Abstract
The V617F activating point mutation in Jak2 is associated with a proportion of myeloproliferative disorders. In normal hematopoietic cells, Jak2 signals only when associated with a growth factor receptor, such as the erythropoietin receptor (EpoR). We sought to identify the molecular requirements for activation of Jak2V617F by introducing a point mutation in the FERM domain (Y114A), required for receptor binding. Whereas BaF3.EpoR cells are readily transformed by Jak2V617F to Epo independence, we found that the addition of the FERM domain mutation blocked transformation and the induction of reactive oxygen species. Further, while cells expressing Jak2V617F had constitutive activation of STAT5, cells expressing Jak2V617F/Y114A did not, suggesting that signaling is defective at a very proximal level. In addition, expression of the Myc and Pim proto-oncogenes by Jak2V617F was found to be FERM domain dependent. An inducible constitutively active STAT5 mutant expressed in BaF3 cells was sufficient to induce Myc and Pim. Finally, the FERM domain in Jak2V617F was also required for abnormal hematopoiesis in transduced primary murine fetal liver cells. Overall, our results suggest that constitutive activation of Jak2 requires an intact FERM domain for a transforming phenotype, and is necessary for activation of the major target of Jak2, STAT5.
Introduction
The nonreceptor tyrosine kinase Jak2 is a frequent target of the activating V617F point mutation in patients with polycythemia vera, essential thrombocythemia, idiopathic myelofibrosis, as well as in several myeloproliferative disorders and infrequently in myelodysplastic syndromes.1-7 Additional genetic abnormalities have been found in the Jak2 gene in a variety of hematologic malignancies but with lower frequencies, such as a T875N activating point mutation in an acute megakaryoblastic leukemia (AMKL) cell line.8 Additional oncogenic Jak2 proteins include the Etv6-Jak2 fusions, as a consequence of a t(9;12)(p24;p13) or variant translocations in patients with a chronic myeloproliferative disease or acute lymphoblastic leukemia.9,10 The Pcm1-Jak2 fusion, as a consequence of a recurrent t(8;9)(p21;p24), was found in patients presented with clinical and hematologic features resembling chronic-phase chronic myeloid leukemia (CML), chronic eosinophilic leukemia, acute leukemia similar to blast crisis of CML, or Bcr-Abl–positive acute lymphoblastic leukemia.11 The Bcr-Jak2 fusion gene results from the t(9;22)(p24;q11) translocation and has been described in only one patient with a CML-like phenotype.12 Jaks are thought to primarily tyrosine phosphorylate and thus activate signal transducer and activator of transcription (STAT) transcription factors, even though it is clear that there are other activities. It has been suggested that activated Jak2 transforms in part through the STAT5 transcription factor. A constitutively active form of STAT5 is sufficient to support growth of murine BaF3 cells,13 and STAT5 expression is required for immature hematopoiesis and fetal erythropoiesis in vivo.14,15 STAT5 may therefore help to support growth of hematopoietic cells transformed by Jak2. The exact transcriptional targets that contribute to this phenotype are not known, but previous data suggest that STAT5 can directly regulate the expression of proteins known to support growth and viability, such as the serine/threonine kinase Pim1.16 Additional growth-associated proteins, such as Myc, are possibly regulated by STAT5 through a distal mechanism.16 Nevertheless, the expression of Pim1 and Myc may require similar structural elements within growth factor receptors, including the erythropoietin receptor (EpoR).17
A general mechanism of Jak2 activation and autophosphorylation includes recruitment of the kinase to a membrane proximal region of ligand-activated cytokine receptors. The 4 mammalian Jak family members (Jak1, Jak2, Jak3, and Tyk2) contain several regions with significant sequence homology among the kinases (see for review Yamaoka et al18 ). The tyrosine kinase domain is adjacent to the pseudokinase domain, which is thought to regulate its kinase activity. This mechanism of regulation is likely to be compromised in Jak2V617F. The amino-terminal conserved FERM domain (also known as the protein 4.1, ezrin, radixin, moesin domain) regulates binding to box 1 and box 2 regions in the cytoplasmic portion of membrane-associated receptors and may be involved in stabilizing receptor expression, such as the EpoR.19 Thus, full transcriptional activation of STAT5 downstream of Jak2 is expected to require the interaction of the box 1/box 2 motif in the EpoR with the Jak2 FERM domain. The receptor requirements for transformation by Jak2V617F in vitro and in vivo are still being worked out and may be different, depending on the disease outcome. However, erythroid progenitors in polycythemia vera show hypersensitivity to erythropoietin (Epo) or Epo independent growth, suggesting that in vivo Jak2V617F can signal, at least in part, through this pathway.
The detailed molecular requirements for Jak2V617F activation are of interest and may require the interaction with multiple proteins and phosphorylation of regulatory sites within Jak2. Our goal was to further define signaling mechanisms activated by Jak2V617F and identify targets that would aid in the development and implementation of targeted therapies for the treatment of Jak2V617F-related disorders. Using an EpoR-expressing BaF3 cell line model system, we found that a functional FERM domain was required to inhibit p27Kip and induce cyclin D2 expression as well as maintain Jak2V617F-induced factor-independent cell growth. The Y114A mutation in the FERM domain, known to inhibit receptor box 1/2 motif binding,19 also regulated the phosphorylation of Jak2 and STAT5 as well as the induction of Myc, Pim, and elevated reactive oxygen species levels. Therefore, Jak2V617F is likely to require interaction with a cytokine receptor through the FERM domain for full activation. Our data also establish an important role for Myc and Pim in transformation by Jak2V617F and their regulation through the FERM domain. In addition, whereas Jak2V617F readily induces cytokine-dependent and-independent colony formation, Jak2V617F.Y114A does not. These data are further supported by our findings, demonstrating that Jak2V617F.Y114A fails to induce splenomegaly in a murine model, indicative of extramedullary hematopoiesis. Finally, these results suggest small molecules that target the FERM domain of Jak2, or Myc and Pim, as new therapies in diseases associated with Jak2V617F mutations.
Methods
The animal experiments were in compliance with institutional protocols and were approved by the Institutional Animal Care and Use Committee.
Cells
The human erythroleukemia cell line HEL was grown in RPMI1640 medium (Mediatech, Herndon, VA) containing 10% fetal calf serum (Tissue Culture Biologicals, Tulare, CA), supplemented with 1 mM sodium pyruvate (Gibco/Invitrogen, Carlsbad, CA). 293T cells were maintained in Dulbecco modified eagle medium (Mediatech) containing 10% fetal calf serum. BaF3 cells with doxycycline-inducible constitutively active STAT5 were maintained as described before.20 STAT5 expression was induced by treatment with 1 μg/mL doxycycline (Sigma-Aldrich, St Louis, MO) in the absence of murine interleukin-3. BaF3.EpoR cells (expressing the erythropoietin receptor, EpoR) were kindly provided by Dr A. D'Andrea (Dana-Farber Cancer Institute, Boston, MA). BaF3 and BaF3.EpoR cell lines were kept in interleukin-3–containing medium unless otherwise indicated. Additional clonal BaF3 cell lines were generated by infection with derivatives of pMSCVJak2.IRES.EGFP1 or pMSCVJak2V617F.IRES.CD419 containing ecotropic retroviruses. In some experiments, cells were treated with Jak inhibitor 1 (Calbiochem, San Diego, CA) and N-acetylcysteine (Sigma-Aldrich) or were transfected with specific human siRNA (siGenome; Dharmacon, Lafayette, CO). Cell growth and viability were determined by trypan blue exclusion or by a colorimetric method (CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay; Promega, Madison, WI).
Electroporation of siRNA
Human Myc-, Pim1-, and Pim2-specific and control siRNA were used to transfect HEL cells in 100 μL Nucleofector solution R (Amaxa, Gaithersburg, MD) according to the manufacturer's instructions using the Nucleofector device (Amaxa). In some experiments, the pmaxGFP vector (Amaxa) was transfected to survey transfection efficiency. Cells were transferred to culture medium after electroporation and incubated at 37°C in 5% CO2.
Immunoblotting
Proteins were extracted from whole cells in buffer containing Tris (50 mM, pH 8.0; Invitrogen), NaCl (150 mM; Fisher Scientific, Pittsburgh, PA), NP40 (1% vol/vol; Calbiochem), deoxycholic acid (0.5% wt/vol; Fisher Scientific), sodium dodecylsulfate (0.1% wt/vol; Bio-Rad, Hercules, CA), NaF (1 mM; Sigma-Aldrich), Na3VO4 (1 mM; Sigma-Aldrich), and glycerol (10% vol/vol; Invitrogen) supplemented with protease inhibitor cocktail (Complete; Roche Diagnostics, Indianapolis, IN). Polyclonal rabbit antibodies against STAT5 (C-17), cyclin D2 (M-20), p27Kip (C-19), Jak2 (C-20), EpoR (M-20; Santa Cruz Biotechnology, Santa Cruz, CA); Jak2 (Upstate Biotechnology, Lake Placid, NY); c-Myc (Cell Signaling, Beverly, MA); or phosphorylated STAT5 (Tyr694; Cell Signaling); and mouse monoclonal antibodies against Pim1 (12H8), Pim2 (1D12; Santa Cruz Biotechnology); or phospho-tyrosine (4G10, Upstate Biotechnology) or β-actin (Sigma-Aldrich) were used for immunoblotting or immunoprecipitation.
Expression vectors and viral supernatant
Retroviral expression vectors pMSCV.IRES.CD4 and pMSCV.IRES.EGFP containing the murine Jak2 cDNA were used. The V617F and the Y114A mutations in Jak2 were generated by site-directed mutagenesis (Quikchange-XL; Stratagene, La Jolla, CA) and confirmed by full-length Jak2 DNA sequencing. Ecotropic retroviruses using the pEcopak packaging plasmid were generated in 293T cells to transduce BaF3 cells or hematopoietic stem cells with standard methods. Infected cells were sorted for GFP expression by flow cytometry or CD4 labeling using magnetic beads (Invitrogen).
Cell-cycle analysis
Cells were fixed with 70% (vol/vol) ethanol in phosphate-buffered saline and incubated on ice for 30 minutes. RNAse-treated samples (10 μg RNAse/mL for 20 minutes at 37°C) were stained with propidium iodide (5 μg/mL; Sigma-Aldrich) at 4°C for at least 10 minutes. Cell-cycle parameters were determined by fluorescence-activated cell sorting (FACS) analysis using a Beckman-Coulter Cytomics FC500 flow cytometer (Beckman-Coulter, Miami, FL). The DNA analysis program MultiCycle for Windows (Phoenix Flow Systems, San Diego, CA) was used to determine cell-cycle distribution.
Measurement of intracellular reactive oxygen species
Fluorescence image analysis was used to determine the relative levels of reactive oxygen species (ROSs) in response to various stimuli. Cells were treated before FACS analysis, and the relative levels of intracellular ROSs analyzed using the cell-permeable redox-sensitive fluorochrome DCF-DA (2′,7′-dichlorofluorescein diacetate; Fisher Scientific/Acros Organics, Pittsburgh, PA). Cells were incubated with DCF-DA (5 μM) for 5 minutes at 37°C and subsequently washed twice in cold PBS before analysis using a Coulter Epics XL flow cytometer (Beckman-Coulter).
Fetal liver transplantation and analysis of mice
Murine fetal liver transplantation experiments were performed as previously described.21 Briefly, the viral titer of pMSCV-GFP retroviral supernatants (Jak2V617F, Jak2V617F.Y114A) was determined by transducing BaF3 cells with supernatant (plus polybrene, 10 μg/mL) and analyzed for the percentage of GFP-positive cells by flow cytometry 2 days after transduction. E13.5 fetal livers of C57BL/6 mice (Taconic, Germantown, NY) were harvested; single-cell suspensions were made by flushing through mesh graft; and cells were cultured for 24 hours in transplant medium (RPMI + 10% FBS + 6 ng/mL IL-3, 10 ng/mL IL-6, and 10 ng/mL stem-cell factor). Cells were treated by spin infection with retroviral supernatants (1 mL supernatant per 4 × 106 cells, plus polybrene, plus HEPES 30 μL/4 mL) and centrifuged at 1000g for 90 minutes at 24 hours before and on the day of the transplantation. Whole fetal liver cells (106) were resuspended in Hanks balanced salt solution, and injected into lateral tail veins of lethally irradiated (2 × 550 cGy) C57BL/6 recipient mice. Mice were maintained on acidified water.
Analysis of disease in mice
C57/Bl6 mice were killed for analysis at 9 weeks. Peripheral blood was collected from the retro-orbital cavity using EDTA glass capillary tubes and analyzed by automated complete and differential blood cell counts and blood smears (stained with Wright and Giemsa). Spleen weights were assessed upon necropsy. Single-cell suspensions of spleen were prepared by pressing tissue through a cell strainer, or of bone marrow by flushing bones, followed by red blood cell lysis.
Colony assays
To detect blast-forming unit erythroid (BFU-e) colonies, 105 peripheral blood and bone marrow and 105 spleen cells from Jak2V617F or Jak2V617F.Y114A mice were plated in duplicate in methylcellulose (M3434, M3234; Stem Cell Technologies, Vancouver, BC) containing 3 U/mL Epo, 10 ng/mL recombinant murine interleukin-3 (rmIL-3), 10 ng/mL rmIL-6, and 50 ng/mL recombinant murine stem cell factor or no additional cytokines according to the manufacturer's protocol. BFU-erythroid colonies were counted after 8 days in culture.
Results
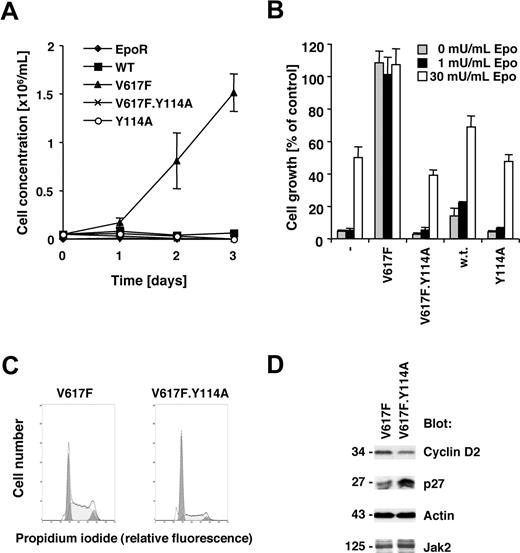
Jak2V617F requires an intact FERM domain for transformation of BaF3.EpoR cells
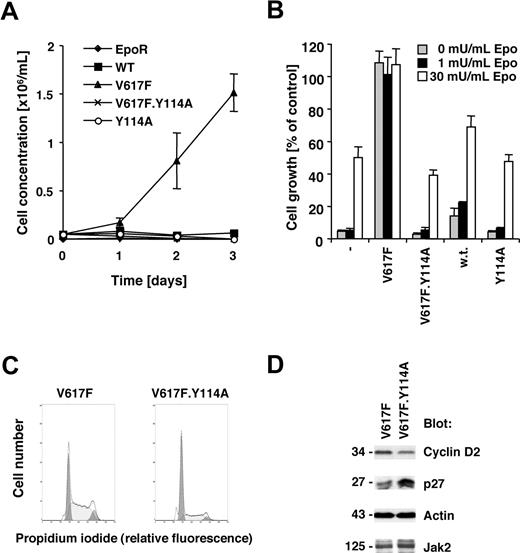
The V617F activating point mutation of the Jak2 tyrosine kinase is sufficient to transform the murine cell line BaF3.EpoR, expressing the EpoR, to growth factor independence.1,22 The EpoR itself or related receptors have been suggested to be required for transformation by Jak2V617F in this cell line model22,23 and may have an important role in Jak2V617F-positive polycythemia vera. We sought to identify molecular requirements within Jak2V617F for transformation. The V617F point mutation or a Y114A point mutation within the FERM domain as well as a combination thereof were introduced in Jak2 and expressed in BaF3.EpoR cells. The Y114A substitution has been previously shown to disrupt the recruitment of Jak2 to the box 1 motif within the thrombopoietin receptor,19 but is not a target of tyrosine phosphorylation.24 Expression of Jak2V617F in BaF3.EpoR cells led to factor-independent growth, in contrast to control cells or wild-type Jak2–transfected cells (Figure 1A). In addition, the Y114A mutation in Jak2 did not alter the growth factor requirement compared with wild-type Jak2–expressing cells. However, introduction of the Y114A mutation in Jak2V617F restored the Epo requirement for cell growth (Figure 1B). Similar results were obtained when BaF3.EpoR cells were stimulated with interleukin-3 (not shown).
Jak2V617F requires an intact FERM domain for transformation of BaF3.EpoR cells. Parental BaF3.EpoR cells or cells expressing various mutants of Jak2 (as indicated) were used. (A,B) Relative growth of cell lines was determined in a 72-hour culture. Cell growth is shown as a change in cell concentration (A) or was calculated as a percentage compared with cells that were grown in an optimal amount of Epo (B). (C) Cell-cycle distribution was determined in the absence of growth factor (typical experiment). (D) Cells expressing Jak2V617F were used and compared with cells expressing Jak2V617F.Y114A in the absence of growth factors. Cellular proteins were detected by immunoblotting as indicated.
Jak2V617F requires an intact FERM domain for transformation of BaF3.EpoR cells. Parental BaF3.EpoR cells or cells expressing various mutants of Jak2 (as indicated) were used. (A,B) Relative growth of cell lines was determined in a 72-hour culture. Cell growth is shown as a change in cell concentration (A) or was calculated as a percentage compared with cells that were grown in an optimal amount of Epo (B). (C) Cell-cycle distribution was determined in the absence of growth factor (typical experiment). (D) Cells expressing Jak2V617F were used and compared with cells expressing Jak2V617F.Y114A in the absence of growth factors. Cellular proteins were detected by immunoblotting as indicated.
We had previously demonstrated that Jak2V617F expression is associated with release of factor-deprived cells from G1 cell-cycle arrest, likely regulated through increased cyclin D2 and re-duced p27Kip expression.22 Comparing Jak2V617F cells to Jak2V617F.Y114A cells, we found that the Y114A mutations led to an accumulation of cells in the G1 cell-cycle phase (Figure 1C). Cells expressing the additional Y114A mutation showed an increase in G1 phase to 62.4% (± 2.5%; n = 3), compared with the V617F-expressing cells with 35.0% (± 1.3%, n = 3; Table 1). The Y114A mutation also blocked the V617F-dependent expression of cyclin D2 and suppression of p27Kip (Figure 1D). These data demonstrate that a functional FERM domain in Jak2V617F is required for growth factor–independent growth of BaF3.EpoR cells.
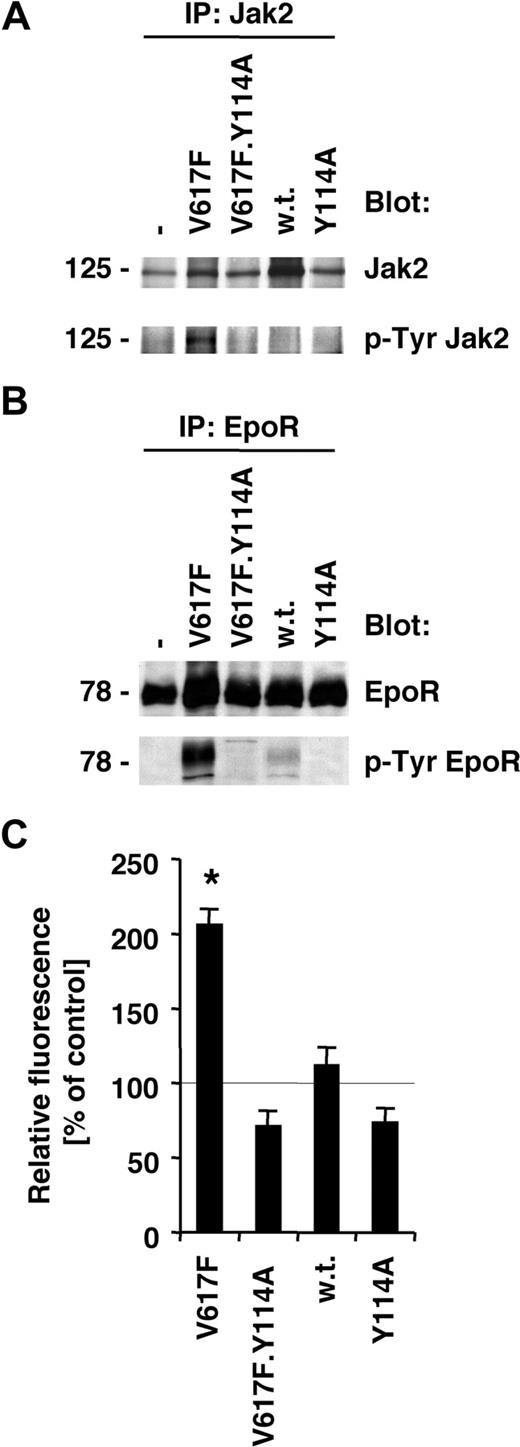
Activation of the Jak2/EpoR pathway by Jak2V617F is inhibited by the Y114A substitution
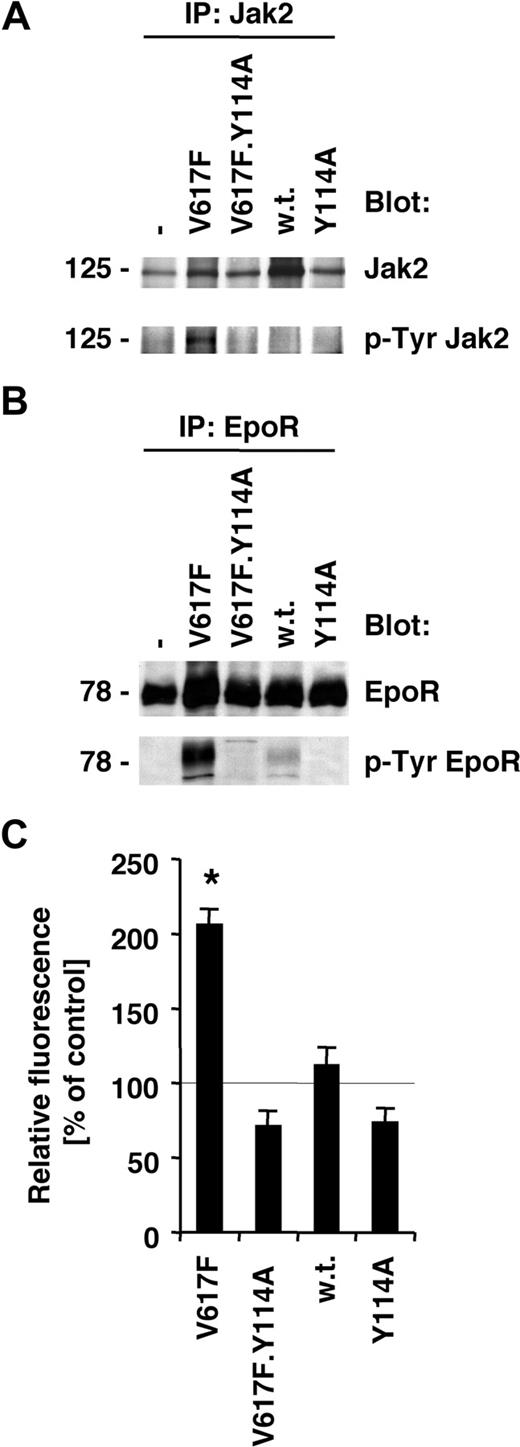
Normal activation of the EpoR by its ligand leads to Jak2-dependent receptor phosphorylation and activation of signaling mechanisms. The V617F mutation in Jak2 itself is thought to release inhibitory constraints, thus partially deregulating ligand-dependent mechanisms. Consistent with our previous data,1,22 we found that Jak2V617F is constitutively tyrosine phosphorylated in BaF3.EpoR cells (Figure 2A), and expression of Jak2V617F leads to tyrosine phosphorylation of the EpoR (Figure 2B). Neither wild-type Jak2 nor Jak2.Y114A increased tyrosine phosphorylation of Jak2 or the EpoR in these cells. Furthermore, a Jak2 mutant that carried both, the V617F and the Y114A mutation, blocked the increase in phosphorylation of Jak2 (Figure 2A) and the EpoR (Figure 2B). We have previously shown that Jak2V617F leads to an increase of ROSs in hematopoietic cells, and this effect was crucial for cell growth and required Jak2 tyrosine kinase activity.22 We therefore also sought to determine whether the Y114A mutation could reduce ROSs in the Jak2V617F-transformed BaF3.EpoR cells. Similar to our previous data,22 there was a significant increase in ROSs in Jak2V617F-expressing cells compared with untransfected cells (Figure 2C). This result was in contrast to wild-type Jak2–expressing cells, which did not increase intracellular ROSs, and neither did Jak2.Y114A-expressing cells. Consistent with an important role of Jak2V617F activity in ROS formation, we did not observe an increase in ROSs in the Jak2V617F.Y114A double mutant–expressing BaF3.EpoR cells (Figure 2C). In additional experiments, we confirmed the biochemical functionality of the EpoR/Jak2 pathway in Epo-stimulated cells (not shown). These data suggest a model whereby signaling through Jak2V617F requires activation through a process that involves FERM domain–dependent interaction with the Epo or related receptors.
Activation of the Jak2/EpoR pathway by Jak2V617F is inhibited by the Y114A mutation. Parental BaF3.EpoR cells or cells expressing various mutants of Jak2 (as indicated) were used. (A,B) Parental BaF3.EpoR cells and cells infected to express Jak2 or various mutants were growth factor deprived. The phosphorylation and protein expression of Jak2 (A) or the Epo receptor (B) were detected after specific immunoprecipitations by immunoblotting as indicated. (C) Intracellular ROS levels were measured by DCF-DA staining (n = 3) in the absence of growth factors. Parental BaF3.EpoR cells were used as a control.
Activation of the Jak2/EpoR pathway by Jak2V617F is inhibited by the Y114A mutation. Parental BaF3.EpoR cells or cells expressing various mutants of Jak2 (as indicated) were used. (A,B) Parental BaF3.EpoR cells and cells infected to express Jak2 or various mutants were growth factor deprived. The phosphorylation and protein expression of Jak2 (A) or the Epo receptor (B) were detected after specific immunoprecipitations by immunoblotting as indicated. (C) Intracellular ROS levels were measured by DCF-DA staining (n = 3) in the absence of growth factors. Parental BaF3.EpoR cells were used as a control.
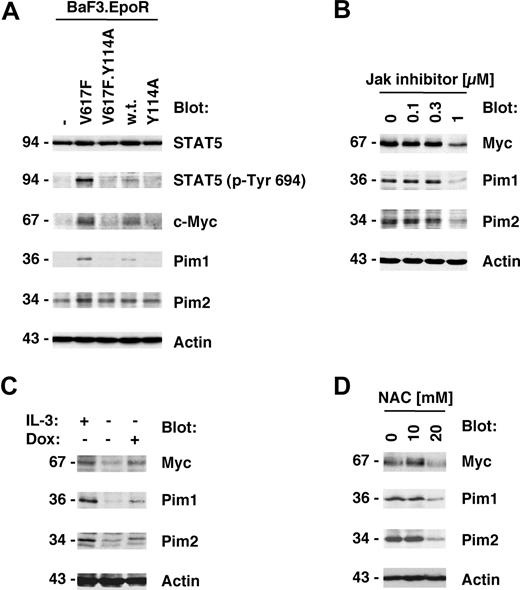
Y114A in Jak2V617F inhibits Myc and Pim expression induced by Jak2V617F
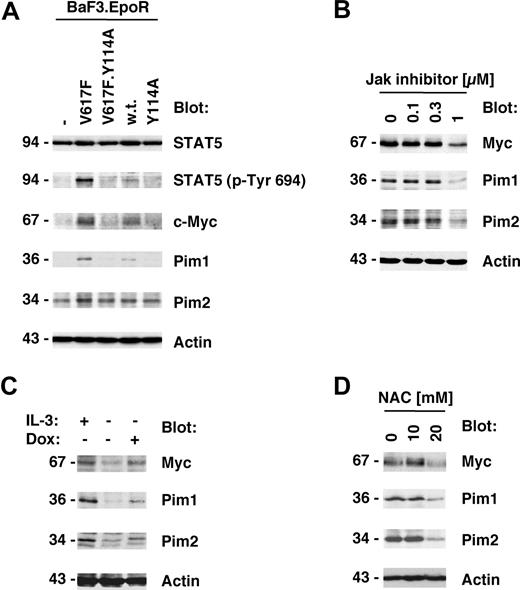
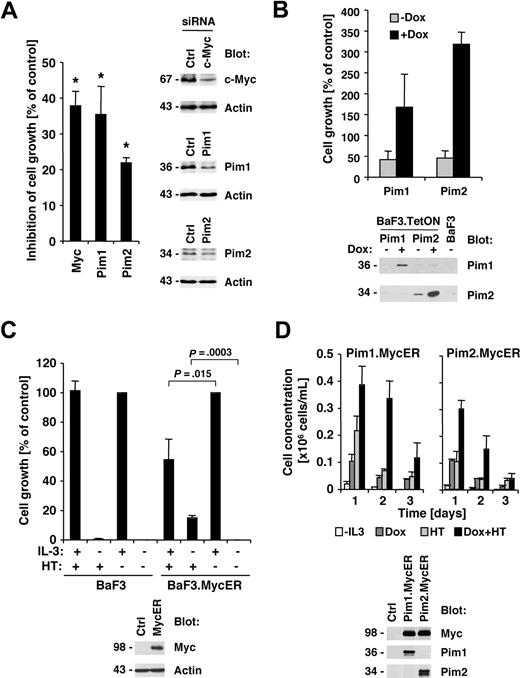
Jak2V617F kinase activity is important for its biologic activity and initiation of downstream signaling events.1,22 We therefore also sought to determine whether the Y114A mutation would affect the phosphorylation of the Jak2 substrate STAT5. As expected, STAT5 phosphorylation on its activation site was elevated in Jak2V617F-expressing cells compared with control cells or cells expressing wild-type Jak2 (Figure 3A top panels). In addition, consistent with the above data, the Y114A mutation was not sufficient to activate STAT5 and inhibited Jak2V617F-mediated activation of STAT5. We next asked if genes that have previously shown to be associated with myeloproliferative disorders25,26 were also regulated by Jak2V617F and if this regulation is dependent on an intact FERM domain. Consistent with the published data, we identified Myc, Pim1, and Pim2 as major induced targets of Jak2V617F in BaF3.EpoR cells compared with control cells (Figure 3A bottom panels). In addition, similar to activation of STAT5, the Y114A mutation in the FERM domain inhibited Jak2V617F-induced expression of these proteins. To confirm that the expression of Myc and the Pim proteins was dependent on Jak2 kinase activity, we treated Jak2V617F-expressing HEL cells with Jak inhibitor I (0-1 μM). The inhibitor was found to reduce Myc, Pim1, and Pim2 expression but did not affect actin levels (Figure 3B), consistent with a role of Jak2 activity in the regulation of these proteins.
Y114A in Jak2V617F inhibits Myc and Pim expression regulated by Jak2V617F. Protein expression was determined in cellular lysates by immunoblotting as indicated. (A) Parental BaF3.EpoR cells and cells infected to express Jak2 or various mutants were growth factor deprived. The phosphorylation of STAT5 on its activation site and the expression of Pim1, Pim2, and c-Myc was detected. (B) HEL cells were treated for 48 hours with the indicated amount of Jak inhibitor I and cellular proteins detected. (C) Untreated BaF3 cells with doxycycline-inducible constitutively active STAT5 were used and compared with cells treated with 1 μg/mL doxycycline (Dox) or cells maintained in interleukin-3 (IL-3)–containing medium. Cellular expression of Pim1, Pim2, and c-Myc was detected. (D) HEL cells were treated for 48 hours with the indicated amount of N-acetylcysteine (NAC) and cellular proteins detected. *Significant differences (P ≤ .05) were observed between treated and control cells.
Y114A in Jak2V617F inhibits Myc and Pim expression regulated by Jak2V617F. Protein expression was determined in cellular lysates by immunoblotting as indicated. (A) Parental BaF3.EpoR cells and cells infected to express Jak2 or various mutants were growth factor deprived. The phosphorylation of STAT5 on its activation site and the expression of Pim1, Pim2, and c-Myc was detected. (B) HEL cells were treated for 48 hours with the indicated amount of Jak inhibitor I and cellular proteins detected. (C) Untreated BaF3 cells with doxycycline-inducible constitutively active STAT5 were used and compared with cells treated with 1 μg/mL doxycycline (Dox) or cells maintained in interleukin-3 (IL-3)–containing medium. Cellular expression of Pim1, Pim2, and c-Myc was detected. (D) HEL cells were treated for 48 hours with the indicated amount of N-acetylcysteine (NAC) and cellular proteins detected. *Significant differences (P ≤ .05) were observed between treated and control cells.
These results demonstrate that activation of the Jak2 pathway is required for optimal induction of Myc and Pim protein levels. To further determine whether activation of the Jak2 substrate STAT5 is sufficient to regulate Myc and Pim protein expression, we used a BaF3 cell line with doxycycline-inducible “constitutively active STAT5.”20 Cells maintained in the absence of growth factor were compared with doxycycline-treated cells or cells stimulated with interleukin-3–containing medium as a positive control. Doxycycline treatment by itself does not alter Myc and Pim levels in BaF3 cells (not shown). Induction of active STAT5 expression led to an increase in Myc and Pim proteins, which was comparable with the increase observed after interleukin-3 treatment (Figure 3C). In control experiments, the induction of STAT5 in these cell lines was determined by immunoblotting (not shown and Walz et al22 ). Overall, these results demonstrate that activation of the Jak2/STAT5 pathway is sufficient for the induction of Myc and Pim proteins. Since activation of the Jak2/Stat5 pathway is associated with elevated reactive oxygen species (ROSs), we also looked at the effect of the antioxidant N-acetylcysteine (NAC) on Myc and Pim protein expression in HEL cells (Figure 3D). HEL cells were treated with 0 to 20 mM NAC and protein expression was measured 2 days later. Myc as well as Pim1 and Pim2 were reduced in expression at a concentration of 20 mM NAC. The same NAC concentration is also sufficient to reduce cell growth in these cells.22 These results suggest that a high oxidative state is important to maintain expression of Myc and Pim proteins in HEL cells and may hint at multiple levels of regulation.
Myc and Pim proteins are required for full transformation of HEL cells
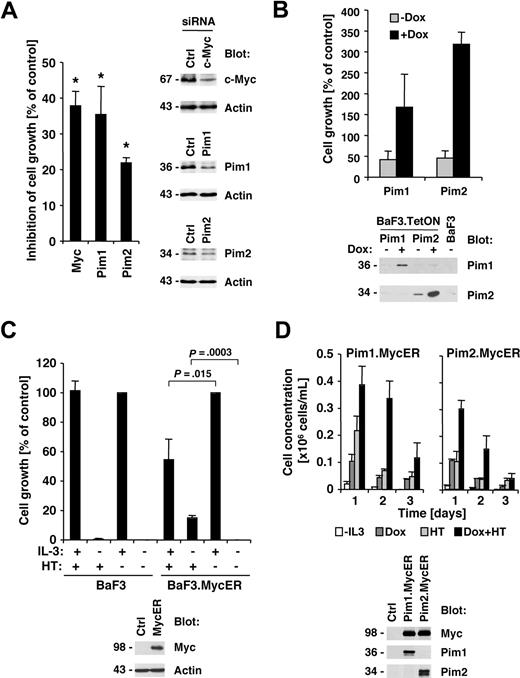
Our previous data demonstrated that Jak inhibitor I efficiently suppressed cell growth of HEL cells,22 and here we demonstrate that the efficacy of the Jak inhibitor also correlates with suppression of Myc and Pim proteins. We therefore asked if reduction in Myc or Pim protein expression would also have an effect on growth in HEL cells. HEL cells were transfected with siRNA pools targeting Myc, Pim1, Pim2, or control siRNA. Whereas Myc and Pim1 were found to be efficiently reduced in expression, the siRNA for Pim2 only slightly reduced expression compared with control siRNA–transfected cells (Figure 4A right panels). Finally, the effect of c-Myc, Pim1, and Pim2 RNA interference on cell growth of HEL cells was determined by comparison with control siRNA–transfected cells. Transfection of Myc (37.9% ± 4.1% inhibition), Pim1 (35.4% ± 7.9% inhibition), and Pim2 (21.9% ± 1.4% inhibition) consistently led to a reduction in cell growth compared with control-transfected cells (Figure 4A left panel). Similar to the protein expression data, the Pim2 siRNA led to a less significant reduction in cell growth. These data suggest that Myc and Pim proteins are required for optimal transformation through Jak2V617F. Based on these findings, we used the BaF3 model with tetracycline-inducible expression to determine whether Pim1 or Pim2 are sufficient to support growth. Expression of murine Pim1 and Pim2 was induced by doxycycline treatment and compared with untreated cells (Figure 4B bottom panel). Pim1 and Pim2 are not expressed in the unstimulated parental BaF3 cells, and some leaky Pim2 expression was found in the untreated BaF3.TetON.Pim2 cells. Interestingly, induction of both Pim1 (1.7 ± 0.8-fold) and Pim2 (3.2 ± 0.3-fold) led to an increase in cell growth. These results are consistent with data previously published by Adam et al27 and support a role for Pim proteins in cell growth. However, we did not find Pim1 or Pim2 to support long-term growth in this model (not shown). To determine whether Myc contributes to cell growth, we expressed a Myc-estrogen receptor (MycER) fusion protein in BaF3 cells (Figure 4C bottom panel), whereby 4-hydroxy-tamoxifen (HT) treatment leads to nuclear translocation of Myc.28 HT treatment did not have an effect on cell growth in the parental BaF3 cells in the presence or absence of interleukin-3 (Figure 4C top left panel). However, in MycER-expressing cells, HT treatment lead to a significant increase in cell growth in the absence of interleukin-3 (15.0% ± 1.6% increase) but also suppressed interleukin-3–induced cell growth (45.6% ± 13.9% reduction; Figure 4C top right panel). Thus, overexpression of Myc inhibits interleukin-3–induced cell growth, but may be sufficient to contribute to cell growth in the absence of growth factor. Finally, we asked if Myc and Pim proteins have the potential to cooperate in the induction of cell growth in this model. We combined the doxycycline-inducible Pim expression system with the MycER system by infecting the Pim-expressing cells with a MycER-expressing construct (Figure 4D bottom panel). The combination of Pim1, Pim2, and Myc led transiently to a larger increase in cell growth, compared with each protein alone (Figure 4D top panel), demonstrating the cooperative effect of both proteins, but coexpression did not support long-term growth.
Myc and Pim proteins are required for full transformation of HEL cells. (A) HEL cells were transfected with either control siRNA or c-Myc–, Pim1-, and Pim2-specific siRNA. Relative growth of HEL cells 72 hours after siRNA transfection was calculated as a percentage compared with control siRNA–transfected cells (n = 3, left panel). The change in expression of c-Myc, Pim1, and Pim2 after siRNA treatment was determined in whole-cell lysates (right panel). *Significant differences (P ≤ .05) were observed between treated and control cells. (B) Untreated BaF3 cells with doxycycline-inducible Pim1 and Pim2 were used and compared with cells treated with 1 μg/mL doxycycline (DOX) or BaF3 cells maintained in interleukin-3 (IL-3)–free medium. Cell growth was measured after 72 hours (top panel) or induction of Pim1 and Pim2 after doxycycline treatment (24 hours) by immunoblotting as indicated (bottom panel). (C) Parental BaF3 cells or cells expressing the MycER fusion protein were left untreated or treated with 4-hydroxy-tamoxifen (HT). The expression of the MycER fusion protein was detected by Myc immunoblotting (bottom panel). (D) Untreated BaF3 cells with doxycycline-inducible Pim1 and Pim2 as well as HT-inducible Myc-ER were used and compared with cells treated with 1 μg/mL DOX, HT, or combinations thereof (0.2 × 106 cells/mL). Cell concentrations were measured over a 3-day period (n = 3) and protein expression was assayed after 24 hours of DOX and HT treatment as indicated.
Myc and Pim proteins are required for full transformation of HEL cells. (A) HEL cells were transfected with either control siRNA or c-Myc–, Pim1-, and Pim2-specific siRNA. Relative growth of HEL cells 72 hours after siRNA transfection was calculated as a percentage compared with control siRNA–transfected cells (n = 3, left panel). The change in expression of c-Myc, Pim1, and Pim2 after siRNA treatment was determined in whole-cell lysates (right panel). *Significant differences (P ≤ .05) were observed between treated and control cells. (B) Untreated BaF3 cells with doxycycline-inducible Pim1 and Pim2 were used and compared with cells treated with 1 μg/mL doxycycline (DOX) or BaF3 cells maintained in interleukin-3 (IL-3)–free medium. Cell growth was measured after 72 hours (top panel) or induction of Pim1 and Pim2 after doxycycline treatment (24 hours) by immunoblotting as indicated (bottom panel). (C) Parental BaF3 cells or cells expressing the MycER fusion protein were left untreated or treated with 4-hydroxy-tamoxifen (HT). The expression of the MycER fusion protein was detected by Myc immunoblotting (bottom panel). (D) Untreated BaF3 cells with doxycycline-inducible Pim1 and Pim2 as well as HT-inducible Myc-ER were used and compared with cells treated with 1 μg/mL DOX, HT, or combinations thereof (0.2 × 106 cells/mL). Cell concentrations were measured over a 3-day period (n = 3) and protein expression was assayed after 24 hours of DOX and HT treatment as indicated.
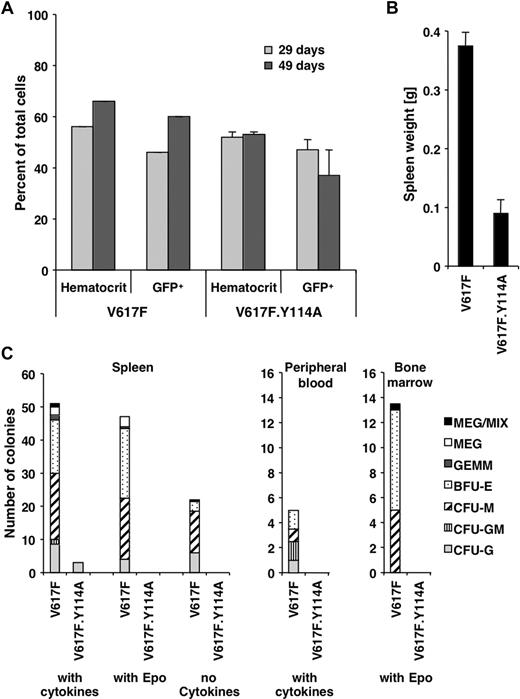
Abnormal hematopoiesis induced by Jak2V617F requires a functional FERM domain
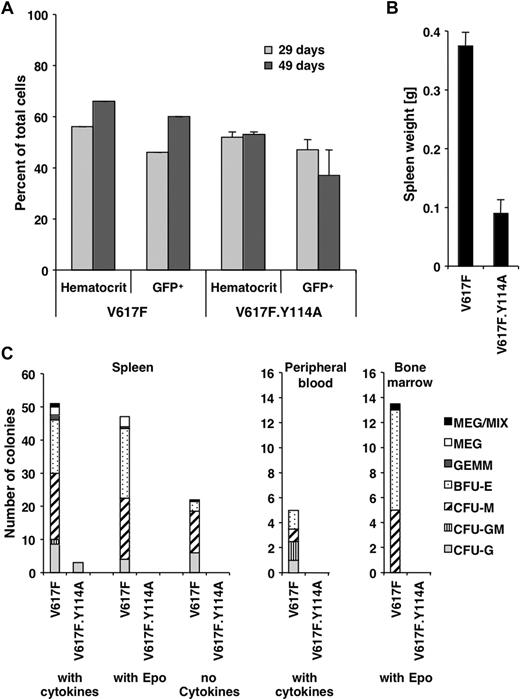
The above data exploring the role of the FERM domain in transformation are mostly limited to the BaF3.EpoR in vitro model. To address the in vivo role of the FERM domain for transformation, we used a previously established mouse model with long latency of the Jak2V617F-associated polycythemia vera–like disease, which is evident by an increased hematocrit and splenomegaly.21 Similar to these previously established parameters, the hematocrit or the number of Jak2V617F- and Jak2V617F.Y114A-expressing hematopoietic cells (GFP positive) in peripheral blood did not change within the first 7 weeks (Figure 5A). Mice were killed after 10 weeks and showed an increased spleen weight for Jak2V617F mice (0.38 ± 0.02 g, n = 3) and a normal spleen weight (0.09 ± 0.02 g, n = 3) compared with previous experiments21 in Jak2V617F.Y114A mice (Figure 5B). Splenomegaly is commonly observed in patients with polycythemia vera and is a sign of extramedullary hematopoiesis.29 The Y114A mutation is therefore likely to inhibit the ability of Jak2V617F-transformed cells to undergo abnormal hematopoiesis. We further found that the formation of different lineage colonies from spleen-, peripheral blood–, and bone marrow–derived cells was greatly reduced in Jak2V617F.Y114A mice compared with Jak2V617F mice (Figure 5C). We observed little or no colonies in Jak2V617F.Y114A cultures in the presence of cytokines (spleen, peripheral blood), the absence of cytokines (spleen), or with Epo alone (spleen, bone marrow). In addition, in control experiments, we did not find a significant impact of the Y114A mutation on colony formation in the presence of cytokines, using bone marrow cells (not shown). Overall, these data confirm a crucial role for the FERM domain in transformation by Jak2V617F and imply an important role for type I cytokine receptors in this process. Future experiments will be necessary to define a link between activation of specific receptors and disease development.
The Y114A mutation within the FERM domain inhibits abnormal hematopoiesis by Jak2V617F. Mice were lethally irradiated and the bone marrow was reconstituted with Jak2V617F- and Jak2V617F.Y114A-expressing fetal liver cells. (A) The hematocrit and the percentage of GFP-positive cells (GFP+) in peripheral blood of Jak2V617F- and Jak2V617F.Y114A-expressing mice were determined, at the times indicated. (B) Mice were killed on day 70 and the weight of the spleens was compared. (C) Spleen, peripheral blood, and bone marrow cells were used to compare the potential for colony-forming units (CFUs) or blast-forming units (BFUs), including megakaryocyte (MEG), mixed (MIX), granulocyte/erythroid/macrophage/megakaryocyte (GEMM), BFU erythroid (BFU-E) or CFU macrophage (CFU-M), CFU granulocyte/macrophage (CFU-GM), and CFU granulocyte (CFU-G). Cells were left untreated or maintained in the presence of growth factors or erythropoietin (Epo; n = 3).
The Y114A mutation within the FERM domain inhibits abnormal hematopoiesis by Jak2V617F. Mice were lethally irradiated and the bone marrow was reconstituted with Jak2V617F- and Jak2V617F.Y114A-expressing fetal liver cells. (A) The hematocrit and the percentage of GFP-positive cells (GFP+) in peripheral blood of Jak2V617F- and Jak2V617F.Y114A-expressing mice were determined, at the times indicated. (B) Mice were killed on day 70 and the weight of the spleens was compared. (C) Spleen, peripheral blood, and bone marrow cells were used to compare the potential for colony-forming units (CFUs) or blast-forming units (BFUs), including megakaryocyte (MEG), mixed (MIX), granulocyte/erythroid/macrophage/megakaryocyte (GEMM), BFU erythroid (BFU-E) or CFU macrophage (CFU-M), CFU granulocyte/macrophage (CFU-GM), and CFU granulocyte (CFU-G). Cells were left untreated or maintained in the presence of growth factors or erythropoietin (Epo; n = 3).
Discussion
The activating Jak2V617F point mutation is found in a variety of myeloproliferative disorders and infrequently in myelodysplastic syndromes.1-6 The molecular mechanisms that lead to the constitutive activation of Jak2 are not entirely understood, but it is thought that the V617F mutation in part disrupts the regulatory function of the pseudokinase domain. This region has previously been implicated in the regulation of Jak2 kinase activity through direct interference with the Jak2 kinase domain.30 An intact Jak2 FERM domain is required for the chaperone effect on trafficking of the EpoR from the endoplasmic reticulum to the Golgi and on to the cell surface.31 The N-terminal domain of Jak2 is required for cell- surface expression of the EpoR and prolactin receptor31 and for the stable localization at the cell surface of the thrombopoietin receptor (Mpl).19 Our goal was to further define molecular mechanisms involved in transformation by Jak2V617F and identify targets that would aid in the development and implementation of targeted therapies for the treatment of Jak2V617F-related myeloproliferative disorders. Previous results by others and our own results suggested that in vitro transformation of the murine pre-B-cell line BaF3 through Jak2V617F was strongly enhanced by certain type I cytokine receptors, including the Epo and the thrombopoietin receptor.1,22,23 In this context, it should be pointed out that one report has shown that parental BaF3 cells can be partially transformed by Jak2V617F,2,4,32 but this may involve clonal effects or require higher expression levels of Jak2V617F, which would then be able to use an endogenous cytokine receptor for scaffolding. To avoid such high expression of Jak2 mutant and to focus on the relevant interactions with type I cytokine receptors, we therefore initially used BaF3 cells expressing the EpoR to evaluate if the Y114A mutation in the FERM domain would interfere with transformation by Jak2. The Y114 residue is not a target of tyrosine phosphorylation itself but is located in a region required for receptor interaction.19,24,33 Consistent with an important role for the Jak2 FERM domain, we found that Jak2V617F.Y114A double mutant–expressing cells failed to transform BaF3 cells to growth factor independence. We had previously shown that a main mechanism of transformation induced by Jak2V617F was to prevent G1 cell-cycle arrest as well as to maintain high cyclin D2 and low p27Kip expression levels of these cell-cycle regulators, which are required for S-phase entry. This effect was found to be disrupted by the Y114A mutation. Furthermore, Jak2V617F.Y114A failed to induce tyrosine phosphorylation of the EpoR as well as Jak2 itself, in contrast to Jak2V617F. Interestingly, in EpoR-expressing cells with a W282R mutation, which fails to activate Jak2 after Epo stimulation, Jak2V617F was also not transforming and not tyrosine phosphorylated, even after ligand stimulation.23 In these experiments, an additional EpoR mutant with all intracellular tyrosines changed to phenylalanine did not show any defects in Jak2V617F activation, suggesting that the Jak2 activation process itself is independent of receptor tyrosine phosphorylation. Our data would imply that the structural requirements for Jak2V617F activation lie within the oncoprotein itself and favor a model, whereby interaction of Jak2V617F with a type I cytokine receptor brings Jak2 molecules within close proximity. The V617F mutation in the regulatory pseudokinase domain lowers or abolishes the threshold for phosphorylation and activation of Jak2, normally induced by receptor ligation. Indeed, overexpression of Jak2 at very high levels in HEK293 cells leads to activation and phosphorylation of Jak2 itself, even in the presence of the Y114A mutation (not shown).
To further define the role of the Y114A mutation in Jak2V617F, we infected fetal liver cells with either Jak2V617F or Jak2V617F.Y114A. Consistent with previous results, a myeloproliferative disorder with long latency evolved in the Jak2V617F-expressing mice. An elevated hematocrit as well as splenomegaly were observed. In contrast, the Jak2V617F.Y114A-expressing mice did not show an abnormal blood count and the spleen had normal weight, although a comparable percentage of mutant cells was detectable, which has been shown to be sufficient to cause disease.21 These data were consistent with the in vitro data using BaF3.EpoR cells and suggest a disruption of the Jak2V617F-activated EpoR pathway by the Y114A mutation. Furthermore, the failure of Jak2V617F.Y114A-expressing cells, from either peripheral blood or spleen, to produce abnormal colony formation, compared with Jak2V617F, hints at a more global function of the FERM domain in activating type I cytokine receptor pathways during transformation by the oncoprotein. These results may open a new venue for indirectly targeting Jak2V617F in related diseases, such as through inhibition of cytokine receptor expression by shRNA. Nevertheless, the direct involvement of any of these receptors in a proliferative phenotype would require further evaluation and correlation to the in vivo disease phenotypes associated with Jak2V617F. It will also be interesting to see in future experiments whether the Y114A mutation fails to support hematopoiesis in a Jak2-null background as well.
Of additional interest are our findings that the Y114A mutation inhibits Jak2 signaling at a very proximal level and disrupted both EpoR as well as STAT5 tyrosine phosphorylation by Jak2V617F. Phosphorylation of the EpoR by Jak2V617F is likely to be required for the recruitment of STAT5 to the receptor and activation by Jak2V617F.23 To further define the role of Jak2V617F, we sought to identify signaling proteins that are regulated through the FERM domain mutation. We found Pim1 and Pim2 kinases and the Myc transcription factor to be increased by Jak2V617F and the expression of either protein was inhibited by the Y114A mutation. The Pim genes have a broad overlap in activity and at least Pim1 is thought to be a transcriptional target of STAT5.16 This is supported by our findings in which constitutively active STAT5 can increase expression of Pim1 itself. We found increased expression of Pim2 as well. In addition, Pim1 is an oxidative stress–induced gene34 and the antioxidant NAC inhibited Pim1 and Pim2 expression in the Jak2V617F-transformed HEL cells.22 This regulation is likely in part through the redox-dependent targeting of the JAK2V617F/STAT5 pathway.22 The mechanism of ROS induction in Jak2V617F-transformed cells is unknown, but activation of the PI3K pathway through constitutively active STAT535 is likely to contribute to elevated levels of ROSs.36
The role of Pim proteins in transformation and abnormal hematopoiesis is not well understood. In Bcr-Abl–transformed cells, Pim1 significantly supports cell growth and viability, but may not be required.27,37 Results from Pim family compound knockout mice, with disruption of the Pim1, the Pim2, and the Pim3 genes, suggest that these kinases are important for growth factor signaling and are crucial for the function of several hematopoietic lineages.38 In vivo, Pim1 has been shown to cooperate with deregulated Myc in transformation.39,40 Pim1 was also found to be transforming by itself and shown to support growth in BaF3 cells.27,41 However, the transforming potential of Pim1 is controversial and may require secondary events.40 To address this, we generated cell lines with inducible Pim1/2 and Myc. Similar to results reported previously,27,41 we observed a weak proliferative effect in response to forced expression of Pim1 and Pim2. Nevertheless, Pim protein expression did not support long-term growth in our model (not shown). In addition, Myc was also sufficient to induce a weak proliferative response and enhanced cell growth in combination with Pim expression (Figure 4). Both Pim1 and Myc have been implicated in genomic instability,42,43 and our results would support a model where genomic instability associated with the long-term expression of either protein would result in a transforming phenotype. Thus, targeting Pim proteins may provide an important additional or primary target for the treatment of Jak2V617F-transformed cells.
Lessons learned from the induction of drug resistance by the ABL kinase inhibitor imatinib and related drugs suggest that oxidative stress and the resulting induction of point mutations may cause a significant problem for the development of targeted therapies.44 It is expected that a Jak2 kinase inhibitor alone may be of significant impact for the treatment of Jak2V617F-related disease but may not be sufficient if point mutations at its binding site in Jak2 occur. In the absence of targeted approaches, there is a clear need to further understand molecular mechanisms that contribute to Jak2V617F transformation. Our data have demonstrated a dramatic effect of functionally inactivating the type I cytokine receptor domain in Jak2V617F, suggesting that drugs that interfere with ligand independent activation or downstream effectors of Jak2V617F, such as Pim1/2 or Myc, may be of potential therapeutic value.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Rachel Okabe for excellent technical assistance.
This work was supported in part by National Institutes of Health grant DK66996, a Leukemia and Lymphoma Society SCOR grant (J.D.G.), and an American Cancer Society Research Scholar grant (M.S.).
National Institutes of Health
Authorship
Contribution: G.W. designed and performed research and revised the paper; J.R.G., B.J.C., M.S.R., M.M.R., H.E.H., C.W., and A.R. performed research; Y.R., S.N.C., and M.H.T. provided vital reagents and revised the paper; K.P., J.D.G., and D.G.G. designed research and revised the paper; and M.S. designed and performed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Sattler, Department of Medical Oncology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: martin_sattler@dfci.harvard.edu.