model. Enhanced expression and activation of STAT1 and decreased claudin-5 were observed in microvessels from autopsied brains of patients with HIV-1–associated dementia. These data support the notion that STAT1 plays an integral role in HIV-1–induced BBB damage and is relevant to viral neuropathogenesis. Inhibition of STAT1 activation could provide a unique therapeutic strategy to attenuate HIV-1–induced BBB compromise and as such improve clinical outcomes.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection of the central nervous system (CNS) commonly results in behavioral, motor, and cognitive impairments.1,–3 Although disease severity and progression has slowed, in part, as a result of antiretroviral therapy, 76% to 83% of brain autopsies continue to show observable neuropathological abnormalities.4,–6 Disease pathology ranges from mild brain atrophy and gliosis to robust viral replication, multinucleated giant cell formation, astrogliosis and microgliosis, myelin pallor, and neuronal loss.7,8 These pathological findings are collectively termed HIV-1 encephalitis (HIVE). HIVE is a common correlate to the later stages of behavioral, motor, neuropsychiatric, and neurologic consequences of disease termed HIV-1–associated dementia (HAD) (for recent reviews, see McArthur1 ; Grant et al2 ; Ghafouri3 ). HIVE is fueled by viral infection and immune activation of brain mononuclear phagocytes (MPs: blood-derived perivascular macrophages and microglia).9 Such MP- and virus-associated neuroinflammation promotes monocyte trafficking across the blood-brain barrier (BBB), MP infiltration into the CNS, and neurodegeneration.10,–12 Thus, dysfunction of the BBB is one critical feature of HIV-1 neuropathogenesis
Brain microvascular endothelial cells, a major component of BBB function and integrity, are connected by tight junctions (TJs) that limit paracellular flux and restrict permeability.13 Indeed, under normal physiologic conditions, the brain endothelium functions as an interface between the blood and the brain parenchyma, strictly regulating influx of ions, molecules, and leukocytes into the CNS. Nonetheless, in disease, a variety of environmental, toxic, degenerative, and microbial insults could cause BBB breakdown.12,14 Such a breakdown occurs during progressive HIV-1 infection12,15,–17 and was documented in laboratory, animal models, human clinical observations, and autopsy studies.10,16,,,–20 Underlying mechanisms of BBB dysfunction and how it affects ongoing disease are incompletely understood.1,12,14 Dysfunction of the BBB enhances penetration of cell-free virus, ingress of activated HIV-1–infected monocytes across the BBB, accumulation of MP in the CNS, and spread of the virus to neighboring microglia and astrocytes.15,21 Thus, BBB breech is commonly associated with accelerated disease and the development of behavioral and cognitive deficits that are signatures of HAD.1,16 Based on these observations, the elucidation of the signaling pathways mediating BBB compromise can prove important for understanding disease mechanisms and development of new therapies.
Signal transducers and activators of transcription (STATs) proteins are latent cytoplasmic transcription factors that are phosphorylated by Janus kinases (JAKs) in response to proinflammatory and regulatory factors.22 STAT1 encodes a 91-kDa protein that is activated by type I (alpha and beta) and type II interferons (IFNs), epidermal growth factor, platelet-derived growth factor, and interleukin-6 (IL-6).23,,–26 Moreover, STAT1 activation often correlates with cellular proinflammatory, antiproliferative, and apoptotic activities.27,28 The JAK/STAT pathway plays a prominent role in cytokine-mediated inflammatory responses and, as such, has been implicated in the pathogenesis of HIV infection.29,,–32 Indeed, relationships between neuroinflammatory activities and specific signaling pathways are well appreciated.27 Mechanistic studies reported herein arise from microarray analyses performed in our laboratories demonstrating that HIV-1 activates proinflammatory and IFN-inducible genes in human brain microvascular endothelial cells (HBMECs).33 We now demonstrate that HIV-1 and HIV-1–infected monocyte-derived macrophages (MDMs) activate STAT1 and induce IL-6 expression in HBMECs through STAT1. A specific STAT1 inhibitor, fludarabine (FLUD), blocked HIV-1–induced STAT1 activation, and decreased IL-6 expression and secretion. Moreover, FLUD diminished HIV-induced down-regulation of TJs and prevented HIV-1– and IL-6–induced monocyte migration across in vitro BBB models. Most importantly, enhanced expression and activation of STAT1 was shown in microvessels acquired from autopsied brains of HIV-infected people, as well as in autopsied brains of patients who died with advanced HAD. These data strongly support the notion that STAT1 plays an integral role in HIV-induced BBB damage and is relevant to the pathogenesis of viral infection. Inhibition of STAT1 activation could provide a unique therapeutic strategy to prevent HIV-1–induced BBB compromise and as such improve clinical outcomes in infected people.
Methods
Endothelial cell culture
Primary HBMECs were isolated from brain tissue obtained during surgical removal of epileptogenic cerebral cortex in adult patients34 and were provided by Drs Marlys Witte and Michael Bernas (University of Arizona, Tucson, AZ). Routine evaluation for von Willebrand factor (VWF), Ulex europeus lectin, and CD31 demonstrated that HBMECs were more than 99% pure. HBMECs were seeded in the upper chamber of collagen-I–coated 6- or 24-well tissue culture inserts (with 0.4-μm pore size) or 6-well tissue culture plates and cultured to confluence in EGM-2 BulletKit media (Cambrex, Walkersville, MD) supplemented with 5% fetal bovine serum (FBS). Only cells at passage 1 to 4 were used in this study.
Monocyte isolation, macrophage culture, and HIV-1 infection
Monocytes were obtained from HIV-1–, HIV-2–, and hepatitis-B–seronegative donor leukopaks, separated by countercurrent centrifugal elutriation and characterized as previously described.20,35 Freshly elutriated monocytes were differentiated into MDMs by 7-day culture in Dulbecco modified Eagle media containing 2 mM l-glutamine (Invitrogen, Carlsbad, CA), 1 U/mL human recombinant macrophage colony-stimulating factor (a generous gift from Wyeth, Cambridge, MA), 10% heat-inactivated human serum, 100 μg/mL gentamicin, and 10 μg/mL ciprofloxacin (Sigma-Aldrich, St Louis, MO). MDMs were infected with HIV-1ADA at a multiplicity of infection (MOI) of 0.01 then used for cocultures with HBMECs at day 5 after viral inoculation. All reagents were prescreened for endotoxin (< 10 pg/mL; Associates of Cape Cod, Woods Hole, MA) and mycoplasma contamination (Gen-probe II; Gen-probe, San Diego, CA). Levels of productive viral replication were measured in culture fluids by HIV-1 p24 enzyme-linked immunosorbent assay (ELISA; Beckman Coulter, Miami, FL).
Cocultures of HIV-1–infected MDMs and HBMECs
In triplicate, HBMECs in transwell inserts (0.4-μm pore size) were cocultured for 12 hours with HIV-1ADA–infected MDMs in endothelial cell media (EGM-2 BulletKit media with 5% FBS). Triplicate samples of HBMECs cocultured for 12 hours with noninfected MDMs or untreated HBMECs served as controls. Following coculture, media from the top (HBMEC side) and bottom (MDM side) wells of each insert were collected and centrifuged for 15 minutes at 1930g. The recovered fluids were cryopreserved at −70°C. In parallel experiments, HBMECs in 6-well plates were exposed directly to purified HIV-1ADA (MOI: 0.01) for 5 minutes to 24 hours, and the fluids cryopreserved at −70°C.
Monocyte migration through an in vitro BBB model
For migration assays, 2 × 104 HBMECs were seeded on collagen-coated FluoroBlok-tinted tissue culture inserts (with 3-μm pore size; BD Biosciences, Franklin Lakes, NJ). Because monolayers are not visible on these inserts, manual readings of transendothelial electric resistance were taken with a voltmeter (EVOM; World Precision Instruments, Sarasota, FL) to confirm monolayer formation and cell confluence. Freshly elutriated, infected or noninfected monocytes were labeled with calcein-AM (Invitrogen) at 5 μM/1 × 106 cells for 45 minutes and washed to eliminate residual label. Then, 2.5 × 105 labeled monocytes were placed on HBMECs (upper chamber of the FluoroBlok insert) and allowed to migrate for 2 hours (37°C, 5% CO2). Migrated monocytes were quantified with a fluorescence plate reader (absorbance: 494 nm; emission: 517 nm), with a standard curve derived from a serial dilution of known numbers of calcein-labeled cells. For migration experiments testing the effects of FLUD, HBMECs were exposed to FLUD (20 μM) (Sigma) for 30 minutes before migration. We previously performed dose-dependent migration experiments with FLUD and determined that 20 μM was optimal in altering monocyte transendo-thelial migration.
Detection of IL-6
IL-6 levels in each sample were quantified using the Human IL-6 ELISA kit (eBioscience, San Diego, CA) according to the manufacturer's instructions. IL-6 levels in media from infected and noninfected MDMs and HBMECs prior to coculture served as controls.
Microvessel isolation
Brain tissues (cortex) from HIV-1–seropositive patients without neurocognitive impairment, HAD patients, and aged-matched seronegative controls were obtained from the National NeuroAIDS Tissue Consortium (NNTC) and our Center for Neurovirology and Neurodegenerative Diseases (CNND) brain bank. The clinical histories of all brain microvessel (MV) donors are detailed in Table 1. The MV isolation procedure was as described by Brooks et al.36 Briefly, brain tissues were homogenized in a buffer containing 103 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 14 mM HEPES, 2.5 mM NaHCO3, 10 mM d-glucose, 1 mM sodium pyruvate, pH 7.4, and 1 × protease inhibitor cocktail. Following homogenization, each sample was passed sequentially through a 350-, 112-, and 40-μm nitex mesh filters (Sefar, Depew, NY). Following the last filtration, the 40-μm mesh filters were removed and rinsed to collect trapped MVs. Isolated MV was pelleted by centrifugation at 5600g for 10 minutes at 4°C. RNA and protein were extracted from each MV sample for real-time polymerase chain reaction (PCR) and Western blot analyses. For immunofluorescence and confocal microscopy, 50 μL MV solution was spread onto poly-l-lysine– or fibronectin-coated glass coverslips, air-dried, fixed for 20 minutes with a 4% paraformaldehyde solution (pH 7.4), rinsed with phosphate-buffered saline (PBS), air-dried, and used for immunostaining with antibodies to STAT1 and phospho-STAT1. Immunostaining with antibodies to the TJ protein claudin-5 and VWF was used to confirm the vascular identity of the isolated MV as previously described.37,38
RNA isolation and real-time PCR
HBMECs and brain MVs were lysed using the Trizol reagent (Invitrogen); total RNA was extracted from each sample using RNAeasy mini-kit (Qiagen, Valencia, CA) with DNase treatment according to the manufacturer's instructions. RNA yield and quality were checked using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and for all samples, absorbance ratio of 260/280 was more than 2. The purity and quality of extracted RNA were further assessed using the RNA 6000 Nano LabChip Kit and Agilent-2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) according to the manufacturer's instructions.
For each sample, cDNA was generated from 1 μg RNA using random hexamers and Moloney murine leukemia virus reverse transcriptase (Invitrogen). Reverse transcription was carried out for 1 hour at 42°C. The cDNA obtained was used for real-time quantitative PCR (qRT-PCR) using an ABI PRISM 7000 sequence detector (Applied Biosystems, Foster City, CA). A TaqMan gene detection system was used and quantification performed using the standard curve method as described in user bulletin no. 2 supplied with ABI PRISM 7000 sequence detector.39 All PCR reagents and primers were obtained from Applied Biosystems and primer IDs were as follows: chemokine (C-X-C motif) ligand-1 (CXCL1): Hs00236937; IL-6: Hs00174131_m1; interferon-stimulated gene 15 (ISG15 or GIP2): Hs00192713_m1; tumor necrosis factor-alpha–induced protein-3 (TNFAIP3): Hs00234713_m1; TNF (ligand) superfamily 15 (TNFSF15): Hs00353710_s1; ubiquitin-D (UBD): Hs00197374_m1; RelB: Hs00232399_m1; STAT1: Hs00234829_m1; and superoxide dismutase 2 (SOD2): Hs00167309_m1. For endogenous control each gene expression was normalized to GAPDH (Hs99999905_m1).
Immunofluorescence microscopy
Confluent HBMECs grown on glass coverslips were washed with PBS and fixed in methanol/acetone (1:1) for 20 minutes. Fixed MVs and HBMECs were permeabilized with 0.1% triton X-100 and blocked for nonspecific binding with 3% BSA in PBS (10 minutes at 4°C). Cells were incubated with primary antibodies (1:50 dilution) for 1 hour at room temperature, followed by staining (1 hour in the dark at room temperature) with secondary antibodies coupled with Alexa-488 or Alexa-594 (1:500 dilution; Invitrogen). Stained cells were mounted in Prolong Gold (Invitrogen) and examined using an E800 fluorescent microscope (Nikon, Melville, NY) connected to a color MagnaFire digital camera (Optronics, Goleta, CA).
Protein extraction and Western blot analyses
Protein extraction and quantification were performed as we previously described.19,20 Protein (25 μg)was fractionated in a 4% to 15% gradient gel and electrophoretically transferred to nitrocellulose membranes. Membranes were blocked for 1 hour with SuperBlock T-20 (Pierce, Rockford, IL), blotted 2 hours or overnight with the appropriate primary antibodies, 1 hour with the secondary antibody, washed, and visualized using SuperSignal West Pico Substrate (Pierce). All STAT and phospho-STAT antibodies were from Cell Signaling Technology (Danvers, MA). Claudin-5, ZO-1, and ZO-2 antibodies were from Invitrogen. For Western blot assays performed with phosphorylated STAT antibodies (pSTAT1, pSTAT2, pSTAT3, pSTAT5, pSTAT6), membranes were stripped using Restore Western Blot Stripping Buffer (Pierce) and reblotted with antibodies to STAT1, STAT2, STAT3, STAT5, and STAT6, respectively, then stripped again and reblotted with β-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Results were expressed as ratios of relative intensity of the target protein (phospho-STAT) to that of the internal standard, β-actin, or STAT. For experiments with inhibitors, HBMECs were exposed for 30 minutes to HIV-1 with or without FLUD (20 μM), the JAK2 (AG490, 50 μM), and JAK3 (WHI-P154, 30 μM) inhibitors, and protein was extracted and analyzed as above. AG490 and WHI-P154 were from Calbiochem (San Diego, CA). Controls consisted of untreated cells, and cells exposed to FLUD, AG490, and WHI-P145.
Statistical analysis
One- or 2-way analysis of variance and the general linear models procedures were performed using the SAS program (SAS Institute, Cary, NC) followed by Dunnet or Tukey multiple comparison tests. Both statistical procedures test differences among several means for significance without increasing type I error rate.40 The threshold for significance was .05.
Results
HIV-1– and HIV-infected MDMs induce IL-6 expression in HBMECs
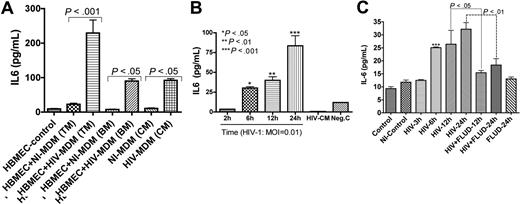
We previously showed that HIV-1 up-regulates mRNA for several proinflammatory cytokines in HBMECs, including IL-6.33 To determine whether increased IL-6 mRNA was associated with increased protein, we quantified IL-6 in cell-culture media. Coculture of HBMECs with infected MDMs increased IL-6 secretion in HBMECs by 10-fold (229.2 ± 38 pg/mL) compared with HBMECs cocultured with uninfected MDMs (23 ± 3 pg/mL) (P < .001; Figure 1A). Endothelial-macrophage coculture did not change MDM IL-6 levels even following HIV-1 infection (Figure 1A). To determine whether the induced HBMEC IL-6 was caused by virus or by secretory products from infected MDMs, we exposed endothelial cells to 0.01 infectious viral particles of HIV-1ADA for 2 to 24 hours and quantified IL-6 levels in culture fluids. Each of 3 experiments was done in quadruplicate. A significant and time-dependent increase in HBMEC IL-6 was observed following exposure to HIV-1 (Figure 1B). IL-6 levels in HBMECs exposed to HIV-1 virions were 3.5 (± 0.2), 30.3 (± 1.7), 40.1 (± 4.4), and 83.5 (± 13.0) pg/mL, respectively, after 2, 6, 12, and 24 hours of viral exposure.
STAT1 modulates HIV-1–induced up-regulation of IL-6 expression in HBMECs. (A) Interaction of HBMECs with HIV-infected MDMs induced IL-6 secretion in HBMECs. Coculture of HBMECs with infected MDMs increased IL-6 expression/secretion in endothelial cells by 10-fold (229.2 ± 38 pg/mL) compared with endothelial cells cocultured with noninfected MDMs (23 ± 3 pg/mL). Endothelial-MDM coculture did not change MDM IL-6 levels regardless of HIV-1 infection. (B) Direct exposure of HBMECs to 0.01 infectious viral particles induced a time-dependent increase in IL-6 secretion. (C) Fludarabine (FLUD, 20 μM) significantly diminished HIV-1–induced IL-6 secretion in HBMECs. Controls are untreated cells; NI-control represents HBMECs exposed for 24 hours to conditioned media from noninfected MDMs. For each experimental condition, the number of replicates is 6. (***P < .001 compared with controls.) TM indicates top media; BM, bottom media; CM, conditioned media; NI-MDM, noninfected monocyte-derived macrophage; HIV-MDM, HIV-infected monocyte-derived macrophage; and MOI, multiplicity of infection.
STAT1 modulates HIV-1–induced up-regulation of IL-6 expression in HBMECs. (A) Interaction of HBMECs with HIV-infected MDMs induced IL-6 secretion in HBMECs. Coculture of HBMECs with infected MDMs increased IL-6 expression/secretion in endothelial cells by 10-fold (229.2 ± 38 pg/mL) compared with endothelial cells cocultured with noninfected MDMs (23 ± 3 pg/mL). Endothelial-MDM coculture did not change MDM IL-6 levels regardless of HIV-1 infection. (B) Direct exposure of HBMECs to 0.01 infectious viral particles induced a time-dependent increase in IL-6 secretion. (C) Fludarabine (FLUD, 20 μM) significantly diminished HIV-1–induced IL-6 secretion in HBMECs. Controls are untreated cells; NI-control represents HBMECs exposed for 24 hours to conditioned media from noninfected MDMs. For each experimental condition, the number of replicates is 6. (***P < .001 compared with controls.) TM indicates top media; BM, bottom media; CM, conditioned media; NI-MDM, noninfected monocyte-derived macrophage; HIV-MDM, HIV-infected monocyte-derived macrophage; and MOI, multiplicity of infection.
STAT1 inhibition attenuates HIV-1–induced IL-6 expression
As IL-6 signals principally through the JAK/STAT pathway, we determined whether STAT1 modulates HIV-1–induced IL-6 production in HBMECs. HBMECs were exposed to HIV-1ADA (MOI = 0.01) for 3, 6, 12, and 24 hours, or to HIV-1 with 20 μM FLUD for 12 and 24 hours; and IL-6 levels in culture supernatant were quantified by ELISA. HIV-1 induced a time-dependent increase in IL-6 levels and FLUD significantly diminished IL-6 secretion (P < .01; Figure 1C). No IL-6 was detected in HBMECs exposed to conditioned media from uninfected MDMs or in untreated HBMECs.
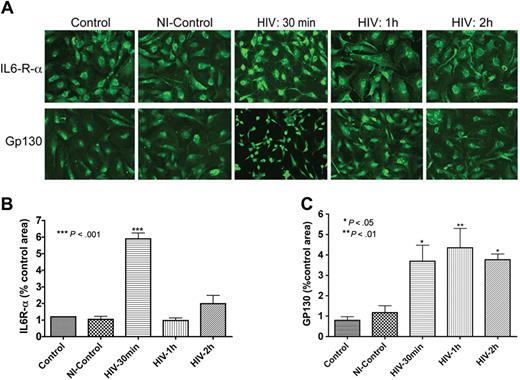
HIV-1 up-regulates HBMEC IL-6 receptor expression
As IL-6 secretion was up-regulated following HIV-1 exposure or coculture with infected MDMs (Figure 1), we evaluated the expression of IL-6 receptor (IL-6R) in HBMECs. Immunofluorescence analyses demonstrated that HBMECs express the IL-6R-α and gp130 (α and β chains of the IL-6 receptor, Figure 2). Next, we determined whether this receptor could be up-regulated by exposure to HIV-1. Indeed, HIV-1 significantly increased the expression of both α and β subunits of the IL-6R (Figure 2A). These observations were confirmed in 2 independent experiments using HBMECs from 2 different donors. We performed semiquantitative analyses of IL-6 expression (percentage of immunostained area) using computer-assisted image analysis (Image-ProPlus; Media Cybernetics, Silver Spring, MD). Image-ProPlus software measures the fluorescence intensity and densitometry of immunostained cells and normalize each measurement to the cell's size. The data showed that 30 minutes of HIV-1 exposure increased IL6-R-α expression 6-fold (P < .001) and gp130 expression 4-fold (P < .05). Conditioned media from uninfected MDMs did not change IL-6R staining (Figure 2). Interestingly, HIV-1–induced up-regulation of IL6-R-α expression was transient and decreased after 30 minutes (Figure 2B), while gp130 expression was sustained for up to 2 hours following HIV-1 treatment (Figure 2C).
HBMECs express the IL-6 receptor and HIV-1 exposure increases receptor expression. Exposure of HBMECs to HIV-1 for 30 minutes significantly increased the expression of both alpha (IL6-R-α; A,B) and beta (Gp130; A,C) subunits of the IL-6 receptor, and altered endothelial cell phenotype (A). HIV-1–induced up-regulation of IL6-R-α expression was transient and decreased after 30 minutes (A,B), while HIV-induced increase in Gp130 expression was sustained for up to 2 hours (C). Control represent untreated HBMECs; NI-control consisted of HBMECs exposed for 1 hour to conditioned media from noninfected macrophages. Original magnification of images in panel A: ×200.
HBMECs express the IL-6 receptor and HIV-1 exposure increases receptor expression. Exposure of HBMECs to HIV-1 for 30 minutes significantly increased the expression of both alpha (IL6-R-α; A,B) and beta (Gp130; A,C) subunits of the IL-6 receptor, and altered endothelial cell phenotype (A). HIV-1–induced up-regulation of IL6-R-α expression was transient and decreased after 30 minutes (A,B), while HIV-induced increase in Gp130 expression was sustained for up to 2 hours (C). Control represent untreated HBMECs; NI-control consisted of HBMECs exposed for 1 hour to conditioned media from noninfected macrophages. Original magnification of images in panel A: ×200.
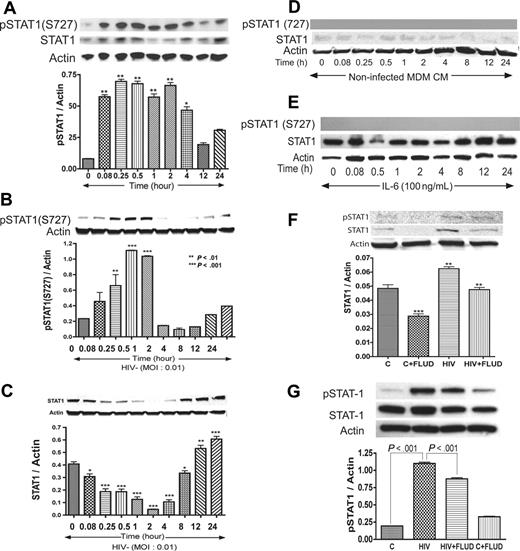
Interactions of HIV-1–infected macrophages with HBMECs activate STAT1 in HBMECs
Based on data obtained regarding the influence of IL-6 and HIV-1 on STAT pathways, we tested whether coculture of HBMECs with HIV-infected MDMs activates STAT1 in HBMECs. In these experiments, HBMECs in 0.4-μm culture inserts were cocultured with HIV-infected MDMs for 5 minutes to 24 hours; HBMEC protein was extracted and analyzed by Western blotting. Interaction of HBMECs with infected MDMs induced phosphorylation of STAT1 at serine 727 (S727) from 5-minutes to 4-hour coculture and also increased total STAT1 levels (Figure 3A). No activation of STAT1 at tyrosine residues was detected.
HIV-1 activates STAT1 at serine-727 in HBMECs, and FLUD inhibits HIV-induced STAT1 expression and activation. Coculture of HBMECs with HIV-1–infected MDMs (A), or direct exposure of HBMECs to infectious viral particles (B) activates STAT1 at S727. Long-term HIV exposure (12–24 hours) also increased total STAT1 levels in HBMECs (C). No activation of STAT1 at tyrosine residues was detected. No phosphorylation of STAT1 was observed in HBMECs exposed to conditioned media from noninfected MDMs, and there was no change in total STAT1 levels (D). Exposure of HBMECs to IL-6 did not phosphorylate STAT1 at serine or tyrosine residues; however, 12 to 24 hours of IL-6 exposure increased total STAT1 levels (E). The STAT1 inhibitor FLUD significantly diminished HIV-1–induced STAT1 activation and expression at 2 hours (F) and 30 minutes (G) of exposure. (*P < .05; **P < .01; ***P < .001.) CM indicates conditioned media; and C, control.
HIV-1 activates STAT1 at serine-727 in HBMECs, and FLUD inhibits HIV-induced STAT1 expression and activation. Coculture of HBMECs with HIV-1–infected MDMs (A), or direct exposure of HBMECs to infectious viral particles (B) activates STAT1 at S727. Long-term HIV exposure (12–24 hours) also increased total STAT1 levels in HBMECs (C). No activation of STAT1 at tyrosine residues was detected. No phosphorylation of STAT1 was observed in HBMECs exposed to conditioned media from noninfected MDMs, and there was no change in total STAT1 levels (D). Exposure of HBMECs to IL-6 did not phosphorylate STAT1 at serine or tyrosine residues; however, 12 to 24 hours of IL-6 exposure increased total STAT1 levels (E). The STAT1 inhibitor FLUD significantly diminished HIV-1–induced STAT1 activation and expression at 2 hours (F) and 30 minutes (G) of exposure. (*P < .05; **P < .01; ***P < .001.) CM indicates conditioned media; and C, control.
HIV-1 virions induce activation of STAT1 at serine 727 in HBMECs
To evaluate the effects of direct HIV-1 exposure to HBMECs, HIV-1ADA was placed onto HBMECs for 5 minutes to 24 hours, followed by protein extraction and Western blot analysis. Direct exposure of HBMECs to HIV-1 phosphorylated STAT1 at S727, with maximal level of activation at 30 minutes and 1 hour (Figure 3B). Increased phospho-STAT1 correlated with decrease in total STAT1, however, long-term HIV-1 exposure (12–24 hours) increased total STAT1 levels (Figure 3C). HIV-1–induced STAT1 activation was specific to serine residues; no activation at tyrosine residues was detected. Treatment of HBMECs with conditioned media derived from uninfected MDMs (5 minutes to 24 hours) did not induce STAT1 phosphorylation or change in total STAT1 levels (Figure 3D). To assess whether IL-6 can activate endothelial STAT1, HBMECs were treated with IL-6 (100 ng/mL) for 5 minutes to 24 hours, followed by protein extraction and Western blot analysis. IL-6 did not phosphorylate STAT1 at serine or tyrosine residues (Figure 3E). Based on our data showing that FLUD prevent HIV-1–induced IL-6 expression (Figure 1) and diminished HIV- and IL-6–induced monocyte transendothelial migration (Figure 4A), we tested the effect of FLUD on HIV-induced STAT1 expression and activation. HBMECs were exposed for 2 hours or 30 minutes to HIV-1, FLUD (20 μM), or HIV + FLUD and extracted proteins analyzed as described. HIV-1 exposure increased expression of STAT1 and phospho-STAT1 (S727). FLUD significantly diminished HIV-1–induced STAT1 activation and expression at 2 hours (Figure 3F) and 30 minutes (Figure 3G).
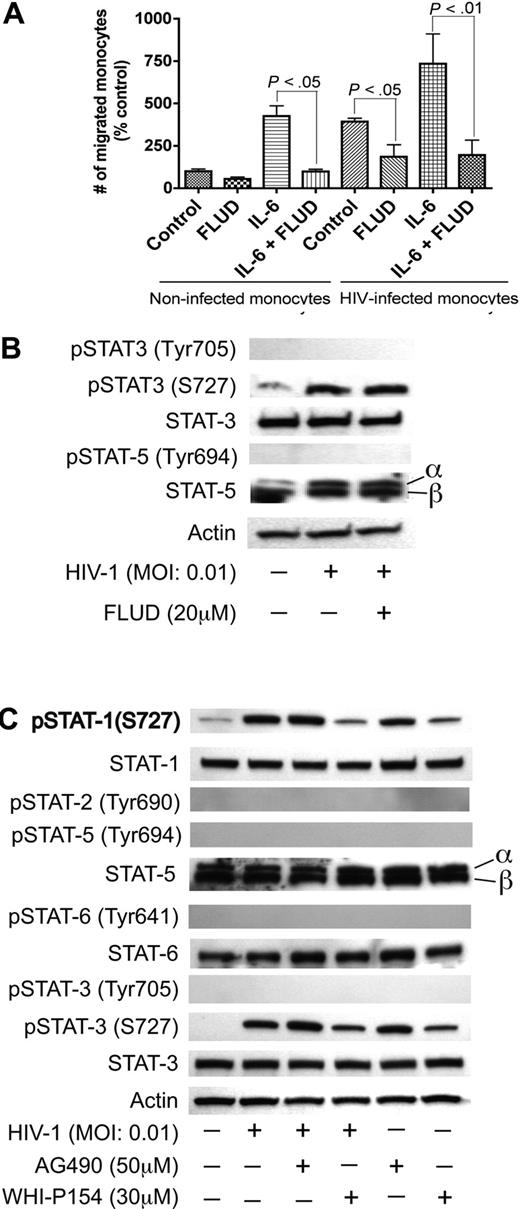
JAK/STAT pathway in HIV-1–induced endothelial dysfunction. (A) Effect of FLUD on monocyte migration across in vitro BBB models in response to HIV-1 and IL-6. HIV-1 infection and IL-6 increased monocyte migration across the BBB and the STAT1 inhibitor (FLUD) significantly diminished HIV-1- and IL-6-induced monocyte migration (*P < .05). NI represent migration of noninfected macrophages. (B,C) HIV-1 phosphorylates STAT1 and STAT3 at serine-727, and the JAK-3 inhibitor WHI-P154 diminished HIV-1–induced phosphorylation of STAT1 and STAT3. HIV-1 induced phosphorylation of STAT-3 at S-727 but did not phosphorylate STAT-5 or STAT-3 at tyrosine residues. FLUD had no effect on HIV-1–induced STAT-3 phosphorylation (panel B). The JAK-3 inhibitor (WHI-P154) significantly diminished HIV-induced serine phosphorylation of STAT1 and STAT3, while the JAK-2 inhibitor (AG490) had no effect (C). No activation of STAT3 at tyrosine residues was detected. HIV-1 did not activate STAT-2, STAT-5 or STAT-6.
JAK/STAT pathway in HIV-1–induced endothelial dysfunction. (A) Effect of FLUD on monocyte migration across in vitro BBB models in response to HIV-1 and IL-6. HIV-1 infection and IL-6 increased monocyte migration across the BBB and the STAT1 inhibitor (FLUD) significantly diminished HIV-1- and IL-6-induced monocyte migration (*P < .05). NI represent migration of noninfected macrophages. (B,C) HIV-1 phosphorylates STAT1 and STAT3 at serine-727, and the JAK-3 inhibitor WHI-P154 diminished HIV-1–induced phosphorylation of STAT1 and STAT3. HIV-1 induced phosphorylation of STAT-3 at S-727 but did not phosphorylate STAT-5 or STAT-3 at tyrosine residues. FLUD had no effect on HIV-1–induced STAT-3 phosphorylation (panel B). The JAK-3 inhibitor (WHI-P154) significantly diminished HIV-induced serine phosphorylation of STAT1 and STAT3, while the JAK-2 inhibitor (AG490) had no effect (C). No activation of STAT3 at tyrosine residues was detected. HIV-1 did not activate STAT-2, STAT-5 or STAT-6.
STAT1 modulates monocyte migration across in vitro BBB models
To evaluate the biologic significance of HIV-1–induced IL-6 up-regulation, we explored whether HIV-1 and IL-6 could affect monocyte transendothelial migration. HIV-1 infection increased monocyte migration 4-fold compared with uninfected monocytes. IL-6 increased transendothelial migration of uninfected monocytes by 4.3-fold and increased migration of HIV-infected monocytes by 7.4-fold (Figure 4A) compared with monocytes without IL-6 application. Since STAT1 mRNA is up-regulated in HBMECs cocultured with HIV-1–infected MDMs and IL-6 signals through the JAK/STAT pathway, we tested whether the STAT1 inhibitor FLUD could prevent HIV-1– and IL-6–induced monocyte transmigration. FLUD significantly diminished HIV-1– and IL-6–induced monocyte migration in our in vitro BBB model (Figure 4A).
HIV-1 virions phosphorylates STAT1 and STAT3 at serine 727 in HBMECs; JAK-3 inhibitor diminished HIV-1–induced phosphorylation of STAT1 and STAT3
To determine whether HIV-1 activates other members of the STAT family in HBMECs, we tested the levels of phosphorylated STAT-2, STAT-3, STAT-5, and STAT-6 in HIV-exposed HBMECs. HIV-1 induced phosphorylation of STAT-1 and STAT-3 at S727; HIV-1 exposure did not induce phosphorylation of STAT-3 at tyrosine residues and did not induce phosphorylation of STAT-2, STAT-5, or STAT-6 (Figure 4B,C). FLUD had no effect on HIV-1–induced STAT-3 phosphorylation, confirming previous findings that FLUD is a specific STAT1 inhibitor.41,–43 To determine which effector upstream of STAT-1 may be involved in HIV-1–induced BBB dysfunction, we analyzed STAT activation in HBMECs exposed to HIV-1 with or without the presence of the JAK-2 inhibitor (AG490, 50 μM) and the JAK-3 inhibitor (WHI-P154, 30 μM). The JAK-3 inhibitor diminished HIV-1–induced activation of STAT-1 and STAT-3 (Figure 4C).
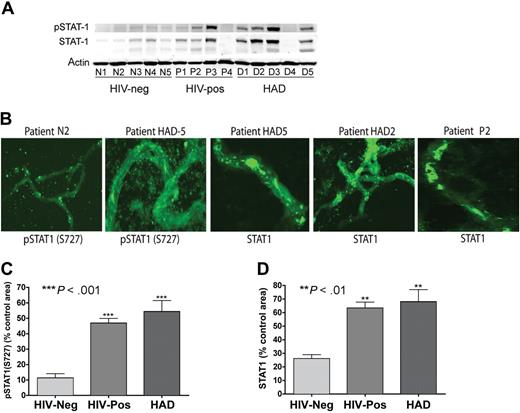
Activation and overexpression of STAT1 and proinflammatory molecules in brain MVs
To determine whether our in vitro findings correlate with changes at the BBB of HIV-1–infected humans, we isolated and analyzed MVs from brain tissues of 5 HIV-1,2–seronegative control subjects, 4 HIV-1–seropositive patients without evidence of HIVE (HIV + no HIVE), and 5 patients with HIVE (HIV-1 dementia scale > 1). Table 1 shows the clinical history, postmortem interval (PMI) between the time of death and autopsy, and a summary of postmortem findings for all 14 individuals. The age ranges were 32 to 46, 27 to 46, and 30 to 50 years, respectively, for seronegative, HIV + no HIVE, and HAD patients. Their PMIs were, respectively, 4 to 8.5 hours, 2.7 to 11 hours, and 6 to 21 hours. HIVE and HAD diagnoses were performed by neuropathologists and neuropsychologists of the NNTC and the CNND rapid autopsy program.44,45 Plasma and cerebrospinal fluid (CSF) viral load (VL) and CD4 counts were available for donors P1 and P4, HAD1, HAD2, and HAD5. For patient P1, plasma VL was 684 444 copies/mL, CD4+ T-cell count was 13/mm3, and CSF VL was 2300 copies/mL. For patient P4, plasma VL was 75 000 copies/mL and CD4 count was 13/mm3. For patient HAD1, plasma VL was 104 300 copies/mL, CD4 count was 8/mm3, and CSF VL was 14 copies/mL. CSF VL was 962 copies/mL for patient HAD2, while for patient HAD5, plasma VL was 570 copies/mL and CD4 count was 361/mm3.
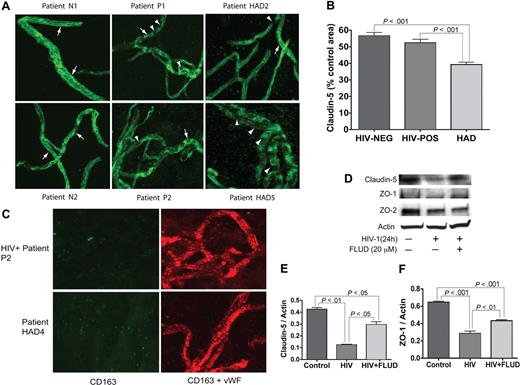
MVs were isolated from the brain cortex as described in “Materials and methods.” The vascular identity of isolated MVs was confirmed by immunostaining and confocal microscopy for the TJ protein claudin-5 and VWF. All MV samples stained positive for claudin-5 and VWF (Figure 5). To assess changes in claudin-5 expression, we performed semiquantitative analyses for all MV samples (percentage of immunostained area) using Image-ProPlus analysis. Claudin-5 levels were lower in MVs from HIV + no HIVE patients, compared with seronegative controls, but the difference was not statistically significant (Figure 5B). Claudin-5 staining was significantly lower in MVs from HAD patients compared with MVs from control or HIV + no HIVE (P < .001, Figure 5B). While continuous TJ strands were found in MVs from seronegative controls, HAD patients with neuropathological evidence of HIVE featured fewer TJ strands with more gaps, or total absence of TJ staining (eg, patient HAD5 in Figure 5A). Because MP migrating through the BBB can be attached to the brain endothelium, we performed double immunostaining of MVs for human CD163 and VWF; we found no macrophage contamination in the isolated MVs (Figure 5C). To determine whether STAT1 can modulate expression of TJ protein in the brain endothelium, we determined the expression of claudin-5, ZO-1, and ZO-2 in HBMECs exposed to HIV-1, with or without FLUD. HIV-1 decreased claudin-5, ZO-1, and ZO-2 expression in HBMECs. FLUD diminished viral-induced down-regulation of claudin-5 and ZO-1 (Figure 5D-F).
Expression of TJ proteins in HBMECs and microvessels from autopsied brains of seronegative controls and HIV-1–infected humans. (A) All MV samples stained positive for claudin-5. MVs from seronegative control donors (eg, patients N1 and N2) show prominent and continuous strands of tight junction (TJ, arrows); TJ strands in MVs from HIV-1–seropositive patients without HIVE (eg, P1 and P2), and HAD HIVE patients (eg, HAD2 and HAD5) were fewer and had more gaps (arrowheads). Patient HAD5 shows very few TJ strands. (B) Computer-assisted semiquantitative analyses of all MV samples show a significant decrease in claudin-5 expression in MVs from HAD patients, compared with MVs from HIV-1–seropositive patients without HIVE (HIV-POS), or seronegative controls (HIV-NEG). MVs from HIV-1–seropositive patients without HIVE also showed a small (nonstatistically significant) decrease in claudin-5 expression compared with seronegative controls. (C) Double immunostaining of MVs for human CD163 (green) and VWF (red) showed that the isolated MVs did not have macrophage contamination. (D-F) Exposure of HBMECs to HIV-1 (MOI: 0.01) for 24 hours decreased the expression of claudin-5, ZO-1 and ZO-2. Densitometry analysis showed that FLUD significantly diminished HIV-induced down-regulation of claudin-5 (E) and ZO-1 (F). Original magnification of images in panels A and C: ×400.
Expression of TJ proteins in HBMECs and microvessels from autopsied brains of seronegative controls and HIV-1–infected humans. (A) All MV samples stained positive for claudin-5. MVs from seronegative control donors (eg, patients N1 and N2) show prominent and continuous strands of tight junction (TJ, arrows); TJ strands in MVs from HIV-1–seropositive patients without HIVE (eg, P1 and P2), and HAD HIVE patients (eg, HAD2 and HAD5) were fewer and had more gaps (arrowheads). Patient HAD5 shows very few TJ strands. (B) Computer-assisted semiquantitative analyses of all MV samples show a significant decrease in claudin-5 expression in MVs from HAD patients, compared with MVs from HIV-1–seropositive patients without HIVE (HIV-POS), or seronegative controls (HIV-NEG). MVs from HIV-1–seropositive patients without HIVE also showed a small (nonstatistically significant) decrease in claudin-5 expression compared with seronegative controls. (C) Double immunostaining of MVs for human CD163 (green) and VWF (red) showed that the isolated MVs did not have macrophage contamination. (D-F) Exposure of HBMECs to HIV-1 (MOI: 0.01) for 24 hours decreased the expression of claudin-5, ZO-1 and ZO-2. Densitometry analysis showed that FLUD significantly diminished HIV-induced down-regulation of claudin-5 (E) and ZO-1 (F). Original magnification of images in panels A and C: ×400.
Our previous works demonstrated that coculture of HBMECs with infected MDMs induces transcriptional up-regulation of inflammatory molecules, including chemokines, cytokines, and IFN-inducible genes.33 To correlate these results with in vivo situation, we performed qRT-PCR with: (1) HBMECs cocultured with infected or noninfected MDMs; and (2) brain MVs from HIV + no HIVE, HAD with HIVE, and seronegative control patients. Data showed similar patterns of HIV-1–induced transcriptional up-regulation of CXCL1, TNFAIP3, TNFSP15, SOD2, UBD, ISG15, and RelB in endothelial cell cultures and brain MVs from HIV-1–infected humans (Figure 6A). Both MVs from HIV + no HIVE and HAD HIVE patients showed transcriptional up-regulation of CXCL1, TNFAIP3, SOD2, UBD, ISG15, and RelB, compared with MVs from seronegative patients, with levels of SOD2 and UBD mRNA higher in HAD HIVE than in HIV + no HIVE (Figure 6A). HAD HIVE patients showed increased levels of TNFSP15 mRNA, while levels in HIV + no HIVE were similar to that of control seronegatives (Figure 6A). Our data also demonstrated similar patterns of HIV-1–induced transcriptional up-regulation of STAT1 and IL-6 in endothelial cell cultures and in MVs of infected humans (Figure 6B-E).
Quantitative RT-PCR of patients' brain MVs and HIV-1–exposed HBMECs. (A) Data showed similar pattern of HIV-1–induced up-regulation of mRNA for inflammatory cytokines, oxidative stress enzymes, ubiquitin-related genes, and the transcription factor RelB in tissue culture and patients' brain MVs. Both coculture of HBMECs with infected MDMs (B) and direct exposure of HBMECs to infectious viral particles (C) induced transcriptional up-regulation of STAT1. Data demonstrated similar pattern of HIV-1–induced transcriptional up-regulation of STAT1 and IL-6 in endothelial cell cultures and in brain MVs of infected humans (B-E). For coculture experiments, controls are untreated endothelial cells. Each group (control, NI, and HIV) had 3 replicate samples; each sample was tested in triplicate and normalized to its GAPDH. (*P < .05; **P < .01; ***P < .001.) NI indicates HBMECs cocultured with noninfected MDMs; HIV, HBMECs coculture with HIV-infected MDMs; HIV-neg, MVs from HIV-seronegative donors [donors N1 to N5]; HIV-pos, MVs from HIV + no HIVE donors [donors P1 to P4]; and HAD, MVs from HAD patients [donors HAD1 to HAD5]).
Quantitative RT-PCR of patients' brain MVs and HIV-1–exposed HBMECs. (A) Data showed similar pattern of HIV-1–induced up-regulation of mRNA for inflammatory cytokines, oxidative stress enzymes, ubiquitin-related genes, and the transcription factor RelB in tissue culture and patients' brain MVs. Both coculture of HBMECs with infected MDMs (B) and direct exposure of HBMECs to infectious viral particles (C) induced transcriptional up-regulation of STAT1. Data demonstrated similar pattern of HIV-1–induced transcriptional up-regulation of STAT1 and IL-6 in endothelial cell cultures and in brain MVs of infected humans (B-E). For coculture experiments, controls are untreated endothelial cells. Each group (control, NI, and HIV) had 3 replicate samples; each sample was tested in triplicate and normalized to its GAPDH. (*P < .05; **P < .01; ***P < .001.) NI indicates HBMECs cocultured with noninfected MDMs; HIV, HBMECs coculture with HIV-infected MDMs; HIV-neg, MVs from HIV-seronegative donors [donors N1 to N5]; HIV-pos, MVs from HIV + no HIVE donors [donors P1 to P4]; and HAD, MVs from HAD patients [donors HAD1 to HAD5]).
We used Western blotting and confocal microscopy to further analyze STAT1 expression in human brain MVs. Significantly, both Western blot (Figure 7A) and confocal microscopy (Figure 7B) showed overexpression of STAT1 and phospho-STAT1 (S727) in brain MVs of HIV-infected individuals compared with MVs obtained from seronegative control subjects. The highest expression and activation of STAT1 were observed in MVs of HAD patients. Semiquantitative analysis of immunostained MVs showed a 4- and 4.8-fold increase in phospho-STAT1 in MVs of HIV-1–seropositive subjects with and without HAD compared with controls (P < .001; Figure 7C). Similarly, there was a 2.4- and 2.6-fold increase in STAT1 in MVs of HIV-1–seropositive and HAD patients, respectively, compared with MVs from HIV-1,2–seronegative controls (P < .01; Figure 7D). Again STAT1 phosphorylation at tyrosine residues was not detected in MV samples. These observations confirmed that HIV-1 activates STAT1 specifically at serine residues.
Increased expression and activation of STAT1 in brain MVs of HIV-1–infected humans. (A) Western blot analysis showed increased expression of STAT1 and phospho-STAT1 (S727) in brain MVs of some HIV-1–seropositive patients and patients with HAD and HIVE. No phosphorylation of STAT1 at tyrosine residues was detected. (B-D) Confocal microscopy showed increased expression of STAT1 and phospho-STAT1 (S727) in brain MVs of HIV-1–seropositive and HAD HIVE patients, compared with MVs from HIV-seronegative controls. Computer-assisted semiquantitative analyses of all MV samples showed a significant increase in phospho-STAT1 (C) and STAT1 (D) in brain MVs of both HIV + no HIVE and HAD patients. (**P < .01; ***P < .001.) Original magnification of images in panel B, ×400.
Increased expression and activation of STAT1 in brain MVs of HIV-1–infected humans. (A) Western blot analysis showed increased expression of STAT1 and phospho-STAT1 (S727) in brain MVs of some HIV-1–seropositive patients and patients with HAD and HIVE. No phosphorylation of STAT1 at tyrosine residues was detected. (B-D) Confocal microscopy showed increased expression of STAT1 and phospho-STAT1 (S727) in brain MVs of HIV-1–seropositive and HAD HIVE patients, compared with MVs from HIV-seronegative controls. Computer-assisted semiquantitative analyses of all MV samples showed a significant increase in phospho-STAT1 (C) and STAT1 (D) in brain MVs of both HIV + no HIVE and HAD patients. (**P < .01; ***P < .001.) Original magnification of images in panel B, ×400.
Discussion
In HIV-1–infected individuals, BBB damage impairs its ability to protect the CNS, enabling infected cells and toxins from the peripheral blood to reach the brain and elicit neuronal injury. Elucidating the mechanisms by which HIV invades the nervous system and induces neurologic deficits and the signaling pathways responsible for BBB compromise are keys to better understand HIV neuropathogenesis and offer opportunities to improve therapies to combat disease. The foundation for the work contained in this report comes from observations made by microarray analysis, which showed a prominent relationship between IL-6, STAT1, and other proinflammatory genes activated in HIV-1–exposed HBMECs.33 In the present study, we demonstrate that HIV-1 induces expression of IL-6 and IL-6R in HBMECs, and STAT1 mediates HIV-induced IL-6 expression and BBB dysfunction. Previous studies suggested that HIV-1 infection dysregulates the expression of IL-6 and IL-6R. HIV-infected individuals showed increased expression of IL-6 and IL-6R in monocytes, and B and T cells.46 Monocytic cell lines expressing IL-6R secreted more IL-6, which up-regulated HIV-1 expression, and antibodies to IL-6R inhibited IL-6–induced HIV-1 expression.47 This suggests that increased expression of IL-6 and IL-6R can potentiate viral-induced effects on the brain endothelium and contribute to HIV-induced BBB dysfunction. In fact, HIV-1 and IL-6 synergistically increase monocyte migration across in vitro BBB models. Higher serum IL-6 in HIV-1–infected humans correlates with increased progression to AIDS and neuroAIDS.48,–50 High levels of IL-6 are found in the CSF and blood cells of HAD patients and are associated with worse cognitive and motor performance.51,,–54
The JAK/STAT pathway plays a prominent role in cytokine-mediated inflammatory responses, and STAT1 has been implicated in the pathogenesis of HIV-1 infection and disease progression.29,30 There is enhanced activation of STAT1 in T cells from HIV-1–infected individuals29 and increased expression of STAT1 in brain tissues of HIV-1–infected mice55 and simian immunodeficiency virus (SIV)–infected monkeys.56 Infection with HIV-1 or HTLV-1 induces STAT1 activation in human lymphocytes.30 The present study demonstrates that IL-6 is involved in HIV-induced endothelial cell dysfunction, and STAT1 mediates these effects. FLUD is an anti-inflammatory compound57 that is clinically approved and currently used in humans for the treatment of hematologic malignancies,58,59 and also shows antiviral effects in SIV-infected monkeys.42 FLUD has been shown to specifically induce loss of STAT1 mRNA and proteins, as well as loss of STAT1-mediated gene activation in vivo and in vitro.41 However, the precise mechanism of FLUD-mediated inhibition of STAT1 has not been elucidated. There is evidence that deletion of DNA methyltransferase in brain precursor cells results in DNA hypomethylation, enhanced phosphorylation of STAT1 and STAT3, and cell differentiation.60 Thus, if HIV-induced up-regulation/activation of STAT1 involves demethylation of STAT1 gene, FLUD could prevent STAT1 activation by inhibiting viral-induced demethylation. There is also evidence that FLUD down-regulates DNA synthesis by inhibiting the primase activity associated with DNA polymerase-alpha, thereby inhibiting RNA-primed DNA synthesis.61 Our future studies will further test these hypotheses to investigate the mechanisms of FLUD-mediated STAT1 inhibition in HIV-induced BBB dysfunction.
HBMEC and MV data presented in this study are significant; and to our knowledge, this is the first demonstration of STAT1 involvement in HIV-1–induced endothelial dysfunction and BBB injury. STAT1 and IL-6 mRNA were higher in MVs from HIV + no HIVE patients, compared with MVs from HAD HIVE patients, while expression of STAT1 and phospho-STAT1 were higher in MVs of HAD HIVE patients. This may suggest that during HIV-1 infection, transcriptional up-regulation of proinflammatory cytokines and STAT1 precedes cytokine expression, STAT1 activation, inflammation, and neuroAIDS. SOD2 and UBD mRNA increased in HIV-1–seropositive MVs, but the levels were much higher in MVs from HAD patients, suggesting increased oxidative stress and impairment of the ubiquitin-proteasome system in HIVE. TNFSF15 is a potent inhibitor of endothelial cell proliferation that induces expression of proinflammatory cytokines, apoptosis, and senescence in endothelial cells.62 Increased levels of TNFSF15 in HIV-exposed HBMECs and HAD HIVE MVs further demonstrate vascular damage in HIVE.
In the JAK/STAT pathway, phosphorylated STATs form dimers and associate with the IFN-stimulated gene factor 3 gamma (ISGF3G) to form a complex transcription factor (ISGF3) that translocates to the nucleus and binds to the IFN-stimulated response element (ISRE) to activate the transcription of interferon- or cytokine-stimulated genes.25 We previously showed that HIV-1 induces transcriptional up-regulation of ISGF3G and IRF7, both downstream effectors of STAT1.33 We demonstrate that STAT1 and STAT3 are the STAT family members activated by HIV-1, which suggests that in HIV-induced BBB dysfunction, the STAT dimers that translocate to the nucleus consist of STAT1-STAT3 heterodimers. The exact mechanism through which HIV-1 activates STAT1 and STAT3 remains to be elucidated. It is possible that HIV-1 up-regulates STAT expression by down-regulating DNA methyltransferase, since hypomethylation enhanced expression and phosphorylation of STAT1 and STAT3.60 STAT1 transactivating domain resides in the C-terminus.63 Phosphorylation of S727 increases the transcriptional activity of STAT1 dimers and greatly increases the ability of STAT1 to interact with ISGF3G.64,65 Mutation of the C-terminal S727 to alanine decreases the transcriptional activity of STAT1,66 and severely reduces STAT1 ability to induce antiproliferative or viability effects.67,68 Furthermore, STAT1 transactivating domain and its phosphorylation at S727 are necessary for the complete induction of gene expression and establishment of antiviral response to type I IFN.26
The main feature of the brain endothelium is the presence of TJs between endothelial cells, which restrict BBB permeability.37 HIV-1 and viral proteins disrupt HBMEC TJ proteins.19,38 In HIV and SIV encephalitis, there is evidence that BBB perturbation is associated with disruption of TJs and increased trafficking of HIV-infected MDMs into the CNS.16,69,70 We now provide new insights into this seemingly complex process. Indeed, the evidence provided shows significant decrease in claudin-5 and TJ strands in MVs from autopsied brains of HAD patients, and clearly demonstrate that STAT1 plays a major role in HIV-induced inflammation and BBB damage. We further demonstrate decreased claudin-5, ZO-1, and ZO-2 expression in HIV-1–exposed HBMECs and show that FLUD diminished viral-induced down-regulation of TJ proteins. Perhaps most importantly, our data suggest that inhibiting STAT1 activation could provide a unique therapeutic strategy to prevent HIV-1–induced BBB dysfunction and as such improve clinical outcomes in infected humans.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the NNTC and the CNND brain bank for providing human brain tissues specimens. We thank Debbie Baer and Robin Taylor for excellent editorial support and critical reading of the paper, and Dr Servio Ramirez for help with the migration procedure.
This work was supported in part by National Institutes of Health grants MH068214 (G.D.K.), NS36126 and NS43985 (H.E.G.), and MH65151 (Y.P.).
National Institutes of Health
Authorship
Contribution: C.A. and B.Y. performed research; H.E.G. and Y.P. provided assistance in data analysis and edited the paper; G.D.K. designed and performed research, collected and analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Georgette D. Kanmogne, Department of Pharmacology and Experimental Neuroscience, Center for Neurovirology and Neurodegenerative Disorders, University of Nebraska Medical Center 985215 NE Medical Center, Omaha, NE 68198-5215; e-mail: gkanmogne@unmc.edu.

![Figure 6. Quantitative RT-PCR of patients' brain MVs and HIV-1–exposed HBMECs. (A) Data showed similar pattern of HIV-1–induced up-regulation of mRNA for inflammatory cytokines, oxidative stress enzymes, ubiquitin-related genes, and the transcription factor RelB in tissue culture and patients' brain MVs. Both coculture of HBMECs with infected MDMs (B) and direct exposure of HBMECs to infectious viral particles (C) induced transcriptional up-regulation of STAT1. Data demonstrated similar pattern of HIV-1–induced transcriptional up-regulation of STAT1 and IL-6 in endothelial cell cultures and in brain MVs of infected humans (B-E). For coculture experiments, controls are untreated endothelial cells. Each group (control, NI, and HIV) had 3 replicate samples; each sample was tested in triplicate and normalized to its GAPDH. (*P < .05; **P < .01; ***P < .001.) NI indicates HBMECs cocultured with noninfected MDMs; HIV, HBMECs coculture with HIV-infected MDMs; HIV-neg, MVs from HIV-seronegative donors [donors N1 to N5]; HIV-pos, MVs from HIV + no HIVE donors [donors P1 to P4]; and HAD, MVs from HAD patients [donors HAD1 to HAD5]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/4/10.1182_blood-2007-05-091207/3/m_zh80060813940006.jpeg?Expires=1767863885&Signature=YuJU5z83Q5wePXeD9YPG7N65uDITs~T~SbqdnrXnT7rNJpxeD-Uk8zPquOLBeb9AB-f51UcleCbnUESRqxtN9VJvtAw9vvTSis-5~LpfDFdvBaSP39N~jAfLARFDwjbo55b6xVRjP9k8xyIVxtkqspk9xg~lGe8ym8m5u4KQ2bQ4WaJTHLFYOQebugui4zrO7zy-Pdy1kB-2wP-Om1qO2skqgmjeZ7LUQc2g5pHbEQcD~uoOU3tV75kyPY64Zdt5iugcWf~lYKJI55NnW~QGet8gwJOPjOJxSL~WdsUaNqEPpStvl0jdFiK1pyrK2-a2Ppi~drymBbG~4jIhCZTMwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)