Abstract
CCL2 (MCP-1) has been shown to enhance HIV-1 replication. The expression of this chemokine by macrophages is up-modulated as a consequence of viral infection or gp120 exposure. In this study, we show for the first time that the phosphatidylcholine-specific phospholipase C (PC-PLC) is required for the production of CCL2 triggered by gp120 in human monocyte-derived macrophages (MDMs). Using a combination of pharmacologic inhibition, confocal laser-scanner microscopy, and enzymatic activity assay, we demonstrate that R5 gp120 interaction with CCR5 activates PC-PLC, as assessed by a time-dependent modification of its subcellular distribution and a concentration-dependent increase of its enzymatic activity. Furthermore, PC-PLC is required for NF-kB–mediated CCL2 production triggered by R5 gp120. Notably, PC-PLC activation through CCR5 is specifically induced by gp120, since triggering CCR5 through its natural ligand CCL4 (MIP-1β) does not affect PC-PLC cellular distribution and enzymatic activity, as well as CCL2 secretion, thus suggesting that different signaling pathways can be activated through CCR5 interaction with HIV-1 or chemokine ligands. The identification of PC-PLC as a critical mediator of well-defined gp120-mediated effects in MDMs unravels a novel mechanism involved in bystander activation and may contribute to define potential therapeutic targets to block Env-triggered pathologic responses.
Introduction
Macrophages are major cell targets for HIV infection in vivo. Infected macrophages serve as reservoirs for viral persistence and play important and well-established roles in multiple aspects of AIDS pathogenesis.1,2 Furthermore, macrophages are highly secretory cells and represent an important source for a variety of soluble immune mediators, including cytokines and chemokines, and strongly contribute to the dysregulation of soluble factor production observed at all stages of HIV infection.3-5
An important aspect of the immunopathogenesis of AIDS is represented by the observation that HIV target cells can also be exposed to viral gene products, expressed at the surface of infected cells, or released in the microenvironment as a consequence of the death of infected cells.6 Soluble HIV proteins have been shown to exert bystander effects on neighboring immune cell populations in the absence of productive infection. Furthermore, they interfere with the induction of protective immune responses by altering the production of soluble mediators as well as the cellular differentiation/activation state.6-8 In particular, HIV-1 gp120 can be detected in tissues,9,10 circulates in the blood of HIV-infected individuals on the surface of virions (both infectious and non infectious) and as a free protein,11 and it is thought to play a role in the progressive immune derangement observed in AIDS patients.7,12 It is now well known that HIV-1 gp120 elicits various intracellular signaling events both in primary cells and cell lines13,14 that are similar but not identical to that caused by chemokines.15,16 In particular, it has been demonstrated that the interaction of virion-associated or soluble gp120 with CCR5 or CXCR4 coreceptors, independent of CD4 engagement, results in receptor-coupled G protein activation and intracellular Ca2+ accumulation, leading to Pyk2 activation.16-19 The Src kinase Lyn and MAP and PI3 kinases are also activated through gp120 engagement of coreceptors,17,20-23 whereas STAT family member activation is specifically triggered through gp120 interaction with CD4.24 Interestingly, the gp120-mediated activation of some of these pathways has been directly correlated with the production of soluble factors.17,21,22
Phospholipase-mediated phospholipid hydrolysis is a widespread response elicited by most growth factors, cytokines, hormones, neurotransmitters, and other extracellular signals.25 Phospholipases are a family of enzymes classified according to the phospholipid bond that is cleaved. Their hydrolysis products include many of the most important second messengers that have been implicated in a wide range of cellular responses. Besides phosphoinositides, phosphatidylcholine (PC) is also known to act as substrate for the production of second messengers, mediated by different stimulated phospholipase activities (ie, PLD, PLC, and PLA2), involved in receptor-mediated cell signal transduction. Among these enzymes, PC-PLC catalyzes PC hydrolysis into phosphocholine (PCho) and diacylglycerol (DAG), whereas PC-PLD hydrolyzes PC to generate phosphatidic acid (PA) and choline (Cho). Interestingly, PC-PLC plays a role in basic mechanisms of the immune response, such as cytokine/chemokine production in different experimental systems26-32 and cytokine-induced natural killer (NK) cell–mediated cytotoxicity.33-35 PC-PLD has been implicated in a variety of cellular processes, including vesicle trafficking, receptor signaling, and cell proliferation and survival.36
In this study, we investigated the role of PC-PLC in gp120-triggered CCL2 secretion in monocyte-derived macrophages (MDMs). We report for the first time that PC-PLC is activated by gp120, but not CCL4, interaction with CCR5 and is required for the downstream NF-kB–driven CCL2 production. Thus, HIV-1 gp120-mediated PC-PLC activation in MDMs is critical for the regulation of CCL2 secretion, a CC-chemokine playing a regulatory role in HIV infection of these cells.37 This finding suggests that this signal transduction pathway can also be relevant for the modulation of viral replication in MDMs as well as for the recruitment of new cell targets. Overall, the identification of PC-PLC as a novel signal transduction pathway mediating some of the gp120 biologic effects unravels a new mechanism by which HIV-1 may deregulate macrophage functions and contribute to AIDS pathogenesis.
Methods
Cell separation and culture
Monocytes were isolated from the peripheral blood of healthy donors by elutriation followed by negative selection of contaminant cells as previously described.38 Monocyte-derived macrophages (MDMs) were obtained after 7 days of in vitro culture in endotoxin-free Iscoves medium (Life Technologies, Gaithersburg, MD) containing 15% FBS as previously described.37
Reagents
Recombinant HIV-1 R5 gp120 preparations were obtained from Dr I. Jones, University of Reading, United Kingdom (primary isolate CN54) or kindly provided by G. Gao, Chinese Academy of Science, Beijing, China (laboratory-adapted strain BaL). Unless otherwise mentioned, similar results were obtained with the 2 different gp120 preparations (data not shown). Tak779 was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Bethesda, MD). Tricyclodecan-9-yl-xanthogenate (D609) and N-p-Tosyl-l-phenylalanine chloromethyl ketone (TPCK) were purchased from Sigma-Aldrich (St Louis, MO). Recombinant CCL4 was purchased from R&D Systems (Minneapolis, MN). The rabbit polyclonal anti–PC-PLC Ab, raised against bacterial (Bacillus cereus) PC-PLC and cross-reacting with mammalian PC-PLC, was obtained, purified, and characterized as previously described.33-35,39,40 The rabbit polyclonal anti–NF-kB p65 Ab was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The Alexa Fluor 594–conjugated goat anti–rabbit IgG, used as fluorochrome-conjugated secondary Ab for indirect immunofluorescence, was purchased from Molecular Probes (Eugene, OR). LPS contamination of reagents was excluded by checking their endotoxin activity by the limulus amebocyte assay (Charles River Endosafe, Charleston, SC; detection limit: 0.125 endotoxin units/mL). The endotoxin content determined in gp120 preparations was less than 0.125 endotoxin unit/mL.
Virus inactivation procedure
The HIV-1BaL virus preparation was purchased from ABI (Columbia, MD) and consisted of pelleted virus obtained after propagation in primary human macrophages. 2,2′-dithiodipyridine (aldrithiol-2; AT-2)–inactivated virions were prepared as previously described.41 At the end of the inactivating procedure, treatment agent was removed by ultracentrifugation at 17 000g for 1 hour at 4°C. Viral pellets were resuspended in endotoxin-free Iscoves medium containing 15% FBS.
Measurement of CC-chemokines
The levels of CCL2 (detection limit: 5 pg/mL), CCL3 (MIP-1α) (detection limit: 10 pg/mL), CCL4 (detection limit: 4 pg/mL) and CCL5 (RANTES) (detection limit: 2 pg/mL) present in culture supernatants were measured by enzyme-linked immunosorbent assay (ELISA) kits purchased from R&D Systems. For some experiments, CCL2 (detection limit: 1.5 pg/mL), CCL3 (detection limit: 3 pg/mL), CCL4 (detection limit: 0.8 pg/mL), and CCL5 (detection limit: 0.2 pg/mL) were measured by multiplex sandwich ELISA (Search Light Human Chemokine Array 1; Pierce Endogen, Rockford, IL) following the manufacturer's instructions.
Confocal laser-scanner microscopy analysis
For confocal laser-scanner microscopy (CLSM) analysis, freshly isolated monocytes were seeded in 24-well cluster plates on cover glasses (diameter: 12 mm; 2 × 105 cells per well in 1 mL). After 6 days, culture medium was removed and replaced with fresh medium lacking serum to maintain the cells in a quiescent state. After 24 hours, cells were exposed to the different stimuli for the indicated periods of time. The cover glasses were then extensively washed with phosphate-buffered saline (PBS), mounted on the microscope slide with Vectashield antifade medium containing DAPI (Vector Laboratories, Burlingame, CA), fixed, permeabilized, and stained with rabbit anti–PC-PLC or rabbit anti-p65 Abs and fluorochrome-conjugated secondary Abs, as previously described.33,34,39 CLSM observations were performed with a Leica TCS SP2 AOBS apparatus, using a 63×/1.40-0.60 numeric aperture oil objective and 405/594 nm, excitation spectral laser lines appropriately tuned by acousto-optical tunable filter. Image acquisition and processing were carried out using the Leica Confocal Software 2.3 (Leica, Lasertechnik, Heidelberg, Germany) and Adobe Photoshop CS2 1.4.10 software programs (Adobe Systems, San Jose, CA). Signals from different fluorescent probes were taken in parallel. Several cells were analyzed for each labeling condition, and representative results are shown.
Cell lysates
Freshly isolated monocytes were seeded in 6-well cluster plates (3 × 106 cells per well in 3 mL). After 6 days, culture medium was removed and replaced with fresh medium lacking serum to maintain the cells in a quiescent state. After 24 hours, cells were subjected to the different treatment for the indicated periods of time. Cells were then lysed in ice-cold lysis buffer (100 mM Tris-HCl, pH 8; 150 mM NaCl; 1% triton X-100; 1 mM MgCl2; 60 mM octyl-glucoside) supplemented with a cocktail of protease inhibitors (Complete Protease Inhibitor Cocktail tablets; Roche Molecular Biochemicals, Mannheim, Germany) and incubated for 20 minutes on ice. Cell lysates were then clarified by centrifugation at 10 000g for 20 minutes at 4°C. Protein concentrations were determined by protein assay (Bio-Rad Laboratories, Hercules, CA).
In vitro PC-PLC activity assay
PC-PLC activity was determined in vitro using the Amplex Red PC-specific PLC assay kit (Molecular Probes) as described by the manufacturer and adapted by Spadaro et al.34 In brief, in this enzyme-coupled assay, PC-PLC activity was detected using 10-acetyl-3,7-dihydrophenoxazine (Amplex Red reagent), a sensitive fluorogenic probe for H2O2, in a reaction cascade comprising the following steps: (a) PC-PLC–mediated conversion of the PC (egg yolk lecithin) substrate to form DAG and PCho; (b) hydrolysis, catalyzed by alkaline phosphatase (AP), of PCho into Cho and phosphate; (c) Cho oxidation by choline oxidase to form betaine and H2O2; and (d) reaction of H2O2 with Amplex Red reagent (in a 1:1 stoichiometry) in the presence of horseradish peroxidase, to generate the highly fluorescent product resorufin. To evaluate the respective contribution of PC-PLC and PC-PLD activity in step c, assays were conducted in the presence or in the absence of AP. Each sample was run in triplicate.
Analysis of CCL2 mRNA by TaqMan real-time reverse-transcription–polymerase chain reaction
Freshly isolated monocytes were seeded in 6-well cluster plates (3 × 106 cells per well in 3 mL). After 6 days, culture medium was removed and replaced with fresh medium lacking serum to maintain the cells in a quiescent state. After 24 hours, cells were subjected to the different treatments for 4 hours. Cells were then lysed and total RNA was isolated with the RNeasy Mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Total RNA was retrotranscribed into cDNA using polyd(N)6 (GE Healthcare, Pittsburgh, PA). Real-time polymerase chain reaction (PCR) was performed on an ABI-Prism 7700 PCR cycler (Applied Biosystems, Foster City, CA) using the TaqMan universal master mix (Applied Biosystems) according to the manufacturer's instructions. Validated PCR primers and TaqMan MGB probe (6FAM-labeled) for CCL2 were used (Hs00234140 m1; Applied Biosystems). As endogenous control, primers with TaqMan probe for the human β actin were used (ACTB RNA; Hs99999903 m1; Applied Biosystems). Thermal cycler conditions were as follows: 1 × 10 minutes at 95°C, 40 cycles of denaturation (15 seconds at 95°C), and combined annealing/extension (30 seconds at 60°C and 30 seconds at 72°C). Relative quantification was performed using the comparative Ct method: arithmetic formulas were used to calculate relative expression levels, and compare with a calibrator (in our conditions, untreated MDMs). The amount of target, normalized to endogenous housekeeping gene (ACTB) and relative to the calibrator (untreated MDMs), is then given by 2−ΔΔCt, where ΔΔCt = ΔCt(target) − ΔCt(calibrator), and ΔCt is the Ct of the target gene subtracted from the Ct of the housekeeping gene. The equation thus represents the normalized expression of the target gene in the unknown sample, relative to the normalized expression of the calibrator sample.
Statistical analysis
Statistical analysis was performed using the unpaired Student t test. Values of P less than .05 were considered significant.
Results
PC-PLC signaling pathway mediates HIV-1 gp120-induced CCL2 production in MDMs
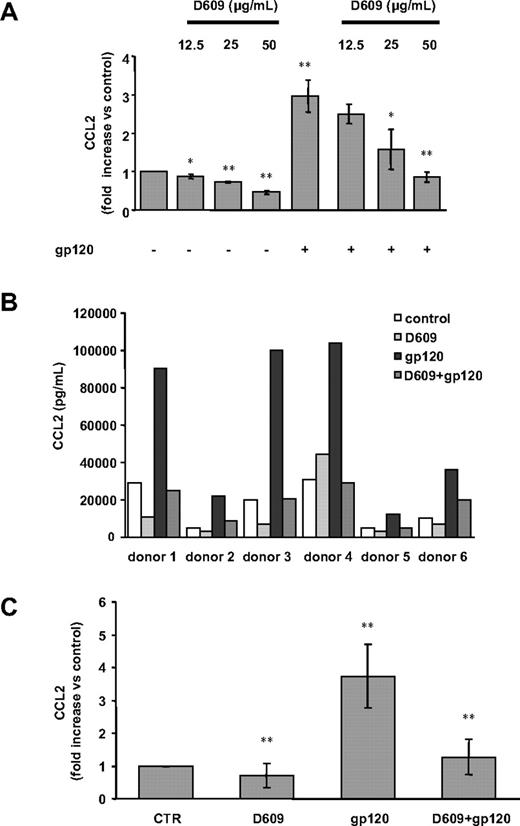
PC-PLC has been previously reported to control cytokine/chemokine production including CCL2 in response to a variety of stimuli in different experimental systems.26-32 On the basis of our previous results indicating that gp120 up-modulates CCL2 production in primary macrophages,42 we therefore evaluated the role of PC-PLC signaling pathway in this phenomena by testing the effect of the PC-PLC–specific inhibitor D609. As shown in Figure 1A, treatment of MDMs with D609 markedly inhibited in a concentration-dependent manner, the secretion of CCL2 induced by R5 gp120. The highest effect was observed at the concentration of 50 μg/mL. Interestingly, D609 also affected CCL2 constitutive secretion in a concentration-dependent manner (Figure 1A). As shown in Figure 1B, a considerable variability in the constitutive and induced expression of CCL2 as well as in the extent of inhibition by D609 was observed among donors. Despite this variability, these effects were consistently observed in all the donors analyzed. As shown in Figure 1C, a 3.8 (± 0.9)-fold (SD) increase in CCL2 secretion was observed in all donors following gp120 stimulation (P < .001 vs control) that was reduced to 1.2 (± 0.5)-fold (SD) upon treatment with D609 (P < .001 versus gp120 alone). We have previously reported that CCL2 secretion is up-modulated upon macrophage exposure to both recombinant gp120 and AT-2 R5 and X4 HIV-1 strains,42 which retain conformational and functional integrity of viral surface proteins.43 Since only a limited fraction (< 0.1%) of circulating viruses is demonstrably infectious,44,45 exposure to inactivated viruses may mimic the most frequent type of macrophage-HIV interactions that occur in vivo. Thus, to further evaluate the role of PC-PLC in the HIV-induced CCL2 up-modulation, we tested the effect of D609 on the secretion of CCL2 triggered upon exposure of MDMs to AT-2–inactivated R5 HIV-1 (BaL strain). As shown in Table 1, D609 exerted a marked inhibitory effect on CCL2 secretion induced by AT-2–inactivated R5 HIV-1 similar to that observed upon stimulation of CCL2 production by recombinant R5 gp120 (Figure 1B,C).
D609 inhibits gp120-mediated CCL2 secretion in MDMs. Monocytes were seeded in 48-well cluster plates (5 × 105 cells/mL per well). After 7 days, culture medium was removed and replaced with 0.5 mL fresh medium. (A) MDMs were treated with different concentrations of D609 (12.5, 25, 50 μg/mL) or left untreated. At all concentrations used, this inhibitor did not affect cell viability (data not shown). After 30 minutes, some cultures were exposed to R5 gp120 (1 μg/mL). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 3 different donors. (B,C) MDMs were treated with D609 (50 μg/mL) or left untreated, and then exposed to R5 gp120 as described in panel A. In panel B, the results obtained with 6 different donors, representative of 12 tested, are shown. In panel C, data are the means (± SD) of the results obtained with all the donors analyzed. *P < .05; **P < .005.
D609 inhibits gp120-mediated CCL2 secretion in MDMs. Monocytes were seeded in 48-well cluster plates (5 × 105 cells/mL per well). After 7 days, culture medium was removed and replaced with 0.5 mL fresh medium. (A) MDMs were treated with different concentrations of D609 (12.5, 25, 50 μg/mL) or left untreated. At all concentrations used, this inhibitor did not affect cell viability (data not shown). After 30 minutes, some cultures were exposed to R5 gp120 (1 μg/mL). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 3 different donors. (B,C) MDMs were treated with D609 (50 μg/mL) or left untreated, and then exposed to R5 gp120 as described in panel A. In panel B, the results obtained with 6 different donors, representative of 12 tested, are shown. In panel C, data are the means (± SD) of the results obtained with all the donors analyzed. *P < .05; **P < .005.
The PC-PLC inhibitor D609 does not affect the gp120-mediated secretion of other CC-chemokines in MDMs
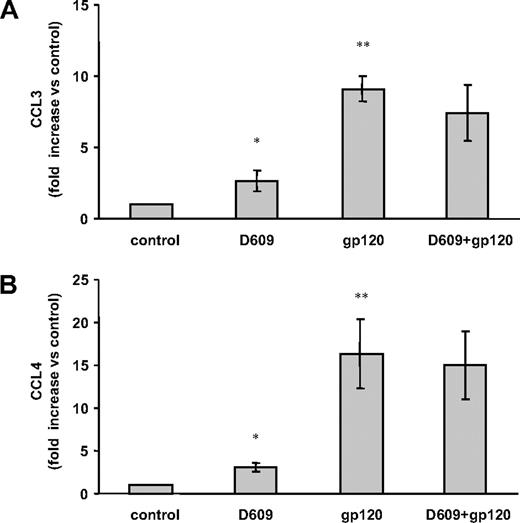
It was reported that gp120-exposed MDMs secrete, in addition to high levels of CCL2, significant amounts of CCL3, CCL4, and CCL5.17,42,46 On the basis of these results, we next evaluated whether the inhibitory effect of D609 could be extended to the gp120-stimulated secretion of these CC-chemokines. To this aim, we measured the secretion of CCL3, CCL4, and CCL5 in gp120-exposed MDMs in the presence or in the absence of D609. As shown in Figure 2, in contrast to its inhibitory effect on constitutive and gp120-induced CCL2 secretion (Figure 1), treatment of MDMs with D609 did not significantly inhibit the gp120-mediated secretion of CCL3 and CCL4, whereas the constitutive production of these chemokines was rather slightly increased in the presence of D609 (Figure 2A,B). CCL3 and CCL4 secretion was induced by both R5 gp120 preparations (BaL and CN54). In contrast, CCL5 secretion was induced only by gp120 derived from the HIV-1Bal strain (86 ± 13-fold [SD] increase vs untreated cells, n = 3, P < .001), and this secretion was not inhibited by D609 (80 ± 5-fold [SD] increase vs untreated cells, P = .45).
Effect of D609 on gp120-mediated secretion of CCL3 and CCL4 in MDMs. Monocytes were seeded, cultured, and treated as described in the legend to Figure 1B. After 24 hours of culture, supernatants were harvested and frozen prior to CCL3 (A) and CCL4 (B) determination. Data are the means (± SD) of the results obtained with 3 different donors. *P < .05; **P < .005.
Effect of D609 on gp120-mediated secretion of CCL3 and CCL4 in MDMs. Monocytes were seeded, cultured, and treated as described in the legend to Figure 1B. After 24 hours of culture, supernatants were harvested and frozen prior to CCL3 (A) and CCL4 (B) determination. Data are the means (± SD) of the results obtained with 3 different donors. *P < .05; **P < .005.
HIV-1 gp120 induces cellular relocalization of PC-PLC and stimulates its enzymatic activity in MDMs
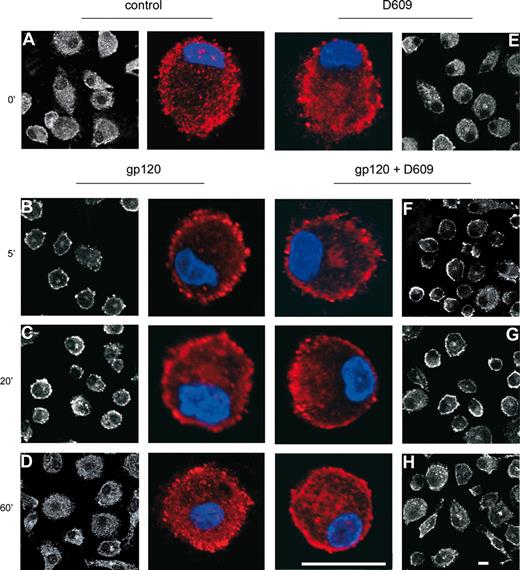
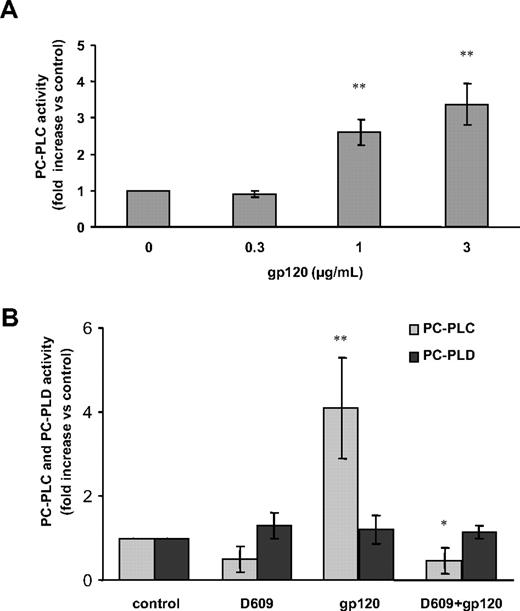
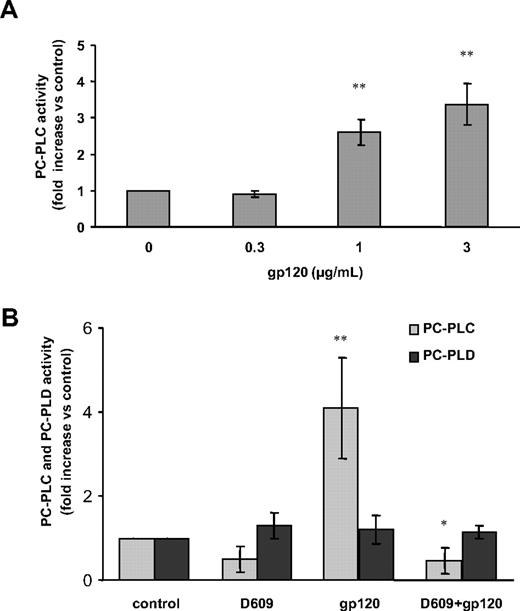
We have previously demonstrated that activation of PC-PLC is associated with its translocation from cytoplasmic regions to the plasma membrane in IL-2–stimulated NK cells33,34 and mitogen-stimulated fibroblasts.39 The novel finding that D609 affects the gp120-induced secretion of CCL2 suggested a role for this viral glycoprotein in PC-PLC activation in MDMs. To explore this possibility, we therefore investigated whether gp120 exposure affected PC-PLC cellular localization in these cells. As shown in Figure 3, CLSM analysis showed that PC-PLC was highly expressed in control MDMs with a wide distribution throughout the cytoplasm (Figure 3A). A short treatment (5 minutes) with gp120 led to a marked accumulation of this enzyme in plasma membrane areas (Figure 3B). This gp120-induced effect was transient since after 20 minutes of treatment the enzyme started to repopulate the cytoplasm (Figure 3C) and at 1 hour, the pattern of PC-PLC cellular localization completely reverted to that observed in control cells (Figure 3D). Interestingly, although D609 did not significantly affect the PC-PLC cellular distribution in control cells (Figure 3E) as well as the initial stimulation-induced membrane redistribution (Figure 3F), it delayed the enzyme repopulation of cytoplasmic areas in gp120-treated cultures (Figure 3G,H). We next investigated whether gp120-induced PC-PLC translocation to the membrane was associated with changes in the activity of this enzyme. To this aim, PC-PLC enzymatic activity was measured in total cell lysates. As shown in Figure 4A, exposure of MDMs to R5 gp120 resulted in a concentration-dependent increase of PC-PLC enzymatic activity. This effect was detected with gp120 concentrations ranging from 1 to 3 μg/mL (P = .001 and P = .002 vs control, respectively), but not at the concentration of 0.3 μg/mL. In keeping with the observed effect on PC-PLC translocation to the plasma membrane, the peak of enzymatic activation was generally observed within 5 minutes upon gp120 exposure and decreased to nearly background levels in approximately 30 minutes (data not shown). Importantly, gp120-mediated PC-PLC enzymatic activation was completely abrogated in MDMs treated with D609 (Figure 4B; n = 4, P = .01 vs gp120 alone). The observation that D609 blocks PC-PLC enzymatic activation, but not the initial stimulation-induced membrane redistribution, suggests that this inhibitor could differentially affect individual events associated with PC-PLC activation/translocation in MDMs. Endogenous PC-PLD, present in cell lysates and responsible for PC conversion into Cho and PA, could also contribute to the final resorufin fluorescence detected by the Amplex Red assay. Thus, its relative contribution was specifically measured by analyzing reaction mixture deprived of AP. As shown in Figure 4B, PC-PLD enzymatic activity did not increase upon gp120 exposure and it was not affected by D609.
HIV-1 gp120 induces cellular relocalization of PC-PLC in MDMs. Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with R5 gp120 (1 μg/mL) for the indicated periods of time (B-D) or left untreated (A). Some cultures were treated with D609 (50 μg/mL) for 30 minutes before gp120 exposure (F-H) or with D609 alone (E). Cells were then fixed, permeabilized, and stained with rabbit anti PC-PLC Abs (detected in white or in red) and DAPI (in blue) as described in “Methods.” PC-PLC expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bars correspond to 20 μm. Color panels show a single cell of the corresponding black and white panels at higher magnification.
HIV-1 gp120 induces cellular relocalization of PC-PLC in MDMs. Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with R5 gp120 (1 μg/mL) for the indicated periods of time (B-D) or left untreated (A). Some cultures were treated with D609 (50 μg/mL) for 30 minutes before gp120 exposure (F-H) or with D609 alone (E). Cells were then fixed, permeabilized, and stained with rabbit anti PC-PLC Abs (detected in white or in red) and DAPI (in blue) as described in “Methods.” PC-PLC expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bars correspond to 20 μm. Color panels show a single cell of the corresponding black and white panels at higher magnification.
HIV-1 gp120 up-modulates PC-PLC enzymatic activity in MDMs. Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated as follows: (A) with different concentrations of R5 gp120 (0.3, 1, 3 μg/mL) for 5 minutes; (B) with D609 (50 μg/mL) for 30 minutes prior to gp120 exposure (3 μg/mL) for 5 minutes. Cells were then lysed and PC-PLC enzymatic activity was measured in cell lysates as described in “Methods.” In panel A, the relative contribution of PC-PLD was subtracted to the total fluorescence. In panel B, PC-PLC and PC-PLD activities are shown separately. Data shown in panels A and B are the means (± SD) of the results obtained with 4 different donors. *P < .05; **P < .005.
HIV-1 gp120 up-modulates PC-PLC enzymatic activity in MDMs. Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated as follows: (A) with different concentrations of R5 gp120 (0.3, 1, 3 μg/mL) for 5 minutes; (B) with D609 (50 μg/mL) for 30 minutes prior to gp120 exposure (3 μg/mL) for 5 minutes. Cells were then lysed and PC-PLC enzymatic activity was measured in cell lysates as described in “Methods.” In panel A, the relative contribution of PC-PLD was subtracted to the total fluorescence. In panel B, PC-PLC and PC-PLD activities are shown separately. Data shown in panels A and B are the means (± SD) of the results obtained with 4 different donors. *P < .05; **P < .005.
PC-PLC activation and CCL2 secretion in MDMs are triggered by R5 gp120 interaction with CCR5
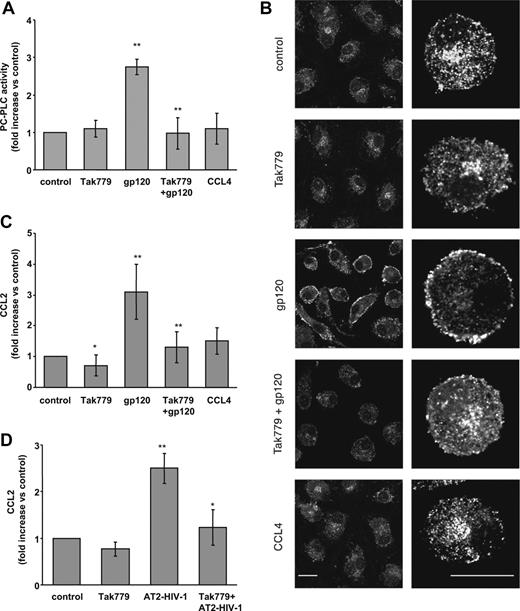
We have previously demonstrated that the gp120-mediated up-modulation of CC-chemokine secretion, including CCL2, occurs through CD4-independent mechanisms and that chemokine receptor engagement triggered chemokine production. This strongly suggested that a specific interaction of gp120 with coreceptors on the cell membrane was required for chemokine induction.17,42 Therefore, we next investigated whether CCR5 was responsible for the gp120-elicited PC-PLC activation and CCL2 secretion in MDMs. To this aim, the CCR5-specific pharmacologic antagonist Tak779, blocking both the HIV-1 coreceptor function and the agonist-induced signaling,47,48 was used. As shown in Figure 5, MDM treatment with Tak779 prior to the addition of R5 gp120 resulted in a clear-cut inhibition of PC-PLC enzymatic activity (Figure 5A; 0.9 ± 0.4-fold [SD] increase vs 2.8 ± 0.2-fold [SD]; P < .001 vs gp120 alone) as well as of its cellular relocalization induced by R5 gp120 (Figure 5B). In contrast, Tak779 exposure did not affect both the enzymatic activity and cellular localization of PC-PLC in control cells (Figure 5A,B). In keeping with these results, blocking gp120 interaction with CCR5 by Tak779 markedly reduced the gp120-induced up-modulation of CCL2 (1.3 ± 0.5-fold [SD] increase vs 3.1 ± 0.9-fold [SD]; P = .002 vs gp120 alone; Figure 5C). Furthermore, Tak779 significantly reduced the constitutive level of CCL2 secretion (Figure 5C). Likewise, Tak779 also significantly reduced CCL2 production triggered by MDM exposure to AT-2–inactivated R5 HIV-1 virions. As shown in Figure 5D, the 2.5 (± 0.3)-fold (SD) increase in CCL2 secretion observed upon AT-2–inactivated R5 virus stimulation (P = .004 versus untreated cells) was reduced to 1.2 (± 0.3)-fold (SD) in the presence of Tak779 (P = .03 vs AT-2 R5 HIV-1 alone). It has been previously shown that macrophage activation by gp120 results in CCR5-mediated signals that resemble in some, but not all, respects those elicited by natural ligands.15 We thus investigated whether CCR5 stimulation by the chemokine ligand would elicit PC-PLC activation and CCL2 secretion at a similar extent than that observed with R5 gp120. To this aim, we exposed MDMs to CCL4, the most specific of several chemokines that signal through CCR5. As shown in Figure 5, a short treatment with CCL4 did not induce any modification in PC-PLC enzymatic activity (Figure 5A) and cellular distribution (Figure 5B), as well as in CCL2 production (Figure 5C). This suggests that CCR5-mediated PC-PLC activation, leading to CCL2 production, is specific for gp120. The differences observed in the effect of gp120 versus CCL4 are not due to coactivation of CD4 by gp120 but not by CCL4, since costimulation of MDMs with CCL4 and a monoclonal antibody to CD4, mimicking gp120 binding, completely failed to activate PC-PLC (data not shown).
Tak779 inhibits R5 HIV-1–mediated PC-PLC activation and CCL2 secretion in MDMs. (A) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with Tak779 (5 μM) for 30 minutes prior to R5 gp120 exposure (1 μg/mL) for 5 minutes. Some cultures were exposed to CCL4 (200 ng/mL) for 5 minutes. Cells were then lysed and PC-PLC enzymatic activity was measured in cell lysates as described in “Methods.” The relative contribution of PC-PLD was subtracted from the total fluorescence. Data are the means (± SD) of the results obtained with 3 different donors. (B) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with Tak779 (5 μM) for 30 minutes prior to R5 gp120 exposure (1 μg/mL) for 5 minutes. Some cultures were exposed to CCL4 (200 ng/mL) for 5 minutes. Cells were then fixed, permeabilized, and stained with rabbit anti–PC-PLC Abs (detected in white) as described in “Methods.” PC-PLC expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bars, indicated in the lower panels, correspond to 20 μm. Right panels show a single cell of the corresponding left panels at higher magnification. (C) Monocytes were seeded and cultured as described in the footnote to Table 1. MDMs were treated with Tak779 (5 μM) or left untreated. After 30 minutes, some cultures were exposed to R5 gp120 (1 μg/mL) or CCL4 (200 ng/mL). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 7 different donors. (D) Monocytes were seeded and cultured as described in the footnote to Table 1. MDMs were treated with Tak779 (5 μM) or left untreated. After 30 minutes, some cultures were exposed to R5 AT-2–inactivated viruses (17 ng HIVp24gag/106 cells). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 3 different donors. *P < .05; **P < .005.
Tak779 inhibits R5 HIV-1–mediated PC-PLC activation and CCL2 secretion in MDMs. (A) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with Tak779 (5 μM) for 30 minutes prior to R5 gp120 exposure (1 μg/mL) for 5 minutes. Some cultures were exposed to CCL4 (200 ng/mL) for 5 minutes. Cells were then lysed and PC-PLC enzymatic activity was measured in cell lysates as described in “Methods.” The relative contribution of PC-PLD was subtracted from the total fluorescence. Data are the means (± SD) of the results obtained with 3 different donors. (B) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with Tak779 (5 μM) for 30 minutes prior to R5 gp120 exposure (1 μg/mL) for 5 minutes. Some cultures were exposed to CCL4 (200 ng/mL) for 5 minutes. Cells were then fixed, permeabilized, and stained with rabbit anti–PC-PLC Abs (detected in white) as described in “Methods.” PC-PLC expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bars, indicated in the lower panels, correspond to 20 μm. Right panels show a single cell of the corresponding left panels at higher magnification. (C) Monocytes were seeded and cultured as described in the footnote to Table 1. MDMs were treated with Tak779 (5 μM) or left untreated. After 30 minutes, some cultures were exposed to R5 gp120 (1 μg/mL) or CCL4 (200 ng/mL). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 7 different donors. (D) Monocytes were seeded and cultured as described in the footnote to Table 1. MDMs were treated with Tak779 (5 μM) or left untreated. After 30 minutes, some cultures were exposed to R5 AT-2–inactivated viruses (17 ng HIVp24gag/106 cells). After 24 hours of culture, supernatants were harvested and frozen before CCL2 determination. Data are the means (± SD) of the results obtained with 3 different donors. *P < .05; **P < .005.
PC-PLC is an upstream regulator of the gp120-triggered NF-kB activation in MDMs
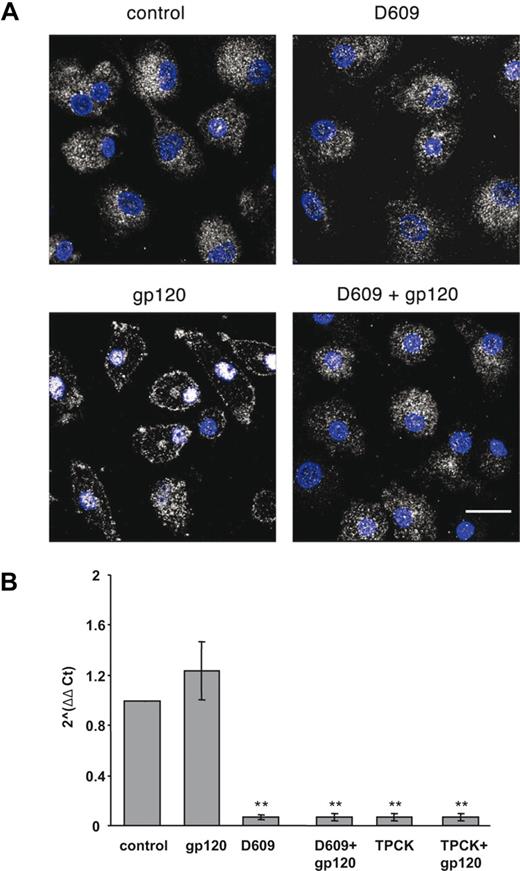
It has been previously reported that HIV-1 induces nuclear translocation of NF-kB p65 subunit in macrophages and that this activation is responsible for cytokine/chemokine induction.49 Furthermore, it has been demonstrated that CCL2 expression is strongly dependent on NF-kB activation in various cell types.28,50 To evaluate whether NF-kB activation was involved in gp120-mediated induction of CCL2 in MDMs, we tested the effect of the protease inhibitor TPCK, which blocks NF-kB activation by preventing IkB proteolytic degradation, on the R5 gp120-induced secretion of CCL2 in these cells. As shown in Table 2, treatment of MDMs with TPCK markedly inhibited R5 gp120-induced CCL2 production in all the donors analyzed. A 3.7 (± 0.6)-fold (SD) increase in CCL2 secretion was observed following gp120 stimulation, that was reduced to 0.9 (± 0.2)-fold (SD) upon pretreatment with TPCK (P = .002 vs gp120 alone). Conversely, the gp120-induced CCL3 and CCL4 secretion (7.2 ± 2.8-fold [SD] increase, n = 4, P = .01; 10 ± 3.2-fold [SD] increase, n = 4, P = .001, vs untreated cells, respectively) was not significantly reduced upon treatment with TPCK (4.5 ± 1.8-fold [SD], P = .23; 5.5 ± 2.4-fold [SD], P = .09 vs gp120 alone, respectively). We therefore performed experiments aimed at evaluating whether PC-PLC activation induced by R5 gp120 could ultimately lead to the downstream activation of NF-kB. As shown in Figure 6A, exposure of MDMs to R5 gp120 induced a marked redistribution of NF-kB p65 subunit leading from a diffuse cytoplasmic localization to an intense staining in nuclei. This redistribution reached its maximum after 1 hour of gp120 exposure (Figure 6A). Interestingly, although D609 did not significantly affect p65 distribution in control cells, it completely prevented p65 nuclear localization triggered by R5 gp120, clearly demonstrating that PC-PLC is a key component in the NF-kB activation induced by gp120. To provide further evidence on the role of PC-PLC in the gp120-mediated NF-kB activation leading to CCL2 secretion, the effect of D609 on the expression of CCL2 mRNA was assessed by TaqMan real-time reverse-transcription–PCR. As shown in Figure 6B, although gp120 does not up-modulate the constitutively expressed CCL2 mRNA, treatment of cells with D609 strongly down-modulates CCL2 transcript accumulation both in the presence and in the absence of gp120. As expected, similar results were obtained by blocking NF-kB activation in the presence of TPCK (Figure 6B).
D609 inhibits the R5 gp120-induced NF-kB p65 subunit nuclear translocation and CCL2 transcript accumulation. (A) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with R5 gp120 (1 μg/mL) for 1 hour or left untreated. Some cultures were treated with D609 (50 μg/mL) for 30 minutes before gp120 exposure or with D609 alone. Cells were then fixed, permeabilized, and double-stained with rabbit anti-p65 Ab (detected in white) and DAPI (in blue) as described in “Methods.” p65 expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bar indicated in the lower panel corresponds to 20 μm. (B) Monocytes were seeded and cultured as described in “Methods.” MDMs were exposed to R5 gp120 (1 μg/mL) or left untreated. Some cultures were treated with D609 (50 μg/mL) or TPCK (10 μM) for 30 minutes before gp120 exposure or with D609 or TPCK alone. After 4 hours, total RNA was extracted, retrotranscribed, and amplified as described in “Methods.” The 2−ΔΔCt values were calculated as described in “Methods.” Data are the means (± SD) of the results obtained with 3 different donors. **P < .005.
D609 inhibits the R5 gp120-induced NF-kB p65 subunit nuclear translocation and CCL2 transcript accumulation. (A) Monocytes were seeded and cultured as described in “Methods.” At day 7, cells were treated with R5 gp120 (1 μg/mL) for 1 hour or left untreated. Some cultures were treated with D609 (50 μg/mL) for 30 minutes before gp120 exposure or with D609 alone. Cells were then fixed, permeabilized, and double-stained with rabbit anti-p65 Ab (detected in white) and DAPI (in blue) as described in “Methods.” p65 expression and cellular localization were detected by CLSM analysis. The results from 1 representative experiment of 3 independently performed are shown. The bar indicated in the lower panel corresponds to 20 μm. (B) Monocytes were seeded and cultured as described in “Methods.” MDMs were exposed to R5 gp120 (1 μg/mL) or left untreated. Some cultures were treated with D609 (50 μg/mL) or TPCK (10 μM) for 30 minutes before gp120 exposure or with D609 or TPCK alone. After 4 hours, total RNA was extracted, retrotranscribed, and amplified as described in “Methods.” The 2−ΔΔCt values were calculated as described in “Methods.” Data are the means (± SD) of the results obtained with 3 different donors. **P < .005.
Discussion
In this study, we have identified a novel signaling pathway, triggered by HIV-1 envelope engagement of CCR5, that is involved in CCL2 production. Our results clearly indicate that PC-PLC, widely distributed throughout the cytoplasm in control macrophages, rapidly and transiently accumulated at the plasma membrane following gp120 treatment. Moreover, we demonstrate that PC-PLC enzymatic activity markedly increased, in a concentration-dependent manner, soon after exposure of MDMs to gp120 and that this activation was completely abrogated in the presence of D609. Conversely, this inhibitor does not affect the initial gp120-induced membrane redistribution but it delays only the enzyme repopulation of cytoplasmic areas. This indicates that D609 differentially affects individual events associated with PC-PLC activation/translocation. Furthermore, it suggests that the mechanisms of PC-PLC activation/inhibition and translocation to/from the plasma membrane are not necessarily coupled, at least in MDMs. Although this peculiar behavior needs further investigation, it is likely that it may depend on cell type–specific interactions with cellular components. D609 is thus far the only available compound acting as a competitive PC-PLC inhibitor.51 Strong evidence for the specificity of the inhibitory effect of this compound on PC-PLC has been provided in our previous studies34,39 as well as in reports by other groups.29 Furthermore, the majority of the studies correlating PC-PLC activation with cytokine/chemokine production relied on the use of D609 as a PC-PLC inhibitor.26-32 However, D609 has also been reported to affect PC-PLD activity52-54 and to inhibit sphingomyelin synthase in some cell types.55 In this respect, it is noteworthy that in our cellular model of primary monocyte-derived macrophages, PC-PLD activity was neither up-modulated by gp120 addition nor reduced in the presence of D609, thus indicating that gp120 selectively activates PC-PLC and D609 specifically inhibits PC-PLC enzymatic activity.
We also report that a PC-PLC–mediated signaling cascade is required for CCL2 production in MDMs, since blocking this pathway by a specific inhibitor completely abolished the capacity of gp120 to stimulate the secretion of this chemokine. Of note, we show that D609 is not a general inhibitor of chemokine production since it does not affect the expression of CCL3, CCL4, and CCL5. These results suggest that, in addition to PC-PLC, alternative pathways activated by gp120 can be responsible for the selective expression of specific chemokines.
Furthermore, our results clearly demonstrate that the gp120-triggered PC-PLC activation is mediated by CCR5, since blocking this chemokine receptor by the specific pharmacologic antagonist Tak779 completely inhibited PC-PLC activation and CCL2 secretion. According to our results, the majority of signaling responses elicited by gp120 in macrophages require its interaction with CCR5 but are independent of CD4.16-18,21,22 Notably, PC-PLC activation through CCR5 appears to be specific for gp120, as treatment with CCL4, the most specific chemokine ligand that signals through CCR5, did not induce any modification of both PC-PLC cellular distribution and enzymatic activity as well as of CCL2 secretion. The differences observed in the effect of gp120 versus CCL4 are not due to coactivation of CD4 by gp120 but not by CCL4, since costimulation of MDMs with CCL4 and a monoclonal antibody to CD4, mimicking gp120 binding, completely failed to activate PC-PLC. In keeping with our results, it has been previously reported that gp120 stimulation of both T cells and macrophages results in CCR5-mediated signals that resemble in some, but not all, respects those elicited by its natural ligand CCL4.16-18,21-23,56 In particular, it has been previously shown that in macrophages both CCL4 and gp120 activated K+, Cl−, and Ca2+ currents, whereas only gp120 activated a nonselective cation current through CCR5.16 Moreover, it has been reported that in glioma cells both R5 gp120 and CCL4 activated p38 and JNK MAP kinases, but only CCL4 activated ERK-1/2.57 Overall, our results further suggest that different signaling pathways can be activated through CCR5 interaction with HIV-1 or natural ligands.
In this study, we also demonstrate that PC-PLC is a key component in the NF-kB–mediated CCL2 production triggered by R5 gp120. In fact, we demonstrate that NF-kB activation is involved in gp120-mediated induction of CCL2 in MDMs, since the NF-kB inhibitor TPCK strongly reduced CCL2 mRNA accumulation and protein secretion. Furthermore, our data show that D609 completely prevented the p65 nuclear translocation mediated by R5 gp120 and strongly reduced CCL2 mRNA accumulation. In this regard, it has been previously reported that in primary endothelial cells PC-PLC–dependent pathways regulate NF-kB–driven TNF-α and NiCl2-mediated CCL2 transcription by regulating the transcriptional activity of NF-kB factors in the transcriptional complex.28 Our observation that D609 inhibits the accumulation of a NF-kB–driven transcript, such as CCL2 mRNA, adds further evidence for a functional role of PC-PLC in the regulation of NF-kB family of transcription factors. In keeping with these results, we found that the gp120-induced secretion of CCL3 and CCL4 was significantly reduced in the presence of neither D609 nor TPCK. How PC-PLC is linked in macrophages to CCR5 upstream and NF-kB downstream remains to be elucidated. Studies are currently under way to determine the relationship between PC-PLC and other signaling pathways, including MAP, PI-3, and Src kinases, activated in gp120-exposed macrophages.
A central question arising from our data concerns the biologic significance of PC-PLC signaling pathway activation by HIV-1 gp120 and its possible involvement in the pathogenesis of HIV infection. In this regard, it is noteworthy that D609 has been previously shown to inhibit the replication of a variety of unrelated DNA and RNA viruses in vitro.58-61 Its broad spectrum of activity suggested that it may affect a fundamental metabolic process required for virus replication. In particular, Mellert et al reported that a D609/undecanoid acid combination inhibited HIV-1 replication in infected lymphoma cells, likely interfering with certain steps of virus assembly.62 Furthermore, Arenzana-Seisdedos et al reported that PC breakdown, initiated by exogenous PC-PLC, induced functional activation of NF-kB and increased HIV-1 replication in chronically infected monocytes and T lymphocytes. This suggests that a cellular transduction pathway dependent on specific PC hydrolysis and activated by physiological and inflammatory inducers may have a major role in the induction of HIV replication in these cells.63 We now demonstrate that PC-PLC activation by HIV-1 gp120 stimulates CCL2 production in MDMs. Previous studies by our and other groups demonstrated a role for CCL2 in the regulation of HIV infection in vitro and in vivo.37,64,65 In particular, we reported that endogenous CCL2 modulates HIV-1 replication and affects cytoskeleton organization in MDMs. This suggest that this CC-chemokine may represent an autocrine factor important for enhancing virion production, likely by affecting the macrophage cytoskeleton.37 On the basis of these previous data and the novel findings described in this paper, we suggest that PC-PLC represents an important cell target for at least some gp120-mediated effects in macrophages. Moreover, the observation that gp120 exposure of MDMs, in the absence of productive infection, is per se sufficient to activate PC-PLC leading to CCL2 secretion suggests that the activation of this signal transduction pathway may contribute to the dysregulation of the chemokine network in noninfected bystander cells. In light of the fundamental role of chemokines as important modulators of HIV infection and replication and as key factors in the regulation of the immune response, we envisage that PC-PLC–mediated signaling pathway may contribute to AIDS pathogenesis by dysregulating uninfected macrophage function, modulating macrophage infection, and controlling the recruitment of new cell targets.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program for providing Tak779 and George Gao for kindly providing the HIV-1 gp120 BaL strain. We are indebted to Pasqualina Bovenzi, Mira van de Velde, and Joanna La Bresh for their technical support in multiplex sandwich ELISA. We thank Anna Maria Fattapposta for excellent editorial assistance. We are indebted to Azzam Maghazachi, Carl Dieffenbach, and Manuela Del Cornò for critically reading the paper.
This work was supported by grants from the NIH (no. R21 AI054215-01), and from the Ministry of Health (Programma Nazionale di ricerca sull'AIDS; 2006, 40G/C) to S.G.
National Institutes of Health
Authorship
Contribution: L.F. designed the research, performed the experiments, and contributed to paper writing; F.S. designed the research and performed the experiments; C.P. and S.C. performed research; C.R., F.B., and F.P. contributed to the intellectual work; S.G. coordinated the research and the paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sandra Gessani, Istituto Superiore di Sanità, Department of Cell Biology and Neurosciences, Viale Regina Elena 299, 00161 Rome, Italy; e-mail: gessani@iss.it.