Abstract
Bortezomib (PS-341), a proteasome inhibitor, has been examined clinically for the treatment of multiple myeloma and several solid tumors. Bortezomib directly induces tumor cell death and has also been reported to inhibit tumor adaptation to hypoxia by functionally inhibiting hypoxia-inducible factor-1α (HIF-1α). However, the mechanism underlying HIF-1 inhibition by bortezomib remains obscure. In the present study, we demonstrated that bortezomib attenuated the hypoxic induction of erythropoietin and vascular endothelial growth factor at subnanomolar concentrations in multiple myeloma and liver cancer cell lines, regardless of cytotoxic concentrations of bortezomib. Bortezomib repressed HIF-1α activity by inhibiting the recruitment of p300 coactivator. Specifically, bortezomib targeted HIF-1α C-terminal transactivation domain (CAD) but not the CAD lacking Asn803, which is a hydroxylation site by the factor inhibiting HIF-1 (FIH). Accordingly, this effect of bortezomib on CAD was augmented by FIH expression and abolished by FIH knock-down. Furthermore, bortezomib stimulated the interaction between CAD and FIH under hypoxic conditions, and FIH inhibition reversed the suppressions of erythropoietin and vascular endothelial growth factor by bortezomib. We propose that the mechanism underlying the inhibitory effects of bortezomib on tumor angiogenesis and hypoxic adaptation involves the repression of HIF-1α transcriptional activity by reinforcing the FIH-mediated inhibition of p300 recruitment.
Introduction
Hypoxia commonly develops in solid tumors because tumor growth outpaces vessel formation and because the blood supply is compromised due to aberrant vasculature formation.1 Tumor hypoxia contributes to angiogenesis and modulates tumor energy metabolism, which are both essential required for tumor growth.2 In multiple myeloma (MM), hypoxia is also an important environmental factor because bone marrow is intrinsically hypoxic in nature.3 Thus, MM cells must survive and grow under such hypoxic conditions, and this requires the expressions of many genes essential for adaptation. Hypoxic adaptation is mainly provided by hypoxia-inducible factor-1 (HIF-1), which orchestrates cellular adaptation to hypoxia by transactivating about 60 genes.4
HIF-1 is composed of HIF-1α and HIF-1β/aryl hydrocarbon nuclear translocator (ARNT),5 and of these, HIF-1α is the key protein that determines the presence of HIF-1 and transactivates genes. Under normoxic conditions, HIF-1α is hydroxylated at its Pro402 and Pro564 residues by HIF-1 prolyl hydroxylases (PHDs), and thus, targeted by von Hippel-Lindau protein (pVHL), ubiquitinated, and finally degraded by 26S proteasomes.6-10 In addition, the C-terminal transactivation domain (CAD) of HIF-1α is hydroxylated at Asn803 by the factor inhibiting HIF-1 (FIH), which represses the transcriptional activity of HIF-1α by blocking the recruitment of p300 coactivator.11,12 However, PHD and FIH activities depend on oxygen tension, and as a result HIF-1α is stabilized and activated under hypoxic conditions, because both proline and asparagine hydroxylation are inhibited. Moreover, HIF-1α associates with ARNT to form HIF-1 in the nucleus, which then transactivates genes such as erythropoietin (EPO), vascular endothelial growth factor (VEGF), and carbonic anhydrase IX (CAIX).
Bortezomib (PS-341; Velcade, Cambridge, MA) is the only proteasome inhibitor that has been examined clinically for the treatment of MM and several solid tumors.13 Bortezomib directly induces tumor cell death by inhibiting the proteasome-mediated regulation of proapoptotic and antiapoptotic proteins.14 In addition, bortezomib indirectly inhibits angiogenesis in MM and neuroblastoma by decreasing VEGF levels, and prevents tumor adaptation to hypoxia by downregulating CAIX.15-17 Mechanistically, these indirect effects of bortezomib might be due to the functional inhibition of HIF-1α,18,19 but little is known about the molecular mechanism underlying HIF-1α inactivation by bortezomib. In the present study, we identified the mechanism underlying bortezomib-induced HIF-1α repression.
Methods
Reagents and antibodies
Dimethyloxalylglycine (DMOG) was obtained from Frontier Scientific (Logan, UT), and [α-32P]CTP (1850 × 1010 Bq [500 Ci/mmol]) was obtained from NEN Life Science (Boston, MA). Culture media and fetal calf serum were purchased from GIBCO/BRL (Grand Island, NY). Other chemicals were purchased from Sigma-Aldrich (St Louis, MO). Anti–HIF-1α antiserum was generated in rabbits20 and anti–OH-P564 HIF-1α antiserum was provided by Dr Huang (University of Utah, Salt Lake City). Other antibodies (anti-ARNT, anti-FIH, anti-p300, anti-CITED2, and anti–β-tubulin) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
Hep3B (human hepatoma), HEK293 (human embryonic kidney), ARH77 (human multiple myeloma), and U299 (human multiple myeloma) cell-lines were obtained from American Type Culture Collection (ATCC; Manassas, VA), and cultured in α-modified Eagle medium, Dulbecco modified Eagle medium, or RPMI-1640, which were supplemented with 10% heat-inactivated fetal calf serum in a humidified atmosphere containing 5% CO2 at 37°C. O2/CO2 levels in the incubator were 20%/5% (normoxic) or 1%/5% (hypoxic).
Preparation of plasmids and siRNAs
Hemagglutinin (HA)–tagged HIF-1α expression plasmid (pcDNA3) was constructed as described previously.21 EPO enhancer-luciferase and VEGF promoter-luciferase reporter genes, HA-FIH plasmid, and FIH siRNA duplex were constructed as previously described.21,22 The Gal4-CAD plasmid was constructed by inserting HIF-1α CAD (aa 776-826) into pCMX-G4(N), and the Gal4-CAD(N803A) plasmid was generated by site-directed mutagenesis.22 The VP16-CH1 plasmid was constructed by inserting the CH1 domain of p300 into the VP16 plasmid (Clontech, Palo Alto, CA).22 For transient transfection with plasmids or siRNA, about 40% of confluent cells in 60-mm cell-culture dishes were transfected with plasmid or siRNA using calcium phosphate or electroporation method. Cells were allowed to stabilize for 48 hours before being used in experiments.
Chemosensitivity assay
To evaluate chemosensitivities of 3 cell lines to bortezomib, the Sigma-Aldrich MTT labeling kit was used. Cells were grown at a cell density of 105 cells/mL in 12-well plates. After incubating with bortezomib for 48 hours, 100 μL of an MTT labeling reagent (5 mg/mL) was added, and incubation was continued in the CO2 chamber for 3 hours. After solubilizing the blue formazan crystal with acidified isopropanol, formazan levels were determined at 570 nm and expressed as percentages of control.
Reporter and mammalian 2-hybrid assays
Cells were cotransfected with EPO or VEGF reporter gene or each of Gal4 reporter gene and Gal4-CAD plasmid. To examine HIF-1α–p300 binding, HEK293 cells were cotransfected with a Gal4-luciferase reporter, Gal4-CAD plasmid, VP16-CH1 plasmid. The β-gal expression plasmid was cotransfected to normalize gene expression. After 16 hours of stabilization, transfected cells were split into 4 aliquots at a cell density of 5 × 104 cells/cm2 and further cultured for 24 hours. The cells were incubated under normoxic or hypoxic conditions, in the absence or presence of bortezomib for 16 hours, and then lysed to determine luciferase and β-gal activities.
Immunoblotting and Immunoprecipitation
Total cell lysates were separated on 6.5% or 8% sodium dodecylsulfate (SDS)/polyacrylamide gels, and transferred to an Immobilon-P membrane (Millipore, Bedford, MA). Membranes were blocked with 5% nonfat milk in Tris/saline containing 0.1% Tween-20 for 1 hour and then incubated overnight at 4°C with a primary antibody diluted 1:1000. Membranes were incubated with a horseradish peroxidase (HRP)–conjugated secondary antibody (1:5000) for 2 hours, and stained using an ECL Plus kit (Amersham Biosciences, Piscataway, NJ). To analyze HIF-1α–FIH interaction, cell lysates were incubated with anti–HIF-1α or anti-FIH antibody for 4 hours, and the immune complexes were precipitated with protein A/G beads. Coprecipitated FIH or HIF-1α was identified by immunoblotting with anti-FIH or anti–HIF-1α antibody, respectively. The HIF-1α–p300 interaction was analyzed by immunoprecipitation with anti-p300 antibody and immunoblotting with anti–HIF-1α antibody.
Semiquantitative RT-PCR
To quantify EPO, VEGF, or β-actin mRNA levels, a highly sensitive, semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed, as previously described.21 Total RNAs were isolated using TRIZOL (Invitrogen, Carlsbad, CA). RNAs (1 μg) were reverse-transcribed at 48°C and the cDNAs were amplified over 18 to 23 PCR cycles in a 20 μL of reaction mixture containing 0.185 MBq (5 μCi) [α-32P]dCTP and 250 nM of each primer set. The PCR products were electrophoresed on a 4% polyacrylamide gel, and dried gels were autoradiographed. Primers were designed as previously described.19
Statistical analysis
Data (means and SDs) and the unpaired Student t test were analyzed using Microsoft Excel 2002 software (Redmond, WA). Differences were considered significant when P was less than .05. All statistical tests were 2-sided.
Results
Bortezomib blunts EPO and VEGF responses to hypoxia, regardless of chemosensitivity
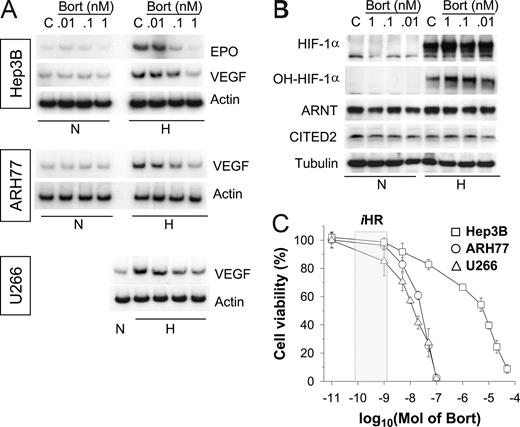
To investigate hypoxic cellular responses, we first used the Hep3B cell line, which is widely used to study the HIF-mediated oxygen-sensing mechanism. Initially, we treated cells with bortezomib at nanomolar concentrations, because bortezomib has been commonly administered in the 1- to 200-nM range during investigations of its apoptotic or antiangiogenic effects.13 Although HIF-1α levels were increased by bortezomib (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), EPO mRNA expression was completely suppressed at concentrations as low as 1 nM (Figure S1B). Therefore, we readjusted bortezomib doses, and subsequently found that bortezomib inhibited EPO and VEGF at subnanomolar levels (Figure 1A; top panel). Moreover, it is likely that the hypoxic induction of VEGF mRNA was also attenuated by low concentrations of bortezomib in 2 MM cell lines, ARH77 and U266 (Figure 1A middle and bottom panels). However, at these subnanomolar concentrations, bortezomib showed little effect on the stabilizations of HIF-1α and hydroxylated HIF-1α (Pro564; Figure 1B). To examine the chemosensitivities of cell lines to bortezomib, cell viabilities were analyzed after treating cells with bortezomib for 48 hours. The bortezomib concentration required for 50% inhibition (IC50) was around 10 nM for the 2 MM cell lines, but exceeded 1 μM in Hep3B, indicating substantial differences between the chemosensitivities of MM and non-MM cells. Nonetheless, VEGF response to hypoxia was blunted by bortezomib at 1 nM or less in both MM and Hep3B cell lines, which suggests that the bortezomib-induced suppressions of EPO and VEGF are not directly related to its cytotoxicity.
Bortezomib inhibits the hypoxic induction of EPO and VEGF in both MM and non-MM cells. (A) Effects of bortezomib on the hypoxic induction of EPO or VEGF mRNA. Hep3B, ARH77, and U266 cells were incubated under normoxic (N) or hypoxic (H) conditions for 16 hours in the presence of various concentrations of bortezomib (Bort). Total RNAs were extracted, and EPO, VEGF, and β-actin mRNA were analyzed by semiquantitative RT-PCR. (B) Effects of bortezomib on expressions of HIF-1 subunits. Under normoxic or hypoxic conditions for 16 hours, Hep3B cells were lysed in a denaturing SDS sample buffer, and HIF-1α, ARNT, and β-tubulin proteins were analyzed by Western blotting. The data shown are representative of 3 separate experiments. (C) Chemosensitivity to bortezomib. Cell viabilities were measured using an MTT-labeling method, as described in “Chemosensitivity assays.” Hep3B (□), ARH77 (○), and U266 (△) cells were treated with various concentrations of bortezomib for 48 hours. Points represent the means plus or minus SD of 6 experiments. The iHR box indicates the concentration range showing inhibited hypoxic responses.
Bortezomib inhibits the hypoxic induction of EPO and VEGF in both MM and non-MM cells. (A) Effects of bortezomib on the hypoxic induction of EPO or VEGF mRNA. Hep3B, ARH77, and U266 cells were incubated under normoxic (N) or hypoxic (H) conditions for 16 hours in the presence of various concentrations of bortezomib (Bort). Total RNAs were extracted, and EPO, VEGF, and β-actin mRNA were analyzed by semiquantitative RT-PCR. (B) Effects of bortezomib on expressions of HIF-1 subunits. Under normoxic or hypoxic conditions for 16 hours, Hep3B cells were lysed in a denaturing SDS sample buffer, and HIF-1α, ARNT, and β-tubulin proteins were analyzed by Western blotting. The data shown are representative of 3 separate experiments. (C) Chemosensitivity to bortezomib. Cell viabilities were measured using an MTT-labeling method, as described in “Chemosensitivity assays.” Hep3B (□), ARH77 (○), and U266 (△) cells were treated with various concentrations of bortezomib for 48 hours. Points represent the means plus or minus SD of 6 experiments. The iHR box indicates the concentration range showing inhibited hypoxic responses.
Bortezomib at subnanomolar concentrations does not induce CITED2, an HIF-1 inhibitor
CITED2 (CBP/p300 interacting transactivator with ED-rich tail 2; alternatively named Mrg1 or p35srj) has been reported to inhibit the transcriptional activity of HIF-1α.23 Mechanistically, CITED2 competes with HIF-1α for binding to the CH1 domain of p300, which results in the repression of HIF-1α activity due to its dissociation from p300.24 Recently, it was demonstrated that CITED2 is up-regulated by MG132 proteasome inhibitor, and that this causes HIF-1 inactivation even under hypoxic conditions.25 Therefore, we tested the possibility that CITED2 induction is responsible for HIF-1 inactivation by bortezomib. However, CITED2 expression was not induced by bortezomib at 1 nM or less (Figure 1B), though higher concentrations of bortezomib successfully induced CITED2 (Figure S1A). These results suggest that CITED2 is not directly related with HIF-1 inactivation by subnanomolar bortezomib.
Bortezomib represses the transcriptional activity of HIF-1 by inhibiting p300 recruitment
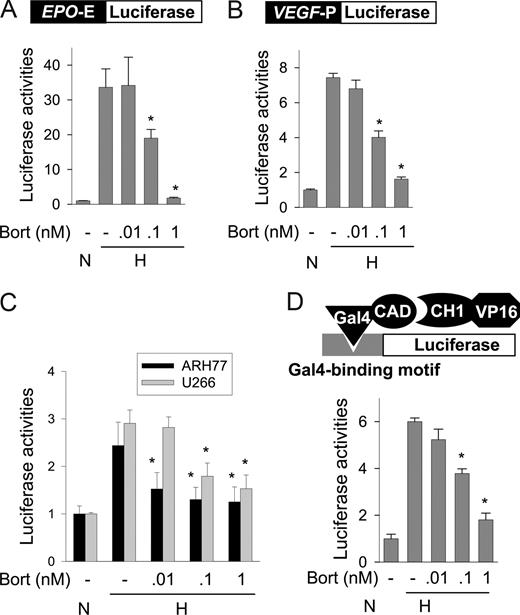
Hypoxia induces EPO and VEGF transcription due to the targeting of EPO 3′ enhancer and VEGF 5′ promoter, respectively, by HIF-1. To examine whether bortezomib inhibits HIF-1–mediated gene transcription, we measured the activities of an EPO reporter and of a VEGF reporter, both of which were designed to analyze HIF-1–specific transcription. In hypoxic Hep3B cells, EPO and VEGF reporter activities were 33- and 7.4-fold higher, respectively, than in normoxic cells. However, bortezomib at 1 nM or less significantly inhibited both EPO enhancer (Figure 2A) and VEGF promoter (Figure 2B) activities. Moreover, VEGF reporter activities in MM cells were increased under hypoxic conditions, and were also repressed by bortezomib at 1 nM or less (Figure 2C). Since p300 binding is essential for HIF-1–dependent transcription, we examined the interaction between HIF-1α and p300 using a mammalian 2-hybrid assay, in which reporter activity was determined by the interaction between Gal4–HIF-1α CAD and VP16-p300 CH1. Figure 2D shows that the HIF-1α–p300 interaction was enhanced by hypoxia and that this interaction was inhibited by bortezomib. These results suggest that bortezomib at subnanomolar concentrations inhibits p300 recruitment by HIF-1α, and thus represses the transcriptional activity of HIF-1α.
Bortezomib represses HIF-1 activity by inhibiting p300 binding to HIF-1α. (A,B) EPO enhancer or VEGF promoter activity in Hep3B cells. Luciferase reporter plasmids (0.5 μg of DNA) containing Epo enhancer (A) or VEGF promoter (B) were cotransfected with 0.5 μg of plasmid cytomegalovirus–β-gal into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured using a Biocounter M1500 luminometer (Lumac, Bad Wildbad, Germany), and transfection efficiencies were normalized versus β-gal activity. (C) EPO enhancer activity in MM cells. Luciferase reporter plasmids (1 μg) containing EPO enhancer were cotransfected with 1 μg of plasmid cytomegalovirus–β-gal into ARH77 or U266 cells. After a 16-hour normoxic or hypoxic incubation with bortezomib, luciferase activity/β-gal activity was measured. (D) Mammalian 2-hybrid assay of HIF-1α CAD-p300 binding. Hep3B cells were cotransfected with 1 μg of Gal4-luciferase reporter, 1 μg of pGal4-CAD, and 0.5 μg of pVP16-CH1. The cells were incubated under normoxic or hypoxic conditions with bortezomib for 16 hours, and then lysed to determine luciferase activity. All results are presented as relative values versus the normoxic control, and are plotted as means plus or minus SEs of 8 experiments. *P < .05 versus the hypoxic control.
Bortezomib represses HIF-1 activity by inhibiting p300 binding to HIF-1α. (A,B) EPO enhancer or VEGF promoter activity in Hep3B cells. Luciferase reporter plasmids (0.5 μg of DNA) containing Epo enhancer (A) or VEGF promoter (B) were cotransfected with 0.5 μg of plasmid cytomegalovirus–β-gal into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured using a Biocounter M1500 luminometer (Lumac, Bad Wildbad, Germany), and transfection efficiencies were normalized versus β-gal activity. (C) EPO enhancer activity in MM cells. Luciferase reporter plasmids (1 μg) containing EPO enhancer were cotransfected with 1 μg of plasmid cytomegalovirus–β-gal into ARH77 or U266 cells. After a 16-hour normoxic or hypoxic incubation with bortezomib, luciferase activity/β-gal activity was measured. (D) Mammalian 2-hybrid assay of HIF-1α CAD-p300 binding. Hep3B cells were cotransfected with 1 μg of Gal4-luciferase reporter, 1 μg of pGal4-CAD, and 0.5 μg of pVP16-CH1. The cells were incubated under normoxic or hypoxic conditions with bortezomib for 16 hours, and then lysed to determine luciferase activity. All results are presented as relative values versus the normoxic control, and are plotted as means plus or minus SEs of 8 experiments. *P < .05 versus the hypoxic control.
HIF-1α repression by bortezomib requires Asn803 and depends on FIH
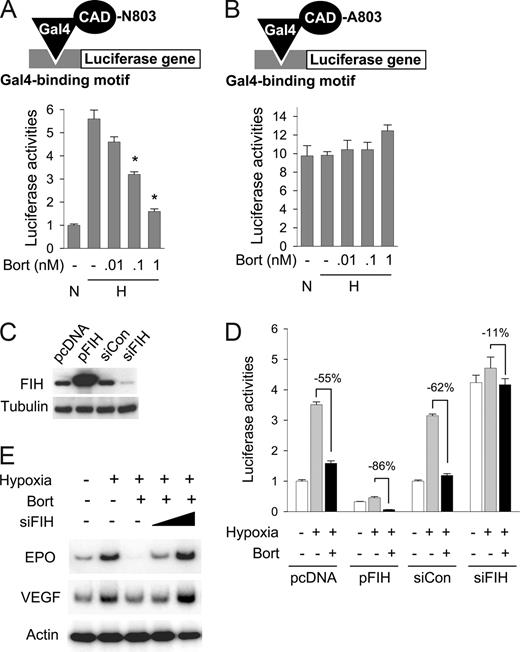
To examine whether bortezomib specifically inhibits the CAD of HIF-1α, we coexpressed a Gal4-CAD fusion protein with Gal4-Luc reporter plasmid in Hep3B cells. Since Gal4 fusion protein constitutively binds to the Gal4 promoter in the reporter plasmid, luciferase expression totally depends on CAD activity. CAD activity was found to be increased under hypoxic conditions by 5.6-fold, and this activation was significantly diminished by bortezomib (Figure 3A). However, when Asn803 in CAD was substituted with alanine, the CAD mutant showed neither hypoxic activation nor inactivation by bortezomib (Figure 3B), suggesting that Asn803 in CAD is required for the bortezomib effect. Moreover, since Asn803 hydroxylation by FIH is a key step in CAD inactivation, we tested the possibility that bortezomib stimulates this hydroxylation process by using FIH overexpression and knock-down, which were verified as shown in Figure 3C. CAD inactivation by bortezomib was augmented more so by FIH overexpression (−86%) than in mock control (−55%), and conversely this effect of bortezomib was reduced by FIH knock-down less so than in the siRNA control (by 11% and 62%, respectively; Figure 3D). Moreover, FIH siRNA was found to effectively rescue EPO and VEGF expressions suppressed by bortezomib (Figure 3E). These results suggest that bortezomib inactivates HIF-1α by augmenting CAD inhibition by FIH under hypoxic conditions, suggesting the possibility that FIH is activated under hypoxic (1% O2) conditions. A recent report demonstrated that CAD activity was regulated by FIH expression or knock-down even at an O2 tension of 1%,18 indicating that FIH regulates CAD even in hypoxia.
FIH is involved in bortezomib-induced HIF-1α repression. (A) Bortezomib inhibited the hypoxic activation of HIF-1α. Gal4-CAD plasmid was cotransfected with Gal4-luc reporter plasmid into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured. The results shown are presented as relative values versus the normoxic control, and are plotted as means plus or minus SEs of 8 experiments. *P < .05 versus the hypoxic control. The reporter system is illustrated in the top panel. (B) Asn803 is required for bortezomib-induced CAD repression. Plasmid for Gal4-CAD mutant, in which Asn803 was substituted with Ala (N803A), was cotransfected with Gal4-luc reporter plasmid into Hep3B cells, and luciferase activities were measured. Results are presented as relative values versus normoxic controls in panel A, and are plotted as means plus or minus SEs of 8 experiments. The reporter system is illustrated in the top panel. (C) Verification of FIH expression and knock-down. pcDNA (0.2 μg), HA-tagged FIH (pFIH) plasmid (0.2 μg) or 20 nM control RNA (siCon), or 20 nM FIH siRNA (siFIH) was transfected into HEK293 cells. After 48 hours, FIH and β-tubulin levels in cell lysates were measured using Western blotting. (D) The effect of bortezomib on HIF-1α activity depended on FIH expression. Indicated plasmids and siRNAs were cotransfected with Gal4-CAD and Gal4-luc reporter plasmids into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured. □, normoxia; ▩, hypoxia; ■, hypoxia plus bortezomib. (E) EPO and VEGF expressions reduced by bortezomib were rescued by inhibiting FIH. Hep3B cells were subjected to normoxia (N) or hypoxia for 16 hours, and then cotreated with bortezomib and FIH siRNA. EPO, VEGF, and β-actin mRNAs were isolated and analyzed by semiquantitative RT-PCR.
FIH is involved in bortezomib-induced HIF-1α repression. (A) Bortezomib inhibited the hypoxic activation of HIF-1α. Gal4-CAD plasmid was cotransfected with Gal4-luc reporter plasmid into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured. The results shown are presented as relative values versus the normoxic control, and are plotted as means plus or minus SEs of 8 experiments. *P < .05 versus the hypoxic control. The reporter system is illustrated in the top panel. (B) Asn803 is required for bortezomib-induced CAD repression. Plasmid for Gal4-CAD mutant, in which Asn803 was substituted with Ala (N803A), was cotransfected with Gal4-luc reporter plasmid into Hep3B cells, and luciferase activities were measured. Results are presented as relative values versus normoxic controls in panel A, and are plotted as means plus or minus SEs of 8 experiments. The reporter system is illustrated in the top panel. (C) Verification of FIH expression and knock-down. pcDNA (0.2 μg), HA-tagged FIH (pFIH) plasmid (0.2 μg) or 20 nM control RNA (siCon), or 20 nM FIH siRNA (siFIH) was transfected into HEK293 cells. After 48 hours, FIH and β-tubulin levels in cell lysates were measured using Western blotting. (D) The effect of bortezomib on HIF-1α activity depended on FIH expression. Indicated plasmids and siRNAs were cotransfected with Gal4-CAD and Gal4-luc reporter plasmids into Hep3B cells. After incubation under normoxic or hypoxic conditions for 16 hours with bortezomib, luciferase activities were measured. □, normoxia; ▩, hypoxia; ■, hypoxia plus bortezomib. (E) EPO and VEGF expressions reduced by bortezomib were rescued by inhibiting FIH. Hep3B cells were subjected to normoxia (N) or hypoxia for 16 hours, and then cotreated with bortezomib and FIH siRNA. EPO, VEGF, and β-actin mRNAs were isolated and analyzed by semiquantitative RT-PCR.
Bortezomib inhibits the interaction between HIF-1α and p300 by stimulating FIH
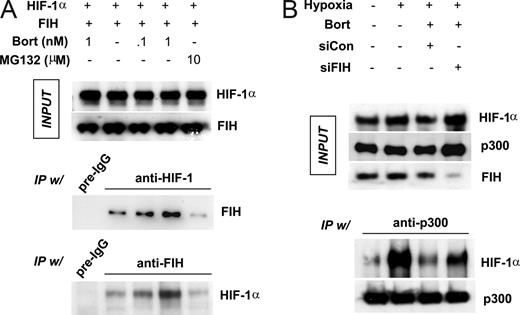
To determine whether bortezomib stimulates the interaction between FIH and HIF-1α, HEK293 cells were cotransfected with HIF-1α and FIH plasmids and incubated in hypoxia. Protein binding was cross-checked by immunoprecipitation and immunoblotting assays using anti–HIF-1α and anti-FIH antibodies. Our observations revealed that expressed protein levels were unchanged by drug treatment (Figure 4A top panel). Interestingly, FIH binding to HIF-1α was significantly and dose-dependently increased by bortezomib (Figure 4A middle and bottom panels). However, FIH binding was not enhanced by 10 μM MG132, even though 10 μM MG132 almost completely abolished proteasomal activity (Figure S2). In contrast, noticeable proteasomal activity remained in cells treated with subnanomolar concentrations of bortezomib. Based on these results, we cannot rule out the possibility that bortezomib specifically stimulates FIH independently of proteasomal inhibition. To re-examine the HIF-1α–p300 interaction in hypoxia, we coexpressed HIF-1α and FIH in HEK293 cells, and then incubated them under hypoxic conditions. Despite the expression of HIF-1α, HIF-1α did not coprecipitate with p300 in normoxia, whereas a large amount of HIF-1α was detected in p300 immunoprecipitates from hypoxic cells, thus confirming the oxygen-dependent nature of the interaction between HIF-1α and p300. Furthermore, the hypoxia-induced HIF-1α–p300 interaction was attenuated by bortezomib and noticeably recovered after knocking-down FIH (Figure 4B).
Bortezomib enhances FIH binding to HIF-1α, and then blocks p300 recruitment by HIF-1α. (A) Bortezomib stimulated the interaction between HIF-1α and FIH. HEK293 cells were cotransfected with FIH and HIF-1α plasmids and then incubated with bortezomib (0.1 and 1 nM) or MG132 (10 μM) under hypoxic conditions for 8 hours. Lysates were prepared and immunoprecipitated using anti–HIF-1α, anti-FIH, or nonimmunized rabbit serum (pre-igG). Coimmunoprecipitated FIH or HIF-1α was identified by Western blotting. (B) FIH knock-down rescues HIF-1α–p300 binding which is inhibited by bortezomib. HEK293 cells were cotransfected with HIF-1α (2 μg) and p300 (1 μg) plasmids, and incubated under normoxic or hypoxic conditions with 1 nM bortezomib for 8 hours. p300 was immunoprecipitated with anti-p300 antiserum and protein G/A beads, and precipitated p300 and coprecipitated HIF-1α was identified by Western blotting.
Bortezomib enhances FIH binding to HIF-1α, and then blocks p300 recruitment by HIF-1α. (A) Bortezomib stimulated the interaction between HIF-1α and FIH. HEK293 cells were cotransfected with FIH and HIF-1α plasmids and then incubated with bortezomib (0.1 and 1 nM) or MG132 (10 μM) under hypoxic conditions for 8 hours. Lysates were prepared and immunoprecipitated using anti–HIF-1α, anti-FIH, or nonimmunized rabbit serum (pre-igG). Coimmunoprecipitated FIH or HIF-1α was identified by Western blotting. (B) FIH knock-down rescues HIF-1α–p300 binding which is inhibited by bortezomib. HEK293 cells were cotransfected with HIF-1α (2 μg) and p300 (1 μg) plasmids, and incubated under normoxic or hypoxic conditions with 1 nM bortezomib for 8 hours. p300 was immunoprecipitated with anti-p300 antiserum and protein G/A beads, and precipitated p300 and coprecipitated HIF-1α was identified by Western blotting.
Discussion
In the present study, we investigated the molecular mechanism underlying HIF-1 repression by bortezomib. At subnanomolar concentrations, bortezomib blocked the hypoxic induction of EPO and VEGF in both drug-sensitive MM cell lines and a resistant Hep3B cell line. Bortezomib also attenuated the HIF-1–mediated hypoxic activations of EPO and VEGF reporter in these cell lines, and prevented p300 binding to HIF-1α CAD. Moreover, transcriptional inhibition by bortezomib was observed only for wild-type CAD containing the FIH target site, Asn803. In addition, CADinhibition and EPO/VEGF suppression by bortezomib did not occur in FIH knock-down cells, and furthermore, bortezomib stimulated the HIF-1α–FIH interaction even under hypoxic conditions and dissociated p300 from HIF-1α. In addition, FIH siRNA rescued the p300–HIF-1α binding inhibited by bortezomib. Given these results, we propose a mechanism for the inhibitory effects of bortezomib on tumor angiogenesis and hypoxic adaptation (ie, that bortezomib inhibits the hypoxic activation of HIF-1α by reinforcing the FIH-mediated inhibition of p300 recruitment).
We emphasize that HIF-1α was found to be inactivated by bortezomib at concentrations as low as 0.1 to 1 nM. Moreover, big differences in bortezomib cytotoxicities were found for different cell types, and in particular, we found that MM cell lines are 1000-fold more sensitive than Hep3B cells. However, higher concentrations of bortezomib were required to induce a cytotoxic effect, even in MM cells, than were required to induce HIF-inhibitory activity. More importantly, the bortezomib concentrations required for HIF-1 inhibition in MM and non-MM cell lines were similar. These findings suggest that HIF-1 inhibition by bortezomib is mechanistically unrelated to its cytotoxicity. Indeed, the HIF-inhibitory effect may not be attributable to proteasome inhibition, which is known to be the main mechanism underlying bortezomib cytotoxicity. Instead, HIF-1α is likely to be inactivated by the reinforcement of the FIH-mediated inhibition of p300 recruitment. Despite its marked inhibition of proteasomal activity, MG132 failed to stimulate FIH action. Moreover, although bortezomib at 1 nM or less weakly inhibited proteasomal activity as compared with 10 μM MG132, it strongly inactivated HIF-1. These results further support the notion that FIH stimulation by bortezomib is not directly linked with proteasome inhibition.
The present study demonstrates that the HIF-inhibitory effect of bortezomib depends on Asn803 in HIF-1α CAD and p300, which are quite different from that previously described.18 In this previous study, N803A-mutated CAD was also inactivated by bortezomib in Saos-2 cells, and the HIF-1α–p300 interaction was not disrupted by bortezomib in MCF-7. Regarding this discrepancy, one possible reason is that the cell lines used differed, and that as the cytotoxicity of bortezomib is highly cell-type specific, the action of bortezomib on FIH could also be cell-type specific. Indeed, Zheng et al26 demonstrated that HIF-1α is differently regulated by proteasome in mouse endothelial cells and HepG2 cells, and that this difference is determined by the compartmentalizations of HIF-1α and proteasome. Likewise, it cannot be ruled out that the FIH regulations of HIF-1α in Saos-2 and MCF-7 cells differ from those in Hep3B and MM cells. Another possible reason is that the bortezomib concentrations used were quite different. In the previous study, bortezomib was used at 500 nM only, which is about 1000 times that used in the present study. However, in view of the fact that 10 nM concentrations of bortezomib are clinically achievable, and enough to inhibit the proliferations of MM and other cells,27 a concentration of 500 nM is probably intolerable physiologically. In addition, we observed that a large proportion of tumor cells underwent apoptosis in the presence of high concentrations of bortezomib (data not shown). Thus, we consider that miscellaneous cellular events other than FIH activation may have interfered with HIF-1α activity in the presence of such a high concerentration of bortezomib.
Despite the accumulation of HIF-1α protein, the transcriptional activity of HIF-1 was inhibited in cells treated with proteasome inhibitors. This paradoxical response of HIF-1α to proteasome inhibitors was originally reported by Kallio et al.28 They also suggested impaired translocation of HIF-1α to the nucleus as a mechanism of HIF-1 inactivation, but other studies clearly showed the nuclear accumulation of HIF-1α stabilized by proteasome inhibitors.19,29,30 We also found that HIF-1α was mainly detected within the nucleus after bortezomib treatment by immunofluorescent imaging (Figure S3). In addition, we found that the cytoplasmic localization of FIH was also unchanged after bortezomib treatment (Figure S4). These observations indicate that the bortezomib inactivation of HIF-1 is unlikely to be related with the localization of HIF-1α or FIH.
FIH was originally identified as a HIF-1α CAD-interacting protein by yeast 2-hybrid screening.11 FIH belongs to the 2-oxoglutarate (2-OG)–dependent dioxygenase superfamily, which characteristically requires molecular oxygen, ferrous iron, and 2-OG.31 Although the enzymatic reaction and the X-ray crystal structure of FIH have been extensively investigated, the molecular entities that regulate the expression and activity of FIH have not been identified. In the present study, we found that bortezomib enhanced the physical interaction between FIH and HIF-1α CAD and functionally stimulated the inhibitory action of FIH on CAD activity, and that this was not accompanied by a change in FIH expression. However, we did not determine the mechanism by which bortezomib stimulates the action of FIH. To understand this mechanism, the molecular mechanism underlying FIH regulation should be addressed.
Solid tumors invariably present hypoxic environments,1 and MM is likely to grow in the natural hypoxic environment of bone marrow.3 Therefore, the inhibition of HIF-mediated hypoxic adaptation presents a strategy for treating MM, and may contribute to the MM-inhibitory effect of bortezomib. Moreover, it should be emphasized that HIF-1α is a master protein that promotes angiogenesis and cell proliferation in a variety of solid tumors,32 which suggests that HIF-1α inhibitors may act as broad spectrum anticancer agents.33 In addition, because bortezomib inhibited HIF-1α more so than proteasome, it may find use as a specific HIF-1α inhibitor, which suggests that bortezomib could be further developed to produce a broad-spectrum anticancer drug. However, the use of bortezomib as an anti-HIF agent remains to be investigated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a grant from The National R&D Program for Cancer Control, Korean Ministry of Health & Welfare Research Fund (no. 0520260-2).
Authorship
Contribution: D.H.S. and J.W.P. performed research. Y.S.C. and J.W.P designed research, analyzed the data, and wrote the paper. L.E.H. and D.S.L provided critical gene materials and cell lines, and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jong-Wan Park, Department of Pharmacology, Seoul National University College of Medicine, 28 Yongon-dong, Chongno-gu, Seoul 110-799, Korea; e-mail: parkjw@snu.ac.kr.