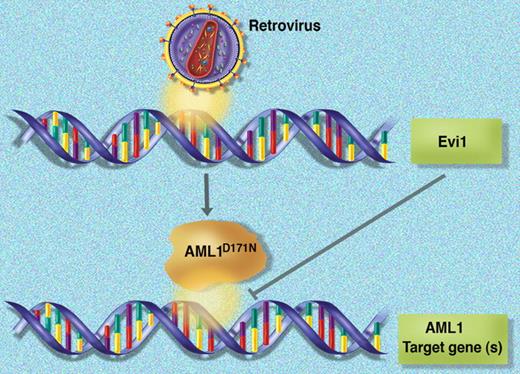
In this issue of Blood, Watanabe-Okochi and colleagues use a mouse bone marrow transplantation model to demonstrate that mutant alleles of AML1 (RUNX1) can initiate a myelodysplastic syndrome (MDS) that progresses to acute myelogenous leukemia (AML) in association with overexpression of Evi1.
The molecular pathogenesis of MDS and the mechanism of its transformation to AML are not well understood. Although several mouse models of MDS have been developed recently, most fail to recapitulate key features of the human disease. In one of the best genetically defined systems, transplantation of bone marrow cells retrovirally transduced with Evi1 results in bone marrow failure, erythroid dysplasia, and evidence of increased apoptosis.1 Despite reproducing these hallmarks of MDS, the mice do not develop AML, suggesting that additional genetic events are required for full transformation.
In a novel mouse model of MDS/AML, retroviral transduction is hypothesized to create two hits (expression of the mutant AML1D171N allele and activation of Evi1 via insertional mutagenesis) that converge on AML1 function.
In a novel mouse model of MDS/AML, retroviral transduction is hypothesized to create two hits (expression of the mutant AML1D171N allele and activation of Evi1 via insertional mutagenesis) that converge on AML1 function.
The AML1 gene is a frequent target of mutations and rearrangements in human leukemia. Although point mutations of AML1 are rare in most AML French-American-British (FAB) subtypes, they are relatively common in M0 AML (12%-33%), MDS (23%), and in therapy-related and radiation-associated MDS/AML (38%-46%).2 In this issue of Blood, Watanabe-Okochi and colleagues use a murine retroviral transduction/bone marrow transplantation model to characterize 2 mutant AML1 alleles previously identified by their group: a frame shift after S291 that results in truncation of the C-terminal transactivation domain, and a D171N missense mutation in the runt homology domain that reduces DNA binding and transcriptional activity. Interestingly, although reconstitution of lethally irradiated mice with cells transduced by either allele die with similar kinetics (latency of ∼150 days and penetrance of 70%-80%), the disease phenotypes are strikingly different. The AML1S291fs mice develop dysplasia and pancytopenia with infrequent progression to AML. The AML1D171N mice have a more proliferative phenotype marked by leukocytosis, hepatosplenomegaly, dysplasia, and more frequent progression to AML. In nearly half the AML1D171N cases, the authors mapped retroviral integration sites to the 5′ flanking region of Evi1 and were able to demonstrate that this was associated with Evi1 overexpression. They went on to show that coinfection of cells with AML1D171N and Evi1 retroviruses shortened disease latency by 50 days and increased penetrance to 100%, providing additional evidence that Evi1 and mutant AML1 can cooperate to induce MDS/AML in mice.
Although the mechanism of Evi1 activation is artificial in this model (retrovirus-mediated insertional mutagenesis), the observations by Watanabe-Okochi and colleagues have important clinical relevance. EVI1 is deregulated by the t(3;3)(q21;q26) and inv(3)(q21;q26) rearrangements in MDS and AML.3,4 Several lines of evidence have also directly linked AML1 and EVI1 in human leukemia biology. First, the genes are fused by the t(3;21)(q26;q22) rearrangement in blast-crisis CML.5 In addition, the proteins physically interact, leading to reduced AML1 DNA binding and transcriptional activity.6 This leads to the prediction, not tested in the current study, that EVI1 and the AML1D171N allele cooperate in vivo because they conspire to reduce AML1 activity. If further studies support this model, there would be a rational basis for developing therapies that target this protein-protein interaction.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■