Abstract
Myelodysplastic syndrome (MDS) is a hematopoietic stem-cell disorder characterized by trilineage dysplasia and susceptibility to acute myelogenous leukemia (AML). Analysis of molecular basis of MDS has been hampered by the heterogeneity of the disease. Recently, mutations of the transcription factor AML1/RUNX1 have been identified in 15% to 40% of MDS–refractory anemia with excess of blasts (RAEB) and MDS/AML. We performed mouse bone marrow transplantation (BMT) using bone marrow cells transduced with the AML1 mutants. Most mice developed MDS and MDS/AML-like symptoms within 4 to 13 months after BMT. Interestingly, among integration sites identified, Evi1 seemed to collaborate with an AML1 mutant harboring a point mutation in the Runt homology domain (D171N) to induce MDS/AML with an identical phenotype characterized by marked hepatosplenomegaly, myeloid dysplasia, leukocytosis, and biphenotypic surface markers. Collaboration between AML1-D171N and Evi1 was confirmed by a BMT model where coexpression of AML1-D171N and Evi1 induced acute leukemia of the same phenotype with much shorter latencies. On the other hand, a C-terminal truncated AML1 mutant (S291fsX300) induced pancytopenia with erythroid dysplasia in transplanted mice, followed by progression to MDS-RAEB or MDS/AML. Thus, we have developed a useful mouse model of MDS/AML that should help in the understanding of the molecular basis of MDS and the progression of MDS to overt leukemia.
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous group of clonal stem-cell disorders characterized by ineffective hematopoiesis and susceptibility to leukemic transformation (MDS/acute myelogenous leukemia [AML]). Progression from MDS-refractory anemia with excess blasts (MDS-RAEB) to AML is frequently observed in the clinical course, which is thought to result from serial acquisition of cytogenetic abnormalities.1-5 According to the 2-hit model of leukemogenesis, one class of mutations (class I), including FLT3-ITD, N-Ras, or K-Ras mutations, confers on cells a proliferative advantage; a second class of mutations (class II), including AML1/ETO, PML/RARα, or MLL-related fusion genes, interferes with hematopoietic differentiation.6 Indeed, it has been reported that a combination of class I and II mutations such as FLT3-ITD plus AML1-ETO or MLL-SEPT6 induced AML in a mouse bone marrow transplantation (BMT) model, while either class I or II mutations alone led to, if anything, myeloproliferative disorders (MPDs), not leukemia.6-13 On the other hand, the precise molecular mechanism underlying development of MDS and MDS/AML remains elusive partly because there are only a few mouse models for MDS and MDS/AML available. So far, 2 distinct models of MDS have been reported: Evi1 induced MDS-like symptoms in a mouse BMT model in which the mice succumbed to fatal peripheral cytopenia,14 while NUP98-HOXD13 transgenic mice developed MDS and died of either various types of acute leukemia or severe anemia and leukocytopenia.15 In the present study, we generated a mouse BMT model of MDS-RAEB and MDS/AML induced by AML1 mutants frequently found in patients with MDS and MDS/AML. Interestingly, the phenotypes of these mice very much resemble those of the human diseases.
The AML1 gene is located on chromosome 21q22 and is the most frequent target for chromosomal translocation in leukemia. Analysis of AML1-deficient mice has shown that AML1 is indispensable for the establishment of definitive hematopoiesis.16-18 As accumulated studies have demonstrated, heterozygous germline mutations in the AML1 gene caused familial platelet disorder with predisposition to AML (FPD/AML),19,20 and sporadic point mutations were frequently found in the development of leukemia: 21% of AML M0, 15.0% to 15.9% of MDS-RAEB and MDS/AML, and 46% of radiation-associated MDS.21-29 The vast majority of AML1 mutations were located in the Runt homology domain (RHD), which mediated its ability to bind to DNA and core-binding factor β (CBFβ). To confirm the involvement of AML1 mutations in hematopoietic disorders, we selected 2 types of AML1 mutants found in patients with MDS/AML: one with a point mutation in RHD (AML1-D171N), and the other with C-terminal truncation caused by a frame-shift (AML1-S291fsX300). After transplantation using bone marrow cells infected with retrovirus vectors harboring AML1 mutants, most of the mice that received transplants died of MDS-RAEB and MDS/AML. Long-term analysis demonstrated that the phenotype of the mice that underwent transplantation depended on the kind of AML1 mutants used in this study and on the integration sites of retroviruses. Considering the recent reports of the effects of retrovirus integration sites on biological results,30-38 identification of integration sites may lead to the discovery of the genes involved in the induction of MDS/AML in concert with AML1 mutants. Intriguingly, the enhanced expression of Evi1 by retrovirus integration seemed to collaborate with AML1-D171N to induce MDS/AML with the same phenotype. Moreover, we confirmed that combination of AML1-D171N and Evi1 induced AML of the same phenotype with shorter latencies in the mouse BMT model. This model will allow valuable insight into the molecular pathogenesis of MDS and MDS/AML.
Methods
Vector construction
We used 2 AML1 mutants, D171N or S291fsX300, identified from case no. 5 or 27, respectively, among patients with MDS/AML.25,26 These mutants are hereafter referred to as AML1-D171N and AML1-S291fs. AML1 wild-type (WT; AML1b), AML1-D171N, or AML1-S291fs, which was fused with a FLAG epitope tag at the N-terminus, was inserted upstream of the IRES-EGFP cassette of pMYs-IG to generate pMYs-AML1 WT, D171N, or S291fs-IG, respectively. pMYs-mouse Evi1-IG were kindly provided by Dr T. Nakamura (The Cancer Institute, Tokyo, Japan).38
Transfection and retrovirus production
Plat-E39 packaging cells maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) were transfected with retroviral constructs by using FuGENE 6 (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's recommendations. The medium was changed 1 day after the transfection, and retroviruses were harvested 48 hours after the transfection as previously described.39,40 Titers of the retroviruses were assessed based on the number of neomycin-resistant colonies of the infected NIH3T3 cells (average: 107 infection U/mL) as described.39
Mouse BMT
Bone marrow mononuclear cells were isolated from the femurs and tibias of C57BL/6 (Ly-5.1) donor mice (9-12 weeks of age) 4 days after intraperitoneal administration of 150 mg/kg 5-fluorouracil (5-FU) and cultured overnight in α minimal essential medium (αMEM) supplemented with 20% FCS and 50 ng/mL of mouse stem cell factor (SCF), mouse FLT3 ligand (FL), human IL-6, and human thrombopoietin (TPO; R&D Systems, Minneapolis, MN). The prestimulated cells were infected for 60 hours with the retroviruses harboring pMYs-AML1 WT, D171N, or S291fs-IG, or an empty vector as a control, using 6-well dishes coated with RetroNectin (Takara Bio, Shiga, Japan) according to the manufacturer's recommendations. Then, 0.2 to 3.5 × 106 of infected bone marrow cells (Ly-5.1) were injected through tail vein into C57BL/6 (Ly-5.2)–recipient mice (8-12 weeks of age) which had been administered a sublethal dose of 5.25 Gy or a lethal dose of 9.5 Gy total-body γ-irradiation (135Cs). For the lethally irradiated mice, a radioprotective dose of 2 × 105 of bone marrow cells (Ly-5.2) was simultaneously injected. Probabilities of overall survival of the mice that received transplants were estimated using the Kaplan-Meiermethod. All animal studies were approved by the Animal Care Committee of the Institute of Medical Science, The University of Tokyo.
Analysis of the mice that underwent transplantation
Engraftment of bone marrow cells infected with retroviruses was confirmed by measuring the percentage of GFP+ and Ly-5.1+ cells in peripheral blood obtained every 1 to 2 months after the transplantation.
After the morbid mice were killed, their tissue samples, including peripheral blood (PB), bone marrow (BM), spleen, liver, and kidney, were analyzed. Circulating blood cells were counted by an analyzer. Morphology of the peripheral blood was evaluated by staining of air-dried smears with Hemacolor (Merck, Darmstadt, Germany). Tissues were fixed in 10% buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Cytospin preparations of bone marrow and spleen cells were also stained with Hemacolor. Percentage of blasts, myelocytes, neutrophils, monocytes, lymphocytes, and erythroblasts was estimated by examination of at least 200 cells. To assess whether the leukemic cells were transplantable, 2 × 105 to 106 total BM cells including blasts were injected into the tail veins of sublethally irradiated mice. A total of 2 or 3 recipient mice were used for each serial transplantation.
Flow cytometric analysis
Red blood cells were lysed by using Ammonium Chloride Lysing Reagent (BD Biosciences, San Jose, CA) in PB or single-cell suspensions of bone marrow and spleen. Washed cells were incubated for 15 minutes at 4°C with 2.4G2 antibody for blocking and then stained for 20 minutes at 4°C with the following monoclonal phycoerythrin (PE)–conjugated antibodies: Ly-5.1, Gr-1, CD11b, B220, CD3, CD41, c-Kit, Sca-1, CD34, and Ter119. Flow cytometric analysis of the stained cells was performed with FACSCalibur flow (BD Biosciences) equipped with CellQuest software (BD Biosciences) and Flowjo software (Tree Star, San Carlos, CA).
Diagnosis
Diagnosis was made according to the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice.41
Real-time RT-PCR
Total RNA was extracted from BM cells using Trizol (Invitrogen, Carlsbad, CA). cDNA was prepared with the Superscript II RT kit (Invitrogen). Real-time reverse transcription–polymerase chain reaction (RT-PCR) was performed using a LightCycler Workflow System (Roche Diagnostics). cDNA was amplified using a SYBR Premix EX Taq (TAKARA). Reaction was subject to one cycle of 95°C for 30 seconds, 45 cycles of PCR at 95°C for 5 seconds, 55°C for 10 seconds, and 72°C for 10 seconds. All samples were independently analyzed at least 3 times. The following primer pairs were used: 5′-CCAGATGTCACATGACAGTGGAAAGCACTA-3′ (forward) and 5′-CCGGGTTGGCATGACTCATATTAACCATGG-3′ (reverse) for Evi1; 5′-TACCTCAACCCCTGACAGCTATGG-3′ (forward) and 5′-TCGGTTGGAGATATCAGAGTGCAG-3′ (reverse) for MN1;42 and 5′-GTTATCCCATCTGCATCAGCATCTGG-3′ (forward) and 5′-GGTCCTCTTCACTCTTCATGAACAGC-3′ (reverse) for MDS1/Evi1.43 Relative gene expression levels were calculated using standard curves generated by serial dilutions of cDNA. Product quality was checked by melting curve analysis via LightCycler software (Roche Diagnostics). Expression levels were normalized by a control, the expression level of GAPDH mRNA.
Western blot analysis
To detect the expression of AML1 WT, mutants, or Evi1, equal numbers of spleen cells were lysed, and Western blotting was performed as described with minor modifications.13 Polyclonal rabbit anti-Evi1 antibody (a kind gift from Dr M. Kurokawa, Tokyo University, Tokyo, Japan), or a monoclonal mouse anti-Flag antibody (Sigma-Aldrich, St Louis, MO) was used for Evi1 or AML1, respectively.
Southern blot analysis
Genomic DNA was extracted from BM or spleen cells. After enzymatic digestion of 10 μg DNA with EcoRI followed by electrophoretic separation, proviruses were probed with a GFP probe.
Bubble PCR
A total of 10 μg of genomic DNA extracted from BM or spleen cells was digested with EcoRI, and the fragments were ligated overnight at 16°C to a double-stranded bubble linker (5′-AATTGAAGGAGAGGACGCTGTCTG-TCGAAGGTAAGGAACGGACGAGAGAAGGGAGAG-3′ and 5′-GACTC-TCCCTTCTCGAATCGTAACCGTTCGTACGAGAATCGCTGTCCTCTCC-TTC-3′).44 Next, PCR was performed on the ligation product using a linker-specific Vectorette primer (5′-CGAATCGTAACCGTTCGTACGAGAATCGCT-3′)44,45 and a long-terminal repeat (LTR)–specific primer (5′-CGAGCTCAATAAAAGAGCCCACAACCCC-3′) under the following conditions: one cycle of 95°C for 5 minutes, 10 cycles of 95°C for 30 seconds, and 67°C for 30 seconds and 72°C for 3 minutes, 10 cycles of 95°C for 30 seconds, and 67°C (this annealing temperature was reduced by 1°C each cycle) for 30 seconds and 72°C for 3 minutes, 15 cycles of 95°C for 30 seconds and 57°C for 30 seconds and 72°C for 3 minutes, and one cycle of 72°C for 90 seconds. Next, nested PCR was performed on 2 μL of PCR products using a linker-specific Vectorette primer and an LTR-specific primer (5′-ATAAAAGAGCCCACAACCCCTCACTCGG-3′) under the following conditions: 1 cycle of 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds and 60°C for 30 seconds and 72°C for 3 minutes, and 1 cycle of 72°C for 90 seconds. The PCR product was electrophoresed using 1.0% agarose gel. Individual bands were excised and purified using PCR clear (Promega, Madison, WI) and were sequenced to identify the integration site of retrovirus. We confirmed inverse repeat sequence “GGGGGTCTTTCA” as a marker of junction between genomic DNA and retrovirus sequence.
Results
The ratio of AML1 mutant-transduced cells gradually increased over several months after transplantation
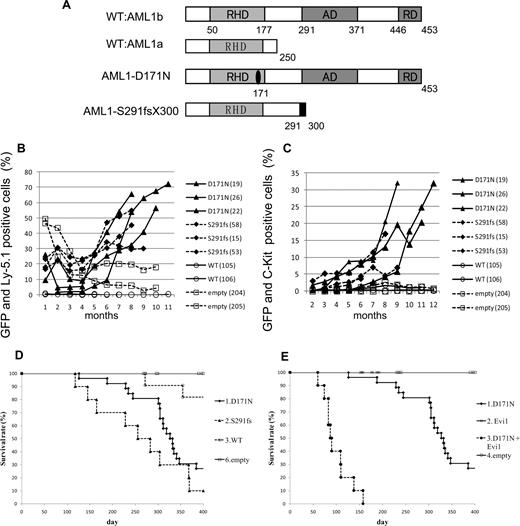
To examine the effect of AML1 mutants on the hematopoietic abnormality, we chose 2 distinct mutants, AML1-D171N and AML1-S291fsX300, which are found in patients with MDS/AML.25,26 The former has a point mutation in RHD, and the latter possesses a frameshift mutation in the C-terminal region, resulting in truncation of the authentic protein (Figure 1A). Ly-5.1 murine BM cells infected with retroviruses harboring AML1 WT, AML1-D171N, AML1-S291fsX300, or empty vector were transplanted into irradiated syngeneic Ly-5.2 mice. In most of mice that received transplants of AML1-D171N– or S291fsX300-transduced cells (hereafter referred to as mice/D171N or mice/S291fs, respectively), the ratio of GFP+ and Ly-5.1+ cells gradually increased over several months after the transplantation (Figure 1B), but not in mice that received transplants of AML1 WT-transduced cells or control retrovirus-infected cells (hereafter referred to as mice/WT or mice/mock, respectively). Gradual increase of c-kit+ cells in the PB was also observed in the mice that received transplants of AML1 mutant-transduced cells during the observation period (Figure 1C). Cells positive for c-kit and GFP—that is, c-kit+ cells transduced with AML1 mutants—were morphologically blasts with high nuclear-cytoplasmic ratios (data not shown). In fact, the percentage of blasts gradually increased in the PB of the mice that received transplants of AML1 mutant-transduced cells, especially the mice/D171N. Finally, most of mice/D171N or mice/S291fs became sick and died with latencies of 4 to 13 months after the transplantation, while mice/mock were healthy over the observation period (Figure 1D). Overall survival of mice/D171N was not significantly different from that of mice/S291fs (P = .218). Expression of the transduced AML1-D171N or AML1-S291fs in spleen cells was confirmed by Western blot analysis (Figure 2A). Two of the mice/WT died during the observation period. BM of the 2 mice was occupied with GFP/Ly5.1 double-negative cells. One of the mice/WT developed leukemia derived from recipient cells at 272 days after transplantation. The remaining one mouse/WT died of anemia with unknown reason at 355 days after transplantation.
MDS and MDS/AML induced by AML1 mutants derived from patients with MDS. (A) Schematics of AML1 WT (AML1a and AML1b) and AML1 mutants (D171N and S291fs). AD indicates transactivating domain; RD, repression domain. (B) Percentages of GFP/Ly-5.1 double-positive cells or (C) c-Kit+ cells in PB. PB was obtained from the tail vein every month after the transplantation. Numbers in parenthesis indicate mouse IDs. (D) Kaplan-Meier analysis for the survival of mice that received transplants of AML1 mutant-transduced BM cells. Average survival days of AML1-D171N (340.6 days) were compared with AML1-S291fs (263.6 days) using the log-rank test; P = .218. AML1 WT (n = 11), D171N (n = 26), S291fs (n = 10), mock (n = 16). (E) Evi1 synergized with AML1-D171N in inducing MDS/AML. D171N (n = 26; same as those in panel D), Evi1 (n = 8), D171N + Evi1 (n = 10), and mock (n = 16) transduced bone marrow cells were transplanted into mice.
MDS and MDS/AML induced by AML1 mutants derived from patients with MDS. (A) Schematics of AML1 WT (AML1a and AML1b) and AML1 mutants (D171N and S291fs). AD indicates transactivating domain; RD, repression domain. (B) Percentages of GFP/Ly-5.1 double-positive cells or (C) c-Kit+ cells in PB. PB was obtained from the tail vein every month after the transplantation. Numbers in parenthesis indicate mouse IDs. (D) Kaplan-Meier analysis for the survival of mice that received transplants of AML1 mutant-transduced BM cells. Average survival days of AML1-D171N (340.6 days) were compared with AML1-S291fs (263.6 days) using the log-rank test; P = .218. AML1 WT (n = 11), D171N (n = 26), S291fs (n = 10), mock (n = 16). (E) Evi1 synergized with AML1-D171N in inducing MDS/AML. D171N (n = 26; same as those in panel D), Evi1 (n = 8), D171N + Evi1 (n = 10), and mock (n = 16) transduced bone marrow cells were transplanted into mice.
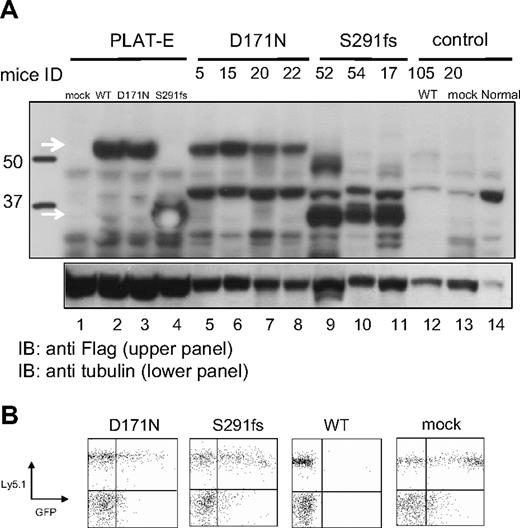
Expression of the transduced AML1-D171N, AML1-S291fs, and AML1 WT in spleen of the transplanted mice. (A) Lysates of spleen cells were immunoblotted with anti-Flag Ab. As a positive control, Plat-E packaging cells were transduced with mock (lane 1), AML1 WT (lane 2), AML1-D171N (lane 3), or AML1-S291fs (lane 4). Spleen cells were derived from mice/D171N (lanes 5-8), mice/S291fs (lanes 9-11), mice/WT (lane 12), mice/mock (lane 13), or control normal mouse (lane 14). White arrows indicate transduced AML1 WT, AML1-D171N, and AML1-S291fs. (B) AML1 WT-transduced cells were undetectable in PB at 1 month after the transplantation. Flow cytometric analysis of PB obtained from mice that received transplants of AML1-D171N, AML1-S291fs, AML1 WT, and mock at 1 month after transplantation.
Expression of the transduced AML1-D171N, AML1-S291fs, and AML1 WT in spleen of the transplanted mice. (A) Lysates of spleen cells were immunoblotted with anti-Flag Ab. As a positive control, Plat-E packaging cells were transduced with mock (lane 1), AML1 WT (lane 2), AML1-D171N (lane 3), or AML1-S291fs (lane 4). Spleen cells were derived from mice/D171N (lanes 5-8), mice/S291fs (lanes 9-11), mice/WT (lane 12), mice/mock (lane 13), or control normal mouse (lane 14). White arrows indicate transduced AML1 WT, AML1-D171N, and AML1-S291fs. (B) AML1 WT-transduced cells were undetectable in PB at 1 month after the transplantation. Flow cytometric analysis of PB obtained from mice that received transplants of AML1-D171N, AML1-S291fs, AML1 WT, and mock at 1 month after transplantation.
Interestingly, in the peripheral blood of mice/WT, GFP+ cell counts were extremely low 1 month after transplantation and thereafter became undetectable, despite the fact that 14% to 27% of the BM cells were positive for GFP before transplantation (Figures 1B,2B; Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Consistently, the expression of transduced AML1 WT was not detected in spleen cells of mice/WT (Figure 2A; mouse ID: 105). These data suggested that forced expression of AML1 WT in the stem cells had a negative effect on the survival and expansion of these cells in the BM. Recently, Tsuzuki et al reported that expression of the full-length isoform AML1b abrogated engraftment potential of murine long-term reconstituting stem cells in a mouse BMT model.46 Their result coincides with our result.
AML1-D171N and AML1-S291fs induced different diseases in mice that underwent transplantation
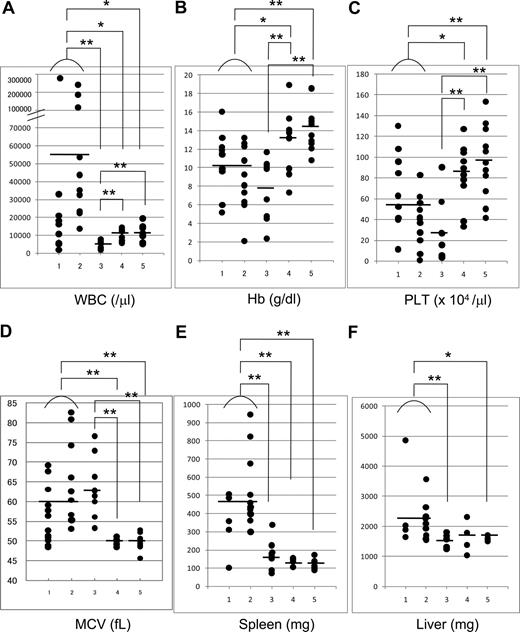
PB cell counts were different between mice/D171N and mice/S291fs; most mice/D171N (Figure 3A; lanes 1,2) showed leukocytosis, while mice/S291fs (Figure 3A; lane 3) showed leukocytopenia. This difference was significant (P = .007). Macroscopic observation of morbid mice revealed that severe hepatosplenomegaly was exclusively found in mice/D171N, but not in mice/S291fs (Figure 3E,F; Table S2).
Peripheral white blood cell counts of mice/D171N showed leukocytosis, while mice/S291fs showed leukocytopenia. (A) Counts of white blood cells (WBCs) in PB. (B) Concentration of hemoglobin (Hg). (C) Counts of platelets (PLT). (D) Red cell MCV. (E,F) Weight of spleen and liver of morbid mice (mice/D171N or S291fs) or 1-year-old healthy mice (mice/WT or mock). Statistical differences were determined by 2-sample t test with Welch correction (*P < .05; **P < .01). Lane 1: mice/D171N without high expression of Evi1 in BM or not examined due to the lack of bone marrow samples (WBC, PLT, Hg, and MCV: n = 11; spleen: n = 5; and liver: n = 4). Lane 2: mice/D171N with high expression of Evi1 (n = 11). Lane 3: mice/S291fs (WBC, PLT, Hg, and MCV: n = 9; spleen and liver: n = 8). Lane 4: mice/WT (WBC, PLT, Hg, and MCV: n = 10; spleen and liver: n = 4). Lane 5: mice/mock (WBC, PLT, Hg, and MCV: n = 12; spleen: n = 6; and liver: n = 5).
Peripheral white blood cell counts of mice/D171N showed leukocytosis, while mice/S291fs showed leukocytopenia. (A) Counts of white blood cells (WBCs) in PB. (B) Concentration of hemoglobin (Hg). (C) Counts of platelets (PLT). (D) Red cell MCV. (E,F) Weight of spleen and liver of morbid mice (mice/D171N or S291fs) or 1-year-old healthy mice (mice/WT or mock). Statistical differences were determined by 2-sample t test with Welch correction (*P < .05; **P < .01). Lane 1: mice/D171N without high expression of Evi1 in BM or not examined due to the lack of bone marrow samples (WBC, PLT, Hg, and MCV: n = 11; spleen: n = 5; and liver: n = 4). Lane 2: mice/D171N with high expression of Evi1 (n = 11). Lane 3: mice/S291fs (WBC, PLT, Hg, and MCV: n = 9; spleen and liver: n = 8). Lane 4: mice/WT (WBC, PLT, Hg, and MCV: n = 10; spleen and liver: n = 4). Lane 5: mice/mock (WBC, PLT, Hg, and MCV: n = 12; spleen: n = 6; and liver: n = 5).
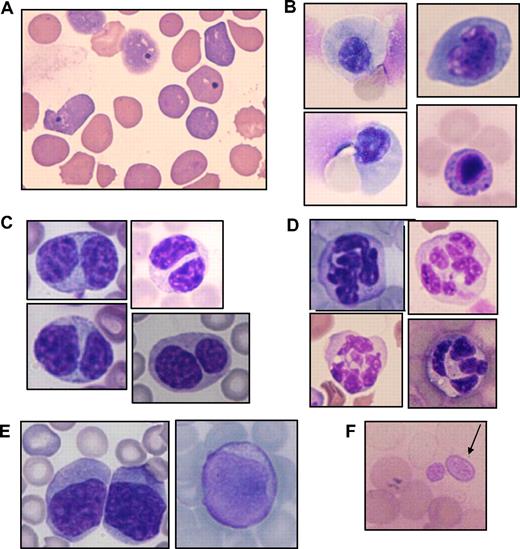
The smear specimens of peripheral blood were obtained every 1 to 2 months. The specimens showed that most of mice/D171N and mice/S291fs suffered from multilineage dysplasia characteristic of MDS. Erythroid dysplasia such as Howell-Jolly bodies, red cell polychromasia, and poikilocytosis (Figure 4A) were frequently detected in both mice. In BM specimens of morbid mice, orthochromatic giant erythroblasts and karyorrhexis were detected (Figure 4B). Erythroid dysplasia was more evident in mice/S291fs than in mice/D171N. As recently described in a mouse MDS model,15 increase of red blood cell mean corpuscular volume (MCV) was also observed in most mice/D171N and mice/S291fs (Figure 3D; Table S2). Myeloid dysplasia such as the pseudo–Pelger-Huet anomaly (Figure 4C) was frequently detected in mice/D171N. Hypersegmented neutrophils (Figure 4D) and giant platelets (Figure 4F) were observed in 2 mice/D171N (mouse IDs 9 and 17). Collectively, AML1 mutants used in this study induced multilineage dysplasia, in particular in erythroid and myeloid lineages. Continuous pancytopenia was observed in 7 of 8 morbid mice/S291fs and 2 of 16 morbid mice/D171N, although BM of the morbid mice was not hypocellular but hypercellular or normocellular. Based on these findings, a final diagnosis was made by the ratio of blasts in the bone marrow according to the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice.41 As a result, MDS/AML was recognized in 13 of 16 morbid mice/D171N and in 5 of 8 morbid mice/S291fs, while MDS-RAEB was recognized in 2 of 16 morbid mice/D171N and 2 of 8 morbid mice/S291fs (Table S2). One mouse/D171N was diagnosed with AML at 4 months after transplantation because we did not examine to see if the MDS phase had preceded AML (mouse ID 5). The leukemic cells derived from either mice/D171N or mice/S291fs were serially transplantable. We confirmed the serial transplantability in 11 mice/D171N (mouse IDs 4, 6, 9, 12-15, 17, 20, 22, and 26) and 6 mice/S291fs (mouse IDs 52, 54-56, 58, and 60). Penetrance of serial transplantation was 100%, except for mouse IDs 9 and 17; that is, 33% and 50%, respectively. Mice that underwent serial transplantation showed more aggressive status than primary mice and died with shorter latencies. Hematologic parameters of mice that underwent serial transplantation are shown in Table S3.
Multilineage dysplasia of hematopoietic cells in mice that received transplants of AML1 mutants. Giemsa-stained PB smears obtained from mice/D171N or S291fs are shown. (A) Howell-Jolly body, polychromasia, and anisopoikilocytosis. (B) Orthochromatic giant erythroblast, karyorrhexis, and nuclear fragments. (C) Pseudo-Pelger-Huet anomaly. (D) Hypersegmented neutrophil. (E) Blasts in peripheral blood. (F) Giant platelet. Images were obtained with a BH51 microscope and DP12 camera (Olympus, Tokyo, Japan); objective lens, UPlanFl (Olympus); magnification, ×1000.
Multilineage dysplasia of hematopoietic cells in mice that received transplants of AML1 mutants. Giemsa-stained PB smears obtained from mice/D171N or S291fs are shown. (A) Howell-Jolly body, polychromasia, and anisopoikilocytosis. (B) Orthochromatic giant erythroblast, karyorrhexis, and nuclear fragments. (C) Pseudo-Pelger-Huet anomaly. (D) Hypersegmented neutrophil. (E) Blasts in peripheral blood. (F) Giant platelet. Images were obtained with a BH51 microscope and DP12 camera (Olympus, Tokyo, Japan); objective lens, UPlanFl (Olympus); magnification, ×1000.
In summary, the pattern and degree of multilineage dysplasia differed among the mice that underwent transplantation. Although both mice/D171N and mice/S291fs died of MDS and MDS/AML within 4 to 13 months after transplantation, a marked difference existed in terms of clinical symptoms, including hematopoietic or macroscopic findings.
A distinct type of disease was identified in mice/D171N
Among the mice that underwent transplantation transduced with AML1-D171N, a distinct group was identified. GFP+ BM cells in 11 of 16 morbid mice/D171N (mouse IDs 4, 6, 7, 12-15, 19, 20, 22, and 26) displayed a similar phenotype, with high percentages of CD11b+ and B220+ cells (Figure 5; data not shown). All these mice/D171N showed dysplasia in myeloid and erythroid lineages for several months and died of MDS/AML with increased number of blasts, anemia, and, in some cases, thrombocytopenia (Table S2). These mice/D171N also showed severe hepatosplenomegaly (Figures 3E,F, 6A; Table S2), and histologic examination showed expansion of blasts and immature myeloid cells in the PB, BM, and spleen, and the invasion of these cells into hepatic portal areas in the liver and spaces among renal tubules in the kidney (Figure 6B,D,F). Giemsa staining of BM showed a high percentage of blasts (Figure 6H), in accordance with a high percentage of both GFP/c-kit double-positive cells by flow cytometolic analysis.
AML1-D171N induced a biphenotypic leukemia in concert with Evi1 in the BMT model. The dot plots show Ly5.1, Gr-1, CD11b, B220, CD3, CD41, c-kit, Sca-1, CD34, or Ter119 labeled with a corresponding PE-conjugated mAb versus expression of GFP. BM cells of morbid mice/D171N with high expression of Evi1 (mouse IDs 6, 20, and 26) and those of morbid mice/D171N + Evi1 (mouse IDs 302 and 305) displayed a similar pattern of surface markers, CD11b+ and B220+.
AML1-D171N induced a biphenotypic leukemia in concert with Evi1 in the BMT model. The dot plots show Ly5.1, Gr-1, CD11b, B220, CD3, CD41, c-kit, Sca-1, CD34, or Ter119 labeled with a corresponding PE-conjugated mAb versus expression of GFP. BM cells of morbid mice/D171N with high expression of Evi1 (mouse IDs 6, 20, and 26) and those of morbid mice/D171N + Evi1 (mouse IDs 302 and 305) displayed a similar pattern of surface markers, CD11b+ and B220+.
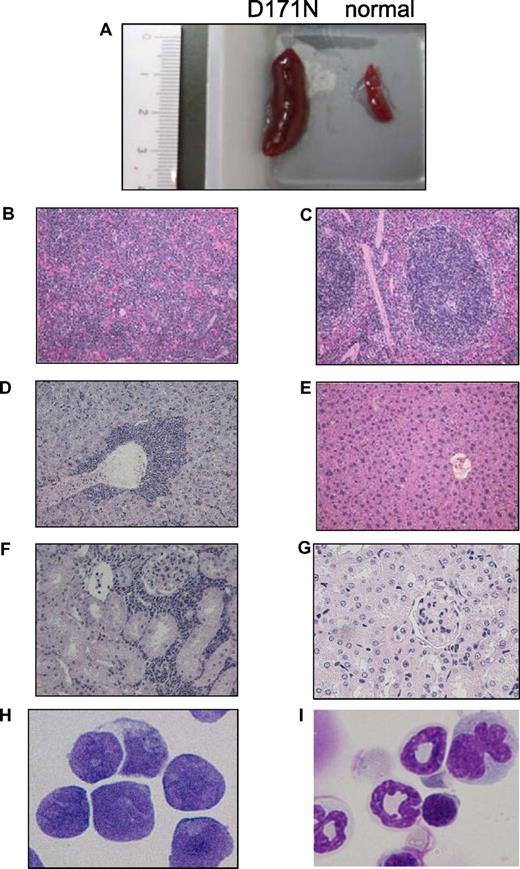
Leukemic cells of mice/D171N with high expression of Evi1 invaded into liver and kidney. (A) Spleen from morbid mice/D171N (left) and from normal mice (right). Histopathologic findings of (B) spleen, (D) liver, and (F) kidney infiltrated with leukemic cells from mice/D171N, stained with H&E. Histopathologic findings of (C) spleen, (E) liver, and (G) kidney from normal mice, stained with H&E. (H) Mice/D171N showed a high percentage of blasts in bone marrow. Cytospin preparations of BM cells from (H) mice/D171N and (I) normal mice, stained with Giemsa. (BX51 microscope, DP12 camera module; objective lens, UPlanFl; magnification, ×200 (B-E); ×100 (F,G); and ×1000 (H,I).
Leukemic cells of mice/D171N with high expression of Evi1 invaded into liver and kidney. (A) Spleen from morbid mice/D171N (left) and from normal mice (right). Histopathologic findings of (B) spleen, (D) liver, and (F) kidney infiltrated with leukemic cells from mice/D171N, stained with H&E. Histopathologic findings of (C) spleen, (E) liver, and (G) kidney from normal mice, stained with H&E. (H) Mice/D171N showed a high percentage of blasts in bone marrow. Cytospin preparations of BM cells from (H) mice/D171N and (I) normal mice, stained with Giemsa. (BX51 microscope, DP12 camera module; objective lens, UPlanFl; magnification, ×200 (B-E); ×100 (F,G); and ×1000 (H,I).
In contrast, the remaining 4 morbid mice/D171N diagnosed as MDS (mouse IDs 9 and 11) or MDS/AML (mouse IDs 10 and 17) showed heterologous phenotypes of GFP+ BM cells (data not shown). Although mouse IDs 9 and 17 displayed hepatosplenomegaly like other mice/D171N, they exhibited leukocytopenia with fewer blasts.
AML1-D171N collaborated with Evi1 in inducing MDS/AML
We then asked why even the same point mutant of AML1 caused different phenotypes of MDS-RAEB and MDS/AML. We assumed the possibility that the integration of retroviruses influenced the outcomes in the BMT model. To explore this, we first performed Southern blot analysis of BM of the morbid mice. A single or several proviral integrations were confirmed (Figure 7A). Next, we used the bubble PCR method to identify the integrated sites.7,44,45 A single or 2 integration sites were identified in each sample (Table 1). Interestingly, integrations near Evi1 site were found in 7 of 15 genomic DNA samples of BM cells derived from mice/D171N, but not from mice/S291fs. Moreover, retrospective examination revealed that these 7 mice presented nearly identical phenotypes, characterized by marked hepatosplenomegaly (Figure 6A), leukocytosis (Figure 3A), and biphenotypic surface markers (CD11b+ and B220+) of the leukemic cells (Figure 5), thus constituting a definite subgroup among mice/D171N. Southern blot analysis showed that all of the leukemic mice with high expression of Evi1 are monoclonal (except for mouse IDs 15 and 19), but the other leukemic mice without high expression of Evi1 are oligoclonal or have several integrations (Figure 7A). Noteworthy was the finding that the Evi1 site was not identified from the genomic DNA samples of mice/S291fs, even though the Evi1 site is a known common integration site of retroviruses.31-38 These led us to postulate that Evi1 collaborated with AML1-D171N in inducing the distinct type of MDS/AML. To test this, we examined whether the expression of Evi1 was enhanced in the BM cells in which the integration into an Evi1 site was identified. Real-time PCR analysis demonstrated that the expression levels of Evi1 were high in all the related samples (Figure 7C). Protein expression levels corresponded to mRNA expression levels of Evi1 (Figure 7D; data not shown). Interestingly, 4 samples derived from mice/D171N harboring no integration near Evi1 also displayed significantly high expression levels of Evi1 (mouse IDs 13, 19, 20, and 22), and the phenotypes of these mice were identical to those induced by AML1-D171N–transduced cells in which retroviruses were integrated into the Evi1 site. In these cases, the expression of Evi1 might have been enhanced secondarily by an unknown mechanism, or we simply failed to detect the integration site. The latter possibility was supported by the fact that multiple integrations were detected in these cases (Figure 7A; mouse IDs 13, 19, and 20). In any case, all the mice/D171N with enhanced expression of Evi1 in their BM cells displayed high percentages of blasts (Figure S1). We also examined whether the expression of MDS1/Evi1 was enhanced by the integration into an Evi1 site. The MDS1 gene is located approximately 240 kb upstream of Evi1, and MDS1/Evi1 is generated from the in-frame splicing of MDS1 to the second exon of Evi1.36,43 Real-time PCR analysis demonstrated that the expression levels of MDS1/Evi1 were low and were not significantly increased when compared with controls (data not shown). The integration sites of Evi1 were focused on two regions (Table 1). One is 15 kb upstream of start site of Evi1 (mouse IDs 12, 14, 15), and another is 107 kb upstream of start site of Evi1 (mouse IDs 4, 6, 7, 26). Morishita et al reported that the retrovirus integrations had occurred near or in 5′ noncoding exons of Evi1 gene.31 The integration site at 15 kb upstream of the start site of Evi1 that we found is near to the site they reported.
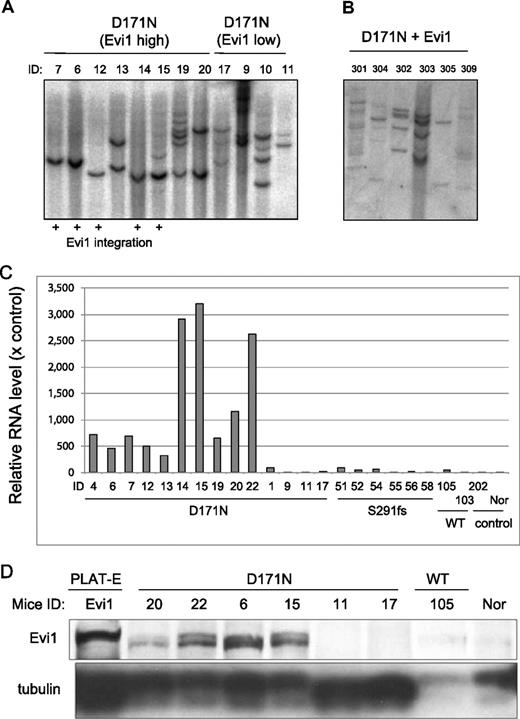
The mice/D171N with integration near Evi1 site were monoclonal. (A) Southern blot analysis of mice/D171N. DNA samples were digested with EcoRI, which cut the retrovirus only once within the multicloning site. Probes used were DNA fragments of the GFP coding sequence. Mouse IDs are shown at the top of the panel. (B) The mice/D171N + Evi1 were polyclonal. DNA samples were digested with EcoRI. Proviruses were probed with a GFP probe. (C) Real-time PCR for Evi1 in BM derived from morbid mice/D171N or mice/S291fs or mice/WT or mice/mock. In addition to 6 samples from mice/D171N harboring integration near Evi1 (mouse IDs 4, 6, 7, 12, 14, and 15), 4 samples derived from mice/D171N without integration near Evi1 display high expression levels of Evi1 (mouse IDs 13, 19, 20, and 22). RNA from normal BM cells served as a control (RNA level = 1). (D) Western blot of lysates from spleen cells of mice/D171N, mice/WT, and normal mice and PLAT-E as controls. Samples from mice/D171N confirmed high expression of Evi1 by RT-PCR showed expression of the protein (mouse IDs 6, 15, 20, and 22), but the other mice without high expression of Evi1 by RT-PCR did not express the protein (mouse IDs 11, 17, and 105).
The mice/D171N with integration near Evi1 site were monoclonal. (A) Southern blot analysis of mice/D171N. DNA samples were digested with EcoRI, which cut the retrovirus only once within the multicloning site. Probes used were DNA fragments of the GFP coding sequence. Mouse IDs are shown at the top of the panel. (B) The mice/D171N + Evi1 were polyclonal. DNA samples were digested with EcoRI. Proviruses were probed with a GFP probe. (C) Real-time PCR for Evi1 in BM derived from morbid mice/D171N or mice/S291fs or mice/WT or mice/mock. In addition to 6 samples from mice/D171N harboring integration near Evi1 (mouse IDs 4, 6, 7, 12, 14, and 15), 4 samples derived from mice/D171N without integration near Evi1 display high expression levels of Evi1 (mouse IDs 13, 19, 20, and 22). RNA from normal BM cells served as a control (RNA level = 1). (D) Western blot of lysates from spleen cells of mice/D171N, mice/WT, and normal mice and PLAT-E as controls. Samples from mice/D171N confirmed high expression of Evi1 by RT-PCR showed expression of the protein (mouse IDs 6, 15, 20, and 22), but the other mice without high expression of Evi1 by RT-PCR did not express the protein (mouse IDs 11, 17, and 105).
Analysis of integration site
| Mutant . | Mouse ID . | Count of Southern blot bands . | Nearest gene . | Chromosome no. . | Gene ID . | Distance to gene (start or end), bp . | Location . | Forward or reverse orientation . | RTCGD hits . |
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | |||||||||
| D171N | 1 | ND | 2010111I01Rik | 13 | 72061 | 238 927 | Intron 10 | F | 0 |
| D171N | 1 | LOC100040862 | 12 | 100040862 | 40 051 | 5′ | F | 0 | |
| D171N | 4 | ND | Evil | 3 | 14013 | 107 412 | 5′ | R | 24 |
| D171N | 4 | Nsmce2 | 15 | 68501 | 221 183 | Intron 5 | R | 2 | |
| Experiment 2 | |||||||||
| D171N | 5 | 3 | Rreb1 | 13 | 68750 | 14 071 | 5′ | R | 9 |
| D171N | 5 | LOC619665 | 6 | 619665 | 123 854 | 5′ | F | 0 | |
| D171N | 6 | 1 | Evil | 3 | 14013 | 106 286 | 5′ | R | 24 |
| D171N | 7 | 1 | Evil | 3 | 14013 | 106 286 | 5′ | R | 24 |
| Experiment 3 | |||||||||
| D171N | 10 | 4 | B4galt6 | 18 | 56386 | 8 961 | Intron 1 | R | 0 |
| D171N | 11 | 2 | Rp1h | 1 | 19888 | 65 523 | 5′ | F | 0 |
| Experiment 4 | |||||||||
| D171N | 12 | 1 | Evil | 3 | 14013 | 14 909 | 5′ | F | 24 |
| D171N | 13 | 2 | Slc38a2 | 15 | 67760 | 41 018 | 5′ | F | 6 |
| D171N | 14 | 1 | Evil | 3 | 14013 | 15 002 | 5′ | F | 24 |
| D171N | 15 | 2 | Evil | 3 | 14013 | 14 850 | 5′ | F | 24 |
| D171N | 15 | Slc38a2 | 15 | 22462 | 11 988 | 5′ | R | 6 | |
| S291fsX300 | 51 | ND | LOC100042800 | 13 | 100042800 | 27 734 | 5′ | R | 0 |
| S291fsX300 | 52 | 2 | P2rx7 | 5 | 18439 | 40 672 | intron 13 | R | 1 |
| S291fsX300 | 52 | Gm pr2 | 14 | 105446 | 17 | intron 1 | R | 0 | |
| S291fsX300 | 54 | 2 | M srb3 | 10 | 320183 | 98 981 | 3′ | F | 0 |
| Empty | 203 | ND | Dph5 | 3 | 13609 | 70 064 | 5′ | R | 1 |
| Experiment 5 | |||||||||
| D171N | 19 | 6 | Gch1 | 14 | 14528 | 16 958 | Intron 1 | R | 0 |
| D171N | 20 | 2 | Gch1 | 14 | 14528 | 16 958 | Intron 1 | R | 0 |
| S291fsX300 | 55 | 2 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| S291fsX300 | 56 | 1 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| S291fsX300 | 58 | 1 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| Experiment 6 | |||||||||
| D171N | 22 | ND | Lrrc8c | 5 | 100604 | 16 056 | 5′ | F | 4 |
| Experiment 7 | |||||||||
| D171N | 26 | 1 | Evil | 3 | 14013 | 106 710 | 5′ | F | 24 |
| S291fsX300 | 60 | 3 | Dock10 | 1 | 210293 | 163 158 | Intron 1 | R | 0 |
| Mutant . | Mouse ID . | Count of Southern blot bands . | Nearest gene . | Chromosome no. . | Gene ID . | Distance to gene (start or end), bp . | Location . | Forward or reverse orientation . | RTCGD hits . |
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | |||||||||
| D171N | 1 | ND | 2010111I01Rik | 13 | 72061 | 238 927 | Intron 10 | F | 0 |
| D171N | 1 | LOC100040862 | 12 | 100040862 | 40 051 | 5′ | F | 0 | |
| D171N | 4 | ND | Evil | 3 | 14013 | 107 412 | 5′ | R | 24 |
| D171N | 4 | Nsmce2 | 15 | 68501 | 221 183 | Intron 5 | R | 2 | |
| Experiment 2 | |||||||||
| D171N | 5 | 3 | Rreb1 | 13 | 68750 | 14 071 | 5′ | R | 9 |
| D171N | 5 | LOC619665 | 6 | 619665 | 123 854 | 5′ | F | 0 | |
| D171N | 6 | 1 | Evil | 3 | 14013 | 106 286 | 5′ | R | 24 |
| D171N | 7 | 1 | Evil | 3 | 14013 | 106 286 | 5′ | R | 24 |
| Experiment 3 | |||||||||
| D171N | 10 | 4 | B4galt6 | 18 | 56386 | 8 961 | Intron 1 | R | 0 |
| D171N | 11 | 2 | Rp1h | 1 | 19888 | 65 523 | 5′ | F | 0 |
| Experiment 4 | |||||||||
| D171N | 12 | 1 | Evil | 3 | 14013 | 14 909 | 5′ | F | 24 |
| D171N | 13 | 2 | Slc38a2 | 15 | 67760 | 41 018 | 5′ | F | 6 |
| D171N | 14 | 1 | Evil | 3 | 14013 | 15 002 | 5′ | F | 24 |
| D171N | 15 | 2 | Evil | 3 | 14013 | 14 850 | 5′ | F | 24 |
| D171N | 15 | Slc38a2 | 15 | 22462 | 11 988 | 5′ | R | 6 | |
| S291fsX300 | 51 | ND | LOC100042800 | 13 | 100042800 | 27 734 | 5′ | R | 0 |
| S291fsX300 | 52 | 2 | P2rx7 | 5 | 18439 | 40 672 | intron 13 | R | 1 |
| S291fsX300 | 52 | Gm pr2 | 14 | 105446 | 17 | intron 1 | R | 0 | |
| S291fsX300 | 54 | 2 | M srb3 | 10 | 320183 | 98 981 | 3′ | F | 0 |
| Empty | 203 | ND | Dph5 | 3 | 13609 | 70 064 | 5′ | R | 1 |
| Experiment 5 | |||||||||
| D171N | 19 | 6 | Gch1 | 14 | 14528 | 16 958 | Intron 1 | R | 0 |
| D171N | 20 | 2 | Gch1 | 14 | 14528 | 16 958 | Intron 1 | R | 0 |
| S291fsX300 | 55 | 2 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| S291fsX300 | 56 | 1 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| S291fsX300 | 58 | 1 | Mn1 | 5 | 433938 | 16 024 | Intron 1 | F | 8 |
| Experiment 6 | |||||||||
| D171N | 22 | ND | Lrrc8c | 5 | 100604 | 16 056 | 5′ | F | 4 |
| Experiment 7 | |||||||||
| D171N | 26 | 1 | Evil | 3 | 14013 | 106 710 | 5′ | F | 24 |
| S291fsX300 | 60 | 3 | Dock10 | 1 | 210293 | 163 158 | Intron 1 | R | 0 |
RTGCD, Retroviral Tagged Cancer Gene Database37 ; and ND, not determined.
In vivo collaboration between Evi1 and AML1-D171N
Next, we tested to see if Evi1 expression collaborates with AML1-D171N in inducing leukemia in the BMT model. Cotransduction of AML1-D171N and Evi1 into BM cells resulted in rapid induction of the disease in the mice that underwent transplantation that was essentially identical with the disease that developed after a long latency in the mice/D171N (Figures 1E,5). In fact, all the mice displayed increased number of blasts in the PB within a month after the transplantation of BM cells transduced with Evi1/D171N. Southern blot analysis showed that these leukemic cells were polyclonal (Figure 7B). These results indicate that AML1-D171N and Evi1 collaborate to induce MDS/AML with a distinct phenotype. On the other hand, cotransduction of AML1-S291fs and Evi1 into BM cells did not induce MDS/AML in 5 months (data not shown). In the present work, mice that received transplants of BM cell–transduced Evi1 alone did not present any abnormalities in 5 months (Figure 1E).
AML1-S291fs induced erythroid dysplasia with pancytopenia
In contrast to mice/D171N, most of mice/S291fs displayed remarkable erythroid dysplasia with continuous pancytopenia (Figures 3A-C,4A). A total of 2 of 8 mice/S291fs died of MDS-RAEB in which the percentage of blasts in the bone marrow was less than 20%, and 5 mice/S291fs developed MDS/AML. The mice displayed severe anemia but not leukocytosis in the PB, and the numbers of blasts were generally lower than those of MDS/AML mice transduced with AML1-D171N (Table S2). Surface markers of leukemic cells derived from mice/S291fs were different from those of mice/D171N (Figure S3).
We found integrations of the retrovirus in the intron of MN1 in 3 of 8 mice/S291fs (Table 1; mouse IDs 55, 56, and 58), and MN1 was overexpressed in the leukemic cells of these mice (Figure S2). The integration site was identical among these leukemic cells, indicating that leukemic cells of the 3 mice were derived from a single hematopoietic progenitor and that overexpression of MN1 induced expansion of the transduced stem cells during the 3-day culture period before the transplantation. Indeed, the mice with the integration at that MN1 site developed MDS/AML with shorter latencies (Table S2).
Discussion
We have established a mouse BMT model for MDS and MDS/AML using AML1 mutants derived from patients with MDS, although previous studies either using similar BMT models or knock-in mice of AML1 mutants failed to do so. There are several potential explanations for this discrepancy. First, because most AML1 mutants work as dominant-negative forms, high expression levels of the mutants would be critical to effectively inhibit WT AML1. In this aspect, our BMT model has an advantage, using the efficient retrovirus vector pMYs40 designed to achieve high expression in hematopoietic progenitor cells and, unlike most other retrovirus vectors, harbors splice donor and acceptor sites derived from the MFG vector to increase expression levels.40,47 Second, using the efficient packaging cell line Plat-E,39 we achieved high titers of retroviruses (average: 107 infection U/mL), which could result in the higher numbers of retrovirus integrations. This also increases the probabilities of up-regulating or disrupting important genes that collaborate with AML1 mutants in inducing MDS and/or MDS/AML. Alternatively, it is also possible that the positions of AML1 mutations are critical for the biological effect. We believe that the combination of these factors has put our system into practice.
In the present MDS model, we used 2 AML1 mutants, D171N and S291fsX300. The latter, a C-terminal–truncated form, is more potent as a dominant-negative form than the former, which harbors a point mutation in the RHD.25,26 In this context, it is reasonable that the S291fs mutant induced the disease in the mice that underwent transplantation with a higher penetrance (Figure 1D). More important, expression of these mutants induced MDS/AML of distinct phenotypes in the mice that underwent transplantation: AML1-S291fs induced pancytopenia associated with dysplasia in the erythroid lineage, while AML1-D171N frequently induced hepatoslenomegaly and leukocytosis associated with marked myeloid dysplasia. This suggests that even different mutations of the same gene could induce heterogeneous diseases. As previously described,25,26 AML1-D171N lost DNA-binding ability and hence transactivation potential because it possessed a point mutation in RHD essential for DNA-binding, while AML1-S291fs had increased DNA-binding ability but lost transactivation potential because it had an intact RHD but lacked a C-terminal transactivation domain. Thus, the different biological outcomes induced by AML1 mutants could be explained in part by structural and functional differences between the mutants. In addition to the dominant-negative functions, these mutants may also have gain of function; the fact that AML1-KO mice did not develop leukemia18 indicates that deletion of AML1 alone is not sufficient to induce leukemia, suggesting the possibility that the AML1 mutants have gain of function as well. Because AML1 associates and forms a ternary complex with other transcriptional factors and cofactors via its specific domains, it is possible that these mutants exert different effects on the proliferation and differentiation of BM cells in the various contexts.
In BMT models using retrovirus-mediated gene transfer, the genes near the retrovirus integration sites are thought to affect the outcomes.30-38 This sometimes obscures the significance of the transduced gene, but simultaneously will give us clues to understanding the collaboration of multiple genes in the development of leukemias. One of the intriguing findings of the present work is that high expression of Evi1, either caused by virus integration or by unknown mechanisms, was able to collaborate with AML1-D171N in inducing the homogeneous disease characterized by leukocytosis, severe myelodysplasia, and marked hepatosplenomegaly that always developed to overt leukemia with high percentages of B220+ and CD11b+ blasts. Together with the recent findings that Evi1 expression was observed in patients with MDS and AML,36,48 and that Evi1 alone did not induce AML in mouse models,14,34,49,50 our result strongly suggested that AML1-D171N collaborated with Evi1 in inducing MDS/AML. It is interesting to note that AML1-S291fs never collaborated with Evi1 during our examination (Table 1), again suggesting that these 2 AML1 mutations transform hematopoietic cells through distinct mechanisms. Importantly, we confirmed the collaboration between AML1-D171N and Evi1 in an in vivo experiment. Cotransduction of AML1-D171N and Evi1 into BM cells resulted in rapid induction of MDS/AML in the mice that received transplants. In addition, the leukemic cells in most of these mice included more clones than those in mice/D171N (Figure 7B), indicating cooperation of Evi1 and AML1-D171N. However, leukemic cells from one mouse (ID 305) seemed to be monoclonal and to contribute to oligoclonal leukemia of mouse 304. In addition, it took 2 to 3 months for leukemias induced by the combination of AML1-D171N and Evi1 to kill the mice that received transplants. Together, these result suggested that while AML1-D171N and Evi1 overexpression collaborated in inducing leukemia, additional steps were required for efficient transformation of hematopoietic progenitors. In the absence of Evi1 high expression, AML1-D171N caused MDS or MDS/AML with low percentages of blasts in BM but still with hepatosplenomegaly. This indicates that hepatosplenomegaly had something to do with AML1-D171N.
In contrast to mice/D171N, most mice/S291fs succumbed to either MDS-RAEB with fatal severe anemia following continuous pancytopenia or MDS/AML without leukocytosis. The integration site in the intron 1 of the MN1 gene found in leukemic cells of 3 mice was derived from the same cell. We also found that MN1 was overexpressed in the leukemic cells of these mice, suggesting that overexpression of MN1 induced effective expansion of leukemic stem cells. Recently, Heuser et al reported that high expression of MN1 correlated with poor outcome in AML with normal cytogenetics.51 Moreover, Slape et al identified MN1 as potential collaborators of NUP98/HOXD13 to induce leukemia.42 Further work will be required to investigate the role of MN1 in MDS/AML.
One fundamental question of this study was whether AML1 mutants alone induce MDS and MDS/AML. In our experiments, 5 of the 6 surviving mice/D171N showed a disappearance of GFP+ cells in time, suggesting that AML1-D171N alone was not able to induce MDS/AML. Previous studies using gene-engineered mice and a BMT model demonstrated that AML1 fusions caused by chromosomal translocations alone were insufficient to induce AML,7-12 except for AML1-MDS1-Evi1, which by itself induced AML with a long latency.52 In addition, several lines of evidence30-38 that implicated the integration site of retroviruses for different biological outcomes led us to consider the same possibility in this BMT model. Indeed, we identified frequent retrovirus integrations near the Evi1 gene in the BM cells derived from mice/D171N whose leukemic cells displayed nearly identical phenotypes and concomitant elevated expression of Evi1. Importantly, coexpression of AML1-D171N and Evi1 induced the same leukemia with shorther latencies, demonstrating the collaboration between AML1-D171N and Evi1 in vivo. These results showed the power of in vivo insertional mutagenesis of retroviruses in a search for genes involved in the pathogenesis of MDS and MDS/AML.
Finally, it is important to relate these in vivo results to clinical data of the human disease. The recent finding25-27 that AML1 point mutations in the C-terminal regions were almost exclusively found in MDS-RAEB and MDS/AML, but not in de novo AML, coincided with our data that AML1-S291fs tended to induce MDS-RAEB–like symptoms in this BMT model. Clinical findings25-27 that the RHD point mutation was often found in de novo AML, mainly AML M0, in addition to MDS-RAEB and MDS/AML, was also in accordance with our data that AML1-D171N induced more progressive MDS/AML with higher percentages of blasts when compared with AML1-S291fs. Classification of MDS and MDS/AML is always controversial because of the heterogeneity of the disease.1,2,27 In the future, this disease will be reclassified based on genetic alterations and their combinations.
In summary, we have generated a mouse BMT model of MDS-RAEB and MDS/AML. The current BMT model, mimicking AML1-related MDS, will be useful for understanding molecular pathogenesis and establishing new therapeutic strategy for MDS and MDS/AML.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We greatly thank Dr Mineo Kurokawa for kindly providing the anti-Evi1 antibody and Dr Takuro Nakamura and Dr Kazuhiro Morishita for kindly providing pMYs-Evi1-IG. We also thank Dr Christopher Slape for kindly giving information about the condition of RT-PCR for MN1. We are grateful to Dr Dovie Wylie for excellent language assistance. We thank Yumi Fukuchi, Fumi Shibata, Miyuki Ito, and Ai Hishiya for technical assistance.
This work was supported by the Grant-in-aid for Cancer Research supported by the Ministry of Health, Labor and Welfare, Japan; a grant from the Vehicle Racing Commemorative Foundation; and a grant from the Japan Society for the Promotion of Science (JSPS). N.W.-O. is a JSPS research fellow.
Authorship
Contributions: N.W.O. did all the experiments and participated in writing the manuscript; J.K. oversaw all the experiments and actively participated in manuscript writing; R.O. provided experimental guidance about and assisted in the BMT model; H.H. provided the general information and made the constructs of AML1 mutants; Y.H. made the constructs of AML1 mutants; Y.K. assisted in the experiments of BMT model; H.N. provided experimental guidance about cell sorting and staining; T.N. provided experimental guidance of the BMT model; T.I. provided the general information and constructs of AML1 mutants; and T.K. conceived and directed the project, secured funding, and actively participated in manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Toshio Kitamura, Division of Cellular Therapy, Advanced Clinical Research Center, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: kitamura@ims.u-tokyo.ac.jp.