Abstract
A mechanism used by Epstein-Barr virus (EBV) for in vitro transformation of B cells into lymphoblastoid cell lines (LCLs) is activation of the NF-κB pathway, which is largely mediated by the EBV latent membrane protein 1 (LMP1). LMP1 is coexpressed with LMP2A in many EBV-associated lymphoid malignancies. Since inhibition of NF-κB leads to apoptosis of EBV-infected LCLs and lymphoma cell lines, we sought to determine whether LMP1 alone, or in combination with other viral proteins, is responsible for initiating NF-κB activation in these cells, thereby playing a role in cell survival. We found that suppression of LMP1 by RNA interference results in inhibition of basal NF-κB and induction of apoptosis. Unexpectedly, knockdown of LMP2A also resulted in comparable decrease of NF-κB activity and apoptosis. We report that LMP2A protein controls the expression of TRAF2 mRNA, which in turn is necessary for signaling by LMP1. Our data contrast with previous studies showing that transfected LMP1 can signal in the absence of LMP2A or TRAF2, and demonstrate that both LMP2A and TRAF2 are required for survival in naturally infected lymphoma cells and LCLs. These results also support LMP1, LMP2A, and TRAF2 as potential therapeutic targets in a subset of EBV-associated lymphoid malignancies.
Introduction
The association of Epstein-Barr virus (EBV) with several specific lymphoid malignancies is quite consistent, indicating an etiopathogenic role in their development. Support for EBV being an oncogenic virus comes from its ability to infect and transform normal human B cells in vitro, resulting in their immortalization and leading to continuously growing lymphoblastoid cell lines (LCLs).1 Infection by EBV in healthy individuals is self-limiting, but a distinct sequence of events occurs that allows this virus to establish life-long latency in memory B cells. To achieve this, EBV expresses 2 latent membrane proteins that mimic signals in B cells that are involved in the germinal center reaction and B-cell differentiation into memory B cells.2 These are the latent membrane protein 1 (LMP1), which functions as a constitutively active CD40 receptor, and LMP2A, containing an immunoreceptor tyrosine-based activation motif (ITAM), thereby mimicking antigen receptor signaling. While these 2 proteins can provide EBV-infected cells with proliferative and antiapoptotic signals, they are also immunogenic, so cells expressing LMP1 and LMP2A are eliminated after acute infection in healthy individuals, and the only surviving EBV-infected B cells that generate the latent reservoir are those that down-regulate these antigenic proteins and differentiate into memory B cells.
EBV is thought to lead to the development of lymphoma as a consequence of alterations in the normal viral life cycle, for example as a consequence of impaired immune responses.3 EBV-associated lymphomas frequently occur in immunodeficient individuals, including those infected with HIV and organ transplant recipients.4,5 In these lymphomas, tumor cells usually express several EBV-encoded genes (latency III), including LMP1 and LM2A and a number of EBV-nuclear antigens (EBNAs).6,7 A large proportion of Hodgkin lymphomas are also associated with infection by EBV.8-10 In positive cases, the Reed-Sternberg and Hodgkin cells express EBNA1, LMP1, and LMP2A (latency II).11
The LMP1 protein is transforming and tumorigenic in vitro12 and in vivo. It is essential for EBV to generate LCLs.13,14 Transgenic mice expressing LMP1 under the control of immunoglobulin gene regulatory elements develop B-cell lymphomas.15 LMP1 functions as constitutively activated member of the tumor necrosis factor receptor (TNFR), of which CD40 is a member. It activates several signaling pathways, predominantly NF-κB and JNK. In particular, the NF-κB pathway, has been shown to be critical for lymphoma cell survival,16,17 and is thought to be a major mechanism by which EBV is transforming. LMP1, like CD40, rescues B cells from apoptosis and induces proliferation of infected cells. Both LMP1 and CD40 require TNF receptor associated factor (TRAF) proteins for signaling to NF-κB. One study showed that while TRAF3 can bind both CD40 and LMP1, it is required only for LMP1 signaling in murine B cells, and that LMP1 can still signal in TRAF2-negative cells.18 A different study looking at LMP1 signaling in knockout mouse embryo fibroblasts showed that TRAF6 is required, but TRAF2 and TRAF5 are not.19 Therefore it appears that TRAF3 and TRAF6 are involved in LMP1 signaling, although they have not yet both been shown to be essential in the same experimental system or in human cells. These requirements are different from those of CD40, which uses TRAF1, TRAF2, and TRAF6, but where TRAF3 has been found to be inhibitory.20,21
LMP2A is a functional homolog of the B-cell antigen receptor (BCR), although it also inhibits antigen-induced activation of the BCR signal transduction cascade. LMP2A is not essential for generation of LCLs,22 but it does contribute to the efficiency of B-cell immortalization in some experimental systems23 and has been found to have several effects on B cells. Experiments with transgenic mice expressing LMP2A targeting expression to the B-cell lineage showed that LMP2A sends survival signals and allows B cells to bypass developmental checkpoints and escape the bone marrow to colonize peripheral lymphoid organs.24-26 LMP2A signaling appears not to affect B-cell proliferation in vivo,27 but rather provide survival signals in BCR-negative B cells.25 These functions are mediated by the ITAM, through which LMP2A activates Ras/phosphatidylinositol 3′-OH kinase (PI3K)/Akt/mTOR signaling in B lymphocytes by activating members of the Src family of tyrosine kinases, and also associates with the BCR signaling effector Syk kinase, a mechanism through which it induces epithelial cell migration.28-32 LMP2A can also engage the JNK mitogen-activated protein (MAP) kinase and beta-catenin signaling pathways.33,34 Two recent studies have shown activation of NF-κB by LMP2A by evaluating the role of this viral protein in conjunction with known antigenic signals in double transgenic mice expressing LMP2A and BCR restricted to hen egg lysozyme (HEL) or ribonucleoprotein Smith (Sm).35,36 Both of these studies found that B cells expressing LMP2A in vivo display constitutive NF-κB activity, compared with LMP2A-negative control B cells.
The interplay between CD40 and antigen receptor signaling has been studied extensively, although it is not yet completely understood. It is clear that BCR and CD40 can both activate the NF-κB pathway, and that they synergize greatly in this activation. Activation of protein tyrosine kinases by BCR triggers a cascade of secondary signals, resulting in activation of several intracellular effector signaling molecules, including members of the protein kinase C (PKC) family of serine-threonine kinases37 that influence activation of NF-κB.38 Recently, protein kinase D (PKD) was found to be activated by BCR and mediate synergy between BCR and CD40. The role of PKD was found to be dependent upon the association of CD40 with TRAF2, and was inhibited by the binding of TRAF3.39 CD40 ligand treatment of normal B cells has also been found to reprogram BCR signaling so it can activate NF-κB through an alternative pathway that is independent of PI3kinase.40 In contrast, studies evaluating the functional relationship between LMP1 and LMP2A are limited, although these 2 proteins are frequently coexpressed. Although usually LMP2A cannot activate NF-κB on its own in transfected cells, its ability to augment signaling by LMP1 has been reported.41 This study found that LMP2A increases the half-life of LMP1, thus increasing LMP1 protein levels when both are transfected into epithelial cell lines. In contrast, a different study found that LMP2A inhibits NF-κB activity in carcinoma cell lines infected in vitro with wild-type recombinant EBV, compared with virus in which LMP2A was deleted.42 Here we evaluated the role of LMP1 and LMP2A in signaling to NF-κB, and thus in the generation of survival signals in LCLs and naturally infected lymphoma cells. We found that, unlike in other experimental systems, LMP1 uses TRAF2 for signaling, and that LMP2A is required for TRAF2 transcription.
Methods
Cell lines and culture conditions
Cell lines used were as follows: 3 EBV-positive cell lines derived from AIDS immunoblastic lymphomas (BCKN-1, IBL-1, and IBL-4: type II/III latency based on LMP1 and LMP2A expression), 2 KSHV+/EBV− primary effusion lymphoma (PEL) cell lines (BC-3 and BCBL1) and 2 lymphoblastoid cell lines (LCL9001 and RPMI-8402). IBL-4 cells were a kind gift from Dr William Harrington. BCKN-1, IBL-1, and BC-3 cells were established in our laboratory. A description and characterization of BCKN-1 and IBL-1 cells, including their karyotype and phenotype, is included as Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition, activation of NF-κB in IBL-1 has been reported,16,17 and the cellular gene expression profile of BCKN-1 cell line has been previously published.43 Cells were grown in RPMI-1640 (Gibco, BRL, Division of Invitrogen, Gaithersburg, MD) supplemented with 20% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 50 mg/mL gentamicin (Sigma-Aldrich, St Louis, MO).
Transient transfection and luciferase assay
Transient transfection of reporter plasmids was performed using FuGene 6 Transfection Reagent (Roche, Indianapolis, IN), TransFectin Lipid Reagent (Bio-Rad Laboratories, Hercules, CA), or Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. The NF-κB firefly luciferase reporter derived from a kappa immunoglobulin promoter was kindly provided by Hsiou-Chi Liou. All the transfections were carried out in the presence of a renilla luciferase reporter under the control of a constitutive promoter, pRL-RSV vector (Promega, Madison, WI), encoding renilla luciferase, in order to normalize the results for the transfection efficiencies. The amounts of DNA transfected were 4 μg for the NF-κB firefly luciferase reporter and 0.2 μg for the renilla luciferase reporter. After 48 hours, lysates were prepared using Cell Culture Lysis Reagent as specified by the manufacturer (Promega). Luciferase assays were performed with a luminometer (Dynex Tech, Chantilly, VA). The baseline (mock transfected) value of firefly luciferase activity prior to normalization ranged from a few hundred to 12 000. Data are presented as relative luciferase units (RLU), which were calculated by dividing the firefly luciferase activity by the renilla luciferase activity.
RNA interference
RNA duplexes were synthesized with 2-nt (2′deoxy) uridine overhangs by Dharmacon Research (Lafayette, CO). The siRNA target sequence to LMP1 mRNA is AAGAGCCUUCUCUGUCCACU and to LMP2A mRNA is AACUCCCAAUAUCCAUCUGCU. The LMP2A siRNA was designed not to overlap with the sequences shared by LMP2B. RNA duplexes for TRAF1, TRAF2, TRAF3, TRAF5, and TRAF6 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). An independent siRNA for TRAF2 was obtained from Dharmacon, and similarly tested to rule out off-target effects. A control scrambled siRNA duplex was obtained from Dharmacon (Scramble II Duplex). Transfection of siRNA duplex was performed using Oligofectamine Reagent (Invitrogen) as reported44 and assayed for silencing 2 days after transfection. All experiments were done in triplicate reactions, and at least in 2 independent experiments were done for each of the cell lines shown.
Immunoblot analysis
BC-3, BCKN1, RPMI, IBL-1, IBL-4, and LCL-9001 cells that were mock transfected or transfected with siRNA were collected 48 hours after transfection. Lysates were prepared in standard radioimmunoprecipitation assay (RIPA) buffer supplemented with 1 μg each of aprotinin, leupeptin, and pepstatin per milliliter, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM each NaVO4 and NaF (Sigma-Aldrich). For isolation of nuclear proteins, the cells were washed in cold phosphate-buffered saline (PBS) before addition of 500 μL cold buffer A (20 mM HEPES [pH 7.9], 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 10% glycerol, 0.2 mM EDTA, 5 μL/mL Protease Inhibitor Cocktail III [Calbiochem]). After 20-minute incubation on ice, nuclei were pelleted by centrifugation. Pellets were washed one more time in 300 μL buffer A, and repelleted before the addition of 300 μL buffer B (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 5 μL/mL Protease Inhibitor Cocktail III). Nuclei were resuspended in buffer C (20 mM HEPES [pH 7.9], 0.4 M NaCl, 20% glycerol, 0.2 mM EDTA, 5 μL/mL Protease Inhibitor Cocktail III) and incubated on ice, vortexing periodically for 60 minutes. Nuclear extracts were collected after a 15-minute centrifugation. The proteins were quantitated by the Bradford method and 40 μg whole cell extract was loaded onto a 10% SDS–polyacrylamide gel. After electrophoresis, semidry transfer to a polyvinylindene difluoride membrane (Millipore, Billerica, MA) was performed. Blots were probed with primary antibody (Ab) overnight at 4°C. Horseradish peroxidase–conjugated secondary Ab was added after washing and was detected by an enhanced chemiluminescence system (ECL; Amersham, Piscataway, NJ). The following primary antibodies were used: anti-LMP1 (Dako, Carpinteria, CA), anti-LMP2A (Ascenion, Munich, Germany), anti-Flag and anti–β-actin (Sigma-Aldrich), anti-p50, anti-p65, anti-p52, anti–RelB, anti-TRAF1, anti-TRAF2, anti-TRAF3, anti-TRAF5, and anti-TRAF6 (Santa Cruz Biotechnology), and phospho-Akt substrate and anti–histone H2A (Cell Signaling Technology, Beverly, MA).
Immunoprecipitation analysis
IBL-1 cells transfected with scrambled and LMP2A siRNA were lysed with Triton buffer (50 mM Tris pH 7.5, 250 mM NaCl, Triton 0.1%, EDTA 1 mM, NaF 50 mM) at a concentration of 5 × 107cells/mL at 4°C for 20 minutes. The lysate was cleared by centrifugation. The proteins were quantitated by the Bradford method and 2 mg total cell lysate was precipitated with 5 μg mouse α-LMP1 (Dako, Carpenteria, CA), for 3 hours. The precipitate was washed 4 times with Triton buffer, resuspended in 5 × loading buffer, separated by SDS–polyacrylamide gel electrophoresis (PAGE), and probed with mouse monoclonal α-LMP1 (Dako), rabbit polyclonal α-TRAF2, and α-TRAF3 (Santa Cruz Biotechnology).
Flow cytometric analysis
Cells were placed in culture at a density of 7.5 × 105/mL. Annexin V analysis was performed as described by the manufacturer (Clontech, Mountain View, CA). Briefly, 2 × 105 cells were washed once with binding buffer and resuspended in 200 μL binding buffer. FITC–annexin V (5 μL) was added and the cells were incubated for 15 minutes in the dark at room temperature and analyzed by flow cytometry using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ).
Real-time quantitative PCR analysis
Real-time quantitative reverse-transcription–polymerase chain reaction (RT-PCR) analysis was performed on an ABI Prism 7000 sequence system with the SYRB Green dye (Applied Biosystems, Foster City, CA). DNase-treated total RNA (1.0 μg) was reverse transcribed with the reverse transcription system (Promega) and resuspended in 100 μL sterile distilled H2O. The real-time PCR contained 1 μL cDNA, 5 pmol of both the forward and reverse primers, and 12.5 μL SYBR Green dye master mix (Applied Biosystems). Sequence-specific primers were designed using Primers Express 2.0 software (Applied Biosystems). The primers used were GAPDH: 5′-CCACCATGGAGAAGGCTGGGGCTCA-3′ and 5′-ATCACGCCACAGTTTCCCGGAGGGG-3′; LMP2A: 5′-TCCCTAGAAA-TGGTGCCAATG-3′ and 5′-GAAGAGCCAGAAGCAGATGGAT-3′; TRAF2: 5′-CAGTTCGGCCTTCCCAGATAA-3′ and 5′-TCGTGGC-AGCTCTCGTATTCTT-3′; TRAF3: 5′-GCGTGTCAAGAGAGCAT-CGTTA-3′ and 5′-TGCTCTGCACAACCTCTGCTT-3′; and TRAF6: 5′-CCACGGAACCCAAAAGGTTT-3′ and 5′-GGAGACCTCACAGCGCACTAAT-3′. No amplification was observed in the no-template control for each primer set. PCR conditions were carried out as described.43 All reactions were done in triplicate, and CT value, the threshold cycle number, was calculated. The higher the value, the lower the mRNA content.45 CT values for GAPDH were used for normalization purposes. ΔΔCT method45 was used to determine the relative gene expression.
Results
Inhibition of LMP1 or LMP2A by RNA interference results in decreased NF-κB activity
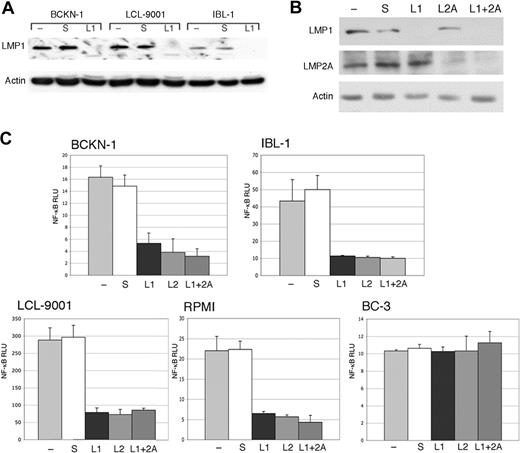
Previous studies have shown that LMP1 can activate NF-κB, and one study showed that LMP2A could enhance activation of this pathway when coexpressed with LMP1.41 However, it was not known whether in actual EBV-positive lymphoma cell lines the constitutive NF-κB activity requires production of one or both of these proteins. Therefore, we developed and tested 2 siRNAs targeting LMP1 and LMP2A. These siRNAs were transfected into 4 EBV-positive (type II/III) cell lines: one immunoblastic lymphoma (IBL-1), one EBV-positive/KSHV-negative effusion lymphoma (BCKN-1), and 2 lymphoblastoid cell lines (RPMI-8402 and LCL-9001). IBL-1 and BCKN-1 were established in our laboratory from patients with AIDS. A significant inhibition of LMP1 and LMP2A proteins was obtained, either singly or in combination, as assessed by immunoblot analysis (Figure 1A,B). LMP1 or LMP2A suppression with siRNA resulted in inhibition of basal NF-κB by 60% to more than 90% in the cell lines examined by 48 hours, indicating that a significant proportion of the constitutive NF-κB activity is being induced by LMP1 and LMP2A (Figure 1C). When the 2 siRNAs where transfected together, NF-κB activity remained almost the same as in the single siRNA transfected cells, so there was no significant additive or synergistic effect. Scrambled siRNA used as a negative control did not have an effect on NF-κB activity and did not substantially affect the level of LMP1 and LMP2A protein. No inhibition was found in BC-3 cells, which have high constitutive NF-κB activity but lack EBV, showing that the inhibition observed in the EBV+ cell lines is not an off-target effect of the LMP1 or the LMP2A siRNA. These results demonstrate that the constitutive NF-κB activity in EBV-infected cell lines requires both LMP1 and LMP2A, and both proteins play a role in the activation of this pathway.
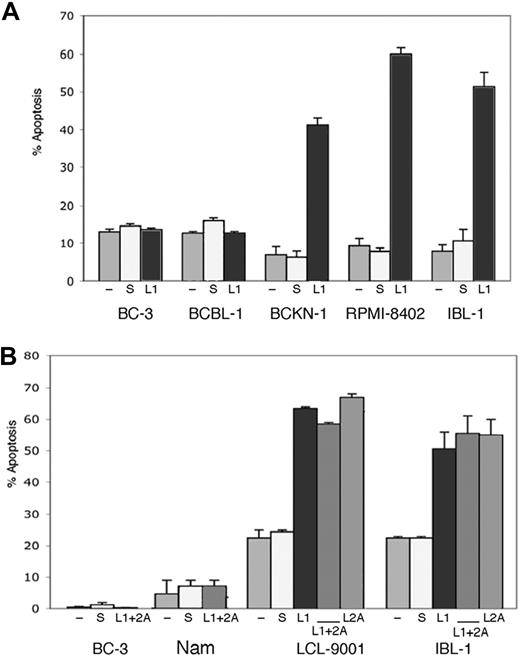
Inhibition of endogenous LMP1 and LMP2A by siRNA results in depletion of constitutive NF-κB activity in EBV+ cells. (A) The indicated cell lines were transfected with a LMP1 siRNA (L1), and scrambled siRNA (s), and compared with mock-transfected cells (−). (B) IBL-1 cells were transfected with an LMP1 siRNA (L1), LMP2A siRNA (L2A), or both, and scrambled siRNA (s), and compared with mock-transfected cells (−). In panels A and B, protein extracts were prepared 48 hours after transfection and probed with antibodies to LMP1 or LMP2A, as indicated. Actin reprobing was performed to assure even protein loading. (C) BCKN-1, IBL-1, LCL-9001, RPMI-8402, and BC-3 cells were transfected with a NF-κB luciferase reporter plasmid, and either scrambled siRNA as a control (S), or siRNA for LMP1 (L1), LMP2A (L2A), or both. Luciferase assays were performed 48 hours after transfection. Values shown are averages (+ SEM) of 1 of 2 independent experiments in which each transfection was performed in triplicate, and are shown as relative luciferase units (RLU), which represent a ratio of NF-κB firefly luciferase to constitutive renilla luciferase. Baseline values correspond to those seen in mock-transfected cells and vary for each cell line.
Inhibition of endogenous LMP1 and LMP2A by siRNA results in depletion of constitutive NF-κB activity in EBV+ cells. (A) The indicated cell lines were transfected with a LMP1 siRNA (L1), and scrambled siRNA (s), and compared with mock-transfected cells (−). (B) IBL-1 cells were transfected with an LMP1 siRNA (L1), LMP2A siRNA (L2A), or both, and scrambled siRNA (s), and compared with mock-transfected cells (−). In panels A and B, protein extracts were prepared 48 hours after transfection and probed with antibodies to LMP1 or LMP2A, as indicated. Actin reprobing was performed to assure even protein loading. (C) BCKN-1, IBL-1, LCL-9001, RPMI-8402, and BC-3 cells were transfected with a NF-κB luciferase reporter plasmid, and either scrambled siRNA as a control (S), or siRNA for LMP1 (L1), LMP2A (L2A), or both. Luciferase assays were performed 48 hours after transfection. Values shown are averages (+ SEM) of 1 of 2 independent experiments in which each transfection was performed in triplicate, and are shown as relative luciferase units (RLU), which represent a ratio of NF-κB firefly luciferase to constitutive renilla luciferase. Baseline values correspond to those seen in mock-transfected cells and vary for each cell line.
Suppression of LMP1 or LMP2A induces apoptosis in EBV-positive cells
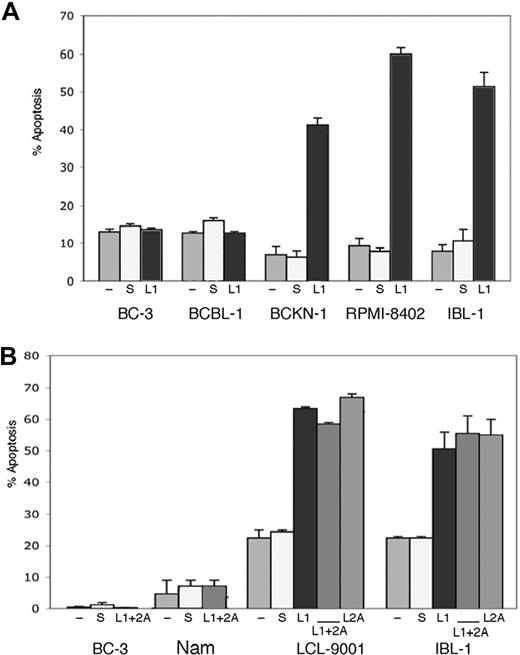
As knockdown of either LMP1 or LMP2A resulted in down-regulation of NF-κB activity and NF-κB is an important survival signal in virus-associated lymphomas, we tested whether elimination of LMP1 or LMP2A is sufficient to induce apoptosis of EBV-infected cells by flow cytometry for annexin V. We transfected siRNA to LMP1, LMP2A, or both into a panel of cell lines that included 2 EBV-negative PEL cell lines (BC-3 and BCBL-1), one cell line that is positive for EBV but lacking LMP-1 expression (Namalwa), and 4 EBV-positive (type II/III) cell lines, 2 corresponding to AIDS lymphomas (BCKN-1 and IBL-1) and 2 LCLs (RPMI-8402 and LCL-9001). Annexin V was evaluated 2, 4, and 6 days after transfection. We found that in all the LMP-1–expressing cell lines, suppression of LMP-1 resulted in induction of apoptosis in 45% to 60% of the cells, while no effect was seen in LMP-1–negative controls. Suppression of LMP2A also induced apoptosis of approximately 50% of EBV+ (type III) cell lines. We evaluated suppression of both simultaneously, which resulted in similar levels of apoptosis, with no significant additive or synergistic effect (Figure 2). No induction of apoptosis was seen in Namalwa cells, or in the 2 EBV-negative PEL cell lines, indicating the effect is specific and due to suppression of the corresponding EBV protein. Apoptosis was seen by day 4 after siRNA transfection in most cells lines, and by day 6 in all of them (shown in Figure 2). As NF-κB down-regulation was clear by day 2, it preceded the onset of apoptosis. This timing is consistent with a causal relationship between NF-κB inhibition and apoptosis, although other signaling pathways affected by LMP1 and LMP2A knockdown could act in conjunction with NF-κB in inducing cell death.
Suppression of endogenous LMP1 and LMP2A by siRNA results in apoptosis of EBV+ cells. (A) The cell lines indicated were transfected with a LMP1 siRNA (L1) or scrambled siRNA (s), or mock transfected (−), at days 0, 2, and 4, and assessment of apoptosis was performed by annexin V staining at day 6. Bars represent the average number of annexin V–positive cells (+ sem) of a representative experiment of 2 independent experiments for each cell line shown, where each transfection and analysis was performed in triplicate. (B) The indicated cell lines were transfected as in panel A, but in addition they were transfected with siRNA to LMP2A (L2) alone or in combination with siRNA to LMP1 (L1 + 2A) as indicated. Suppression of LMP2A was effective on its own in inducing apoptosis of the EBV+/type II/III cell lines tested, but did not enhance the effect seen with LMP1 suppression.
Suppression of endogenous LMP1 and LMP2A by siRNA results in apoptosis of EBV+ cells. (A) The cell lines indicated were transfected with a LMP1 siRNA (L1) or scrambled siRNA (s), or mock transfected (−), at days 0, 2, and 4, and assessment of apoptosis was performed by annexin V staining at day 6. Bars represent the average number of annexin V–positive cells (+ sem) of a representative experiment of 2 independent experiments for each cell line shown, where each transfection and analysis was performed in triplicate. (B) The indicated cell lines were transfected as in panel A, but in addition they were transfected with siRNA to LMP2A (L2) alone or in combination with siRNA to LMP1 (L1 + 2A) as indicated. Suppression of LMP2A was effective on its own in inducing apoptosis of the EBV+/type II/III cell lines tested, but did not enhance the effect seen with LMP1 suppression.
The classical and alternative NF-κB pathways are regulated by LMP1 and LMP2A
It has been shown that LMP1 activates the classical and the alternative NF-κB pathways.46-48 In contrast, LMP2A has not been reported to directly activate NF-κB in transfected cells, but is well known to activate Akt signaling. To get additional insights into the possible mechanism by which LMP2A affects NF-κB, IBL-1 cells were transfected with siRNA to LMP1, LMP2A, or both, and effective suppression was achieved (Figure 3A). Immunoblot analysis of total cell extracts confirmed that suppression of LMP2A results in down-regulation of Akt signaling, as determined by probing with an antibody to phospho-Akt substrates (Figure 3B). To determine whether LMP2A, like LMP1, affects both NF-κB pathways, nuclear extracts were evaluated by immunoblot analysis for different Rel family proteins. Nuclear translocation of p50/p65 (RelA) represents activation of the classical, or canonical, pathway, and nuclear p52 and RelB reflect the alternative, or noncanonical, pathway. We found that elimination of either LMP1 or LMP2A results in marked decrease of nuclear accumulation of all Rel proteins examined (Figure 3A). This decrease in nuclear accumulation occurred by 48 hours after siRNA transfection, at which time point apoptosis has not ensued in the IBL-1 cell line. These data indicate that LMP2A together with LMP1 mediate activation of the canonical and noncanonical pathways in EBV-positive lymphoma cell lines.
LMP2A affects activation of Akt and nuclear translocation of Rel proteins in IBL-1 cells. (A) The IBL-1 lymphoma cell line was transfected with siRNA to LMP1 (L1), LMP2A (L2A), or both (L1 + 2A), as well as scrambled (S) as indicated, or mock transfected (−). Proteins were extracted 48 hours after transfection, and probed with antibodies to LMP1 or LMP2A, confirming suppression of the corresponding protein. Actin reprobing was performed to ensure even protein loading. (B) Extracts as in panel A were evaluated for activation of Akt by immunoblotting with an antibody to phospho-Akt substrates, showing that knockdown of LMP2A affects this pathway more significantly than knockdown of LMP1. (C) Nuclear extracts were prepared from cells 48 hours after transfection as in panel A, and probed for the Rel proteins is indicated. Suppression of both LMP1 and LMP2A resulted in significant decrease of nuclear p65 and p50, as well as RelB and p52, showing down-regulation of both the classical and alternative NF-κB pathways. Reprobing with antibodies to histone H2A was performed to confirm similar amounts of nuclear proteins.
LMP2A affects activation of Akt and nuclear translocation of Rel proteins in IBL-1 cells. (A) The IBL-1 lymphoma cell line was transfected with siRNA to LMP1 (L1), LMP2A (L2A), or both (L1 + 2A), as well as scrambled (S) as indicated, or mock transfected (−). Proteins were extracted 48 hours after transfection, and probed with antibodies to LMP1 or LMP2A, confirming suppression of the corresponding protein. Actin reprobing was performed to ensure even protein loading. (B) Extracts as in panel A were evaluated for activation of Akt by immunoblotting with an antibody to phospho-Akt substrates, showing that knockdown of LMP2A affects this pathway more significantly than knockdown of LMP1. (C) Nuclear extracts were prepared from cells 48 hours after transfection as in panel A, and probed for the Rel proteins is indicated. Suppression of both LMP1 and LMP2A resulted in significant decrease of nuclear p65 and p50, as well as RelB and p52, showing down-regulation of both the classical and alternative NF-κB pathways. Reprobing with antibodies to histone H2A was performed to confirm similar amounts of nuclear proteins.
LMP2A affects signaling by LMP1 by controlling TRAF2 mRNA levels
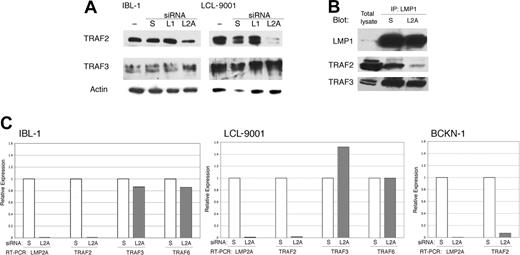
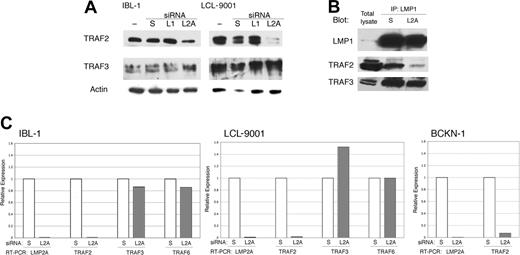
The surprising overlap in signaling between suppression of LMP2A and LMP1, and the lack of a synergistic effect when both proteins are knocked down, led us to hypothesize that LMP2A acts by somehow controlling signaling by LMP1. It has been reported that LMP2A affects LMP1 stability,41 but we did not see any changes in LMP1 protein levels following knockdown of LMP2A (Figures 1B and 3A). As LMP1 signals through TRAFs, we evaluated the presence of TRAF2 and TRAF3 in cell extracts from lymphoma (IBL-1) and LCL cell lines transfected with siRNA to LMP2A. Knockdown of LMP2A resulted in a down-regulation of TRAF2, but TRAF3 was not decreased (Figure 4A). Knockdown of LMP1 did not have any effect on the levels of any of the TRAF proteins examined. We confirmed a decrease of TRAF2, but no decrease in TRAF3 in LMP1-containing complexes after LMP2A suppression by coimmunoprecipitation analysis (Figure 4B).
LMP2A suppression results in transcriptional down-regulation of TRAF2. (A) IBL-1 and LCL-9001 cell lines were transfected with siRNA to LMP1 (L1), LMP2A (L2A), or scrambled (S), or were mock transfected (−), and proteins were extracted 48 hours later. Immunoblot analysis was done with antibodies to TRAF2 and TRAF3. Actin reprobing was performed to assure even protein loading. (B) IBL-1 cells were transfected with siRNA to LMP2A (L2A) or scrambled (S), protein extracts were prepared after 48 hours, and immunoprecipitation (IP) using antibodies to LMP1 was performed. Pulled-down proteins were probed with antibodies to LMP1, TRAF2, and TRAF3 as indicated. (C) IBL-1 and LCL9001 cells were transfected with siRNA to LMP2A (L2A) or scrambled siRNA (S). RNA was extracted 48 hours after transfection, and quantitative real-time RT-PCR was performed in triplicate for LMP2A (L2A), TRAF2 (T2), TRAF3 (T3), and TRAF6 (T6) genes. Two independent experiments were performed for each cell line with similar results. The standard deviation (SD) for the triplicates was 0.1 or less and is not shown. The mean CT value for both cell lines were calculated (avg CT), and the normalized values (ΔCT) were determined from corresponding GAPDH CT values. The average ΔCT values were calculated for scrambled and LMP2A siRNA–transfected cells. The difference between the 2 groups (ΔΔCT) was used to determine the relative gene expression in LMP2A siRNA–transfected cells compared with scrambled siRNA–transfected cells. Efficiency of LMP2A suppression by siRNA is shown by the decrease of LMP2A RNA, which in turn resulted in a marked decrease of TRAF2 RNA, an increase in TRAF3 RNA in the LCL-9001 cell line only, and no change in TRAF6 RNA levels. The decrease of TRAF2 RNA was confirmed in the lymphoma cell line BCKN-1 after knockdown of LMP2A.
LMP2A suppression results in transcriptional down-regulation of TRAF2. (A) IBL-1 and LCL-9001 cell lines were transfected with siRNA to LMP1 (L1), LMP2A (L2A), or scrambled (S), or were mock transfected (−), and proteins were extracted 48 hours later. Immunoblot analysis was done with antibodies to TRAF2 and TRAF3. Actin reprobing was performed to assure even protein loading. (B) IBL-1 cells were transfected with siRNA to LMP2A (L2A) or scrambled (S), protein extracts were prepared after 48 hours, and immunoprecipitation (IP) using antibodies to LMP1 was performed. Pulled-down proteins were probed with antibodies to LMP1, TRAF2, and TRAF3 as indicated. (C) IBL-1 and LCL9001 cells were transfected with siRNA to LMP2A (L2A) or scrambled siRNA (S). RNA was extracted 48 hours after transfection, and quantitative real-time RT-PCR was performed in triplicate for LMP2A (L2A), TRAF2 (T2), TRAF3 (T3), and TRAF6 (T6) genes. Two independent experiments were performed for each cell line with similar results. The standard deviation (SD) for the triplicates was 0.1 or less and is not shown. The mean CT value for both cell lines were calculated (avg CT), and the normalized values (ΔCT) were determined from corresponding GAPDH CT values. The average ΔCT values were calculated for scrambled and LMP2A siRNA–transfected cells. The difference between the 2 groups (ΔΔCT) was used to determine the relative gene expression in LMP2A siRNA–transfected cells compared with scrambled siRNA–transfected cells. Efficiency of LMP2A suppression by siRNA is shown by the decrease of LMP2A RNA, which in turn resulted in a marked decrease of TRAF2 RNA, an increase in TRAF3 RNA in the LCL-9001 cell line only, and no change in TRAF6 RNA levels. The decrease of TRAF2 RNA was confirmed in the lymphoma cell line BCKN-1 after knockdown of LMP2A.
To evaluate the mechanism by which LMP2A regulates TRAF2 expression, we transfected one LCL and one lymphoma cell line (LCL9001 and BCKN-1, respectively) with siRNA to LMP2A, extracted RNA, and performed quantitative real-time RT-PCR for TRAF2, TRAF3, and TRAF6. We found that there is a marked decrease in the levels of TRAF2 mRNA upon suppression of LMP2A (Figure 4C). It is likely that LMP2A is necessary for TRAF2 transcription in these cells, but it remains possible that TRAF2 is regulated by RNA degradation. There was no decrease of TRAF3 or TRAF6 mRNA (Figure 4C), which is consistent with selective regulation of TRAF2 levels.
TRAF2 is essential for NF-κB signaling in EBV-positive lymphoma and lymphoblastoid cell lines
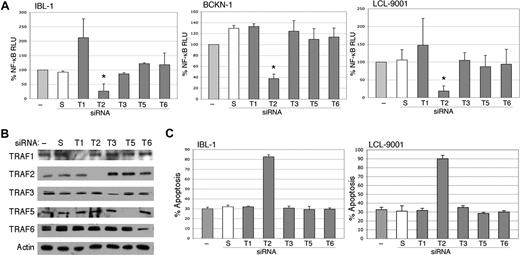
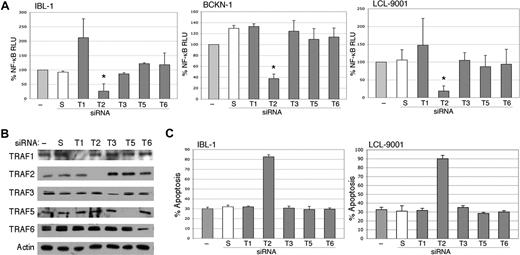
Previous studies have shown that TRAF3 or TRAF6 mediates LMP1 signaling, and that TRAF2 is not required.18,19 However, these studies were done in murine knockout cells with transfected LMP1, and the issue of TRAF requirements has not been evaluated in LCLs or EBV-infected lymphoma cells. Therefore, we transfected siRNA to TRAFs 1, 2, 3, 5, and 6 into 1BL-1, BCKN-1 and LCL-9001 cells. All TRAFs were confirmed to be efficiently knocked down by immunoblot analyses (Figure 5B). Of these, only TRAF2 suppression showed a strong and significant decrease in NF-κB, indicating that it is essential for NF-κB signaling in EBV-infected cell lines (Figure 5A). To confirm these findings, an independent siRNA from a different commercial source was also tested, and shown to efficiently knock down TRAF2 protein and result in similar inhibition of NF-κB (not shown). Knockdown of TRAF1 resulted in the up-regulation of NF-κB in the IBL1 cell line, and also in LCL9001 cells in some experiments, suggesting that in some cellular backgrounds TRAF1 may have an inhibitory activity on NF-κB signaling. We further determined whether suppression of one or more of these TRAF proteins induces apoptosis by performing flow cytometry for annexin V binding following TRAF siRNA transfection. Figure 5C shows that knockdown only of TRAF2 results in apoptosis. These results indicate that TRAF2 is involved in NF-κB signaling and cellular survival of EBV-associated lymphoma and lymphoblastoid cell lines.
Inhibition of endogenous TRAF2 by siRNA abolishes activation of NF-κB. (A) IBL-1, LCL-9001, and BCKN-1 cells were transfected with a NF-κB luciferase reporter plasmid and with either siRNA to TRAF1 (T1), TRAF2 (T2), TRAF3 (T3), TRAF5 (T5), or TRAF6 (T6), or scrambled (S) siRNA, or were mock transfected (−), and protein extracts were prepared 48 hours later. Luciferase assays were performed and values shown are averages (+ sem) of the combination of 2 independent experiments in which each transfection was performed in triplicate, and are presented as percentage change of relative luciferase values compared with untransfected cells. siRNA to TRAF2 resulted in marked reduction of NF-κB activity, but variable or no effect was seen on suppression of TRAF1, TRAF3, TRAF5, or TRAF6. The change of NF-κB activity in TRAF2 siRNA–transfected cells, compared with mock-transfected cells, was statistically significant (*P < .005). (B) To confirm effective and selective knockdown of each TRAF protein, immunoblot analysis using these extracts was performed using antibodies to the TRAFs indicated at the left of each panel. Actin reprobing was performed for all the blots to confirm even protein loading. (C) IBL-1 and LCL9001 cells were transfected with siRNA to the various TRAFs as in panel A at days 0, 2, and 4, and assessment of apoptosis performed by annexin V staining at day 6. Bars represent the average number of annexin V–positive cells (+ SEM) of an experiment in which each transfection and analysis was performed in triplicate.
Inhibition of endogenous TRAF2 by siRNA abolishes activation of NF-κB. (A) IBL-1, LCL-9001, and BCKN-1 cells were transfected with a NF-κB luciferase reporter plasmid and with either siRNA to TRAF1 (T1), TRAF2 (T2), TRAF3 (T3), TRAF5 (T5), or TRAF6 (T6), or scrambled (S) siRNA, or were mock transfected (−), and protein extracts were prepared 48 hours later. Luciferase assays were performed and values shown are averages (+ sem) of the combination of 2 independent experiments in which each transfection was performed in triplicate, and are presented as percentage change of relative luciferase values compared with untransfected cells. siRNA to TRAF2 resulted in marked reduction of NF-κB activity, but variable or no effect was seen on suppression of TRAF1, TRAF3, TRAF5, or TRAF6. The change of NF-κB activity in TRAF2 siRNA–transfected cells, compared with mock-transfected cells, was statistically significant (*P < .005). (B) To confirm effective and selective knockdown of each TRAF protein, immunoblot analysis using these extracts was performed using antibodies to the TRAFs indicated at the left of each panel. Actin reprobing was performed for all the blots to confirm even protein loading. (C) IBL-1 and LCL9001 cells were transfected with siRNA to the various TRAFs as in panel A at days 0, 2, and 4, and assessment of apoptosis performed by annexin V staining at day 6. Bars represent the average number of annexin V–positive cells (+ SEM) of an experiment in which each transfection and analysis was performed in triplicate.
Discussion
The EBV-encoded proteins LMP1 and LMP2A are coexpressed in several malignant lymphoma subtypes, and here we report their interdependence for signaling in this disease. Using a knockdown approach, we were able to examine the function of LMP1 and LMP2A in a relevant experimental system. Specifically, we evaluated the role for these proteins with respect to NF-κB signaling and cellular survival in naturally infected EBV-positive lymphoma cell lines. We found that LMP1 and LMP2A, like their cellular functional homologs CD40 and BCR, have the ability to act in conjunction, affecting each other's signaling properties. Our findings also held true in lymphoblastoid cell lines, which are a model for immunodeficiency-related lymphomas as they have a similar viral gene expression pattern, also called type III latency or growth pattern, that arises from a lack of a competent immune response.
Suppression of LMP1 led to the expected decrease of NF-κB and induction of apoptosis in the examined cell lines. While knockdown of LMP2A led to the anticipated decrease in Akt activation, it also resulted in decrease of NF-κB and induction of cellular apoptosis. An effect of LMP2A in LMP1 signaling was previously reported by Dawson et al, whereupon transfection into carcinoma cells lines LMP2A led to stabilization and increased levels of LMP1.41 However, an opposite result was found upon infection of epithelial cells with a recombinant EBV lacking LMP2 that showed higher NF-κB activity than the wild-type virus-expressing this protein.42 While we did not find that suppression of LMP2A had any effect on LMP1 protein levels, our results did confirm that LMP2A affects LMP1 signaling. We show that suppression of LMP2A leads to an indistinguishable decrease of NF-κB in each of the cell lines from that seen upon suppression of LMP1, and no additive or synergistic effect was observed by suppressing both proteins simultaneously. In addition, knockdown of LMP1 and LMP2A led to inhibition of nuclear translocation of the same Rel proteins. These observations suggest that rather than synergizing, both proteins are interdependent for NF-κB signaling in lymphoma cells.
The finding that LMP1 requires LMP2A was unexpected because transfection of LMP1 alone into multiple cell lines has been consistently shown to result in activation of NF-κB. In addition, recombinant EBV lacking LMP2A can still transform B cells in vitro, and give rise to LCLs.22 This can be explained by the development of addiction to LMP2A in cells that have been highly selected in vitro or in vivo. Our findings are consistent with a model whereby in EBV-associated Hodgkin lymphomas and non-Hodgkin lymphomas expressing LMP2A, tumor cells have become dependent through a process of selection on the signals elicited by this protein for expression of other proteins that are involved in cell survival, including TRAF2. This process of oncogene addiction has been recently described in multiple cancer systems, and is being exploited for therapy.49,50
TRAF proteins are well known to mediate NF-κB activation by LMP1. Here we found that suppression of LMP2A leads to the down-regulation of TRAF2 mRNA. It is possible that this control is transcriptional. The promoter for TRAF2 contains binding sites for multiple transcription factors, including those to cAMP-response element-binding protein (CREB) and NF-κB. CREB plays a key role in the proliferative and survival responses of mature B cells in response to BCR signaling.51 It is likely to be regulated by LMP2A as well, as this viral protein activates many of the same signals as BCR, including protein kinase C (PKC), involved in CREB phosphorylation and activation, although this remains to be verified experimentally. TRAF2 is also controlled by NF-κB, but we did not see any decrease in TRAF2 protein levels after knockdown of LMP1 alone (Figure 4A). A possible explanation is that a decrease in binding of CREB, and perhaps additional transcription factors, is necessary for initial down-regulation of TRAF2 expression, which in turn reduces NF-κB activity leading to a more profound decrease in TRAF2 expression. It also remains possible that TRAF2 RNA levels are controlled indirectly though degradation, which is now known to be a common regulatory process mediated by microRNAs, which can be either cellular or viral and could conceivably be affected by LMP2A.
The control of TRAF2 expression by LMP2A explains the mechanism whereby LMP1 can no longer signal to NF-κB when LMP2A is knocked down. While LMP1 can bind to most members of the TRAF family including TRAF1, 2, 3, and 5,52,53 2 previous studies have evaluated which specific TRAFs are actually involved in signaling by LMP1. In one study from the Kieff laboratory using knockout mouse embryo fibroblasts, TRAF6 was found to be essential, while TRAF2 and TRAF5 were not required (Luftig et al19 ). A second study from Bishop and colleagues using mouse B cells lacking different TRAF proteins reported a requirement for TRAF3, and again TRAF2 deficiency did not affect LMP1 signaling (Xie et al18 ). In view of these 2 studies suggesting that TRAF2 is not required for LMP1 signaling, and our finding that suppression of LMP2A results in down-regulation of TRAF2, we revisited the issue of TRAF requirement by evaluating the effect of knockdown of the various relevant TRAFs in lymphoma and LCL cell lines. In contrast to previous studies, we found that only TRAF2 is completely essential for NF-κB signaling in these cells. This finding underscores the different signaling requirements among species and cell types, and the importance of directly evaluating relevant tumor cells with endogenous expression of the proteins involved in the process being examined.
In this study, we found that suppression of LMP1, LMP2A, or TRAF2 results in the down-regulation of NF-κB, resulting in apoptosis of the cell lines tested, suggesting that these 3 molecules are possible therapeutic targets. While not all EBV-associated lymphomas express LMP1 and LMP2A, a significant fraction does. These include large cell lymphomas in HIV-positive patients, posttransplantation lymphoproliferative disorders, and EBV-positive Hodgkin lymphoma, all of which may benefit from inhibition of one of these viral molecules. It is likely that a combinatorial approach will be essential for the treatment of lymphomas that are not completely dependent on LMP1 or LMP2A expression, and by understanding the interplay of viral and cellular pathways involved in tumor cell survival, we will be able to develop the most effective combinations. The identification of viral proteins that provide survival signals to naturally infected tumor cells provides us with a unique opportunity to develop therapies that are completely specific to the disease process, and not toxic to the host.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grant R01-CA68939 and Leukemia and Lymphoma Society Translational Research grant 6183-02.
National Institutes of Health
Authorship
Contribution: I.G. designed and performed the research and wrote the paper; D.B. performed the real-time quantitative RT-PCR experiments; E.C. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ethel Cesarman, Associate Professor, Department of Pathology and Laboratory Medicine, Weill Medical College of Cornell University, 1300 York Ave, New York, NY 10021; e-mail: ecesarm@med.cornell.edu.