Abstract
The CCAAT enhancer binding protein α (C/EBPα) is an important myeloid tumor suppressor that is frequently mutated in human acute myeloid leukemia (AML). We have previously shown that mice homozygous for the E2F repression–deficient CebpaBRM2 allele develop nonfatal AML with long latency and incomplete penetrance, suggesting that accumulation of secondary mutations is necessary for disease progression. Here, we use SRS19-6–driven retroviral insertional mutagenesis to compare the phenotypes of leukemias arising in Cebpa+/+, Cebpa+/BRM2, and CebpaBRM2/BRM2 mice, with respect to disease type, latency of tumor development, and identity of the retroviral insertion sites (RISs). Both Cebpa+/BRM2 and CebpaBRM2/BRM2 mice preferentially develop myeloid leukemias, but with differing latencies, thereby demonstrating the importance of gene dosage. Determination of RISs led to the identification of several novel candidate oncogenes, some of which may collaborate specifically with the E2F repression–deficient allele of Cebpa. Finally, we used an in silico pathway analysis approach to extract additional information from single RISs, leading to the identification of signaling pathways which were preferentially deregulated in a disease- and/or genotype-specific manner.
Introduction
The CCAAT enhancer binding protein α (C/EBPα) is the founding member of the C/EBP family of transcription factors that also includes C/EBPβ, C/EBPδ, C/EBPϵ, C/EBPγ, and C/EBPζ.1 C/EBPα acts as a lineage instructive factor and mediates differentiation events in several tissues, including liver, lung, fat, and within the hematopoietic system, by a combination of its ability to induce expression of lineage-specific genes and by its potential to promote cell-cycle exit.2-4 Within the hematopoietic system, C/EBPα has been demonstrated to play important roles at differentiation steps along the myeloid lineage. Cebpa-null fetal livers (Cebpa-null mice suffer from perinatal lethality) lack granulocytic cells and are arrested at the transition between the common myeloid progenitor (CMP) and the granulocyte macrophage progenitor (GMP).5-7 Similar observations in an Mx1-Cre–driven conditional knock-out of C/EBPα in adult mice suggest that C/EBPα plays similar roles in fetal and adult hematopoiesis.6 Finally, C/EBPα has been proposed to act as an inhibitor of erythroid differentiation.8
The importance of C/EBPα in regulating differentiation events within the hematopoietic system and its ability to interfere with cell-cycle progression are reflected by its involvement in human acute myeloid leukemia (AML). Mutations within the CEBPA gene are found in approximately 9% of patients with AML with normal karyotype. These mutations are clustered either in the N-terminal third or in the C-terminal part of the protein, where they lead to production of the truncated p30 form of C/EBPα or to C/EBPα variants deficient in DNA binding, respectively.9-18 Very little information is available regarding which genetic lesions collaborate with CEBPA mutants, although an association with 9q deletions has been reported.19 The critical genes have, however, not been identified. C/EBPα levels are also affected by various leukemic fusion proteins through mechanisms that involve transcriptional (RUNX1-ETO20 ) as well as translational (BCR-ABL,21 AML1-MDS1-EVI1,22 and CBFβ-MYH1123 ) repression. Finally, at the protein level, C/EBPα has been found to be inactivated functionally by FLT3-ITD–catalyzed phosphorylation at position S21 and by TRIB2-directed degradation.24,25 These findings suggest that down-regulation of C/EBPα activity and/or levels are at a convergence point in the development of a significant fraction of human AMLs (reviewed in Schuster and Porse,4 Nerlov,26,27 Mueller and Pabst,28 and Rosenbauer and Tenen29 ).
We have previously reported on the hematopoietic phenotypes of mice homozygous for the E2F repression–deficient CebpaBRM2 allele,30,31 which specifically abrogate the growth-inhibitory function of C/EBPα. Young CebpaBRM2/BRM2 mice initially suffer from neutropenia that over time progresses to a myeloproliferative syndrome or to a nonfatal AML-like syndrome with limited peripheral involvement. The stochastic nature of the phenotypic progression as well as the finding that the phenotypes are transplantable suggest that CebpaBRM2/BRM2 mice aquire additional mutations leading to the development of nonfatal AML. Whereas hematopoietic progenitor cells (HPCs) from CebpaBRM2/BRM2 mice displayed increased replating efficiencies in semisolid medium irrespective of their phenotypic progression, leukemic transformation was associated with a massive expansion of primitive Lin−, Sca-1+, cKit+ (LSK) cells. These findings suggest that abrogation of C/EBPα-mediated growth repression leads to increased self-renewal of HPCs that in turn sets the stage for malignant transformation, which is associated with changes in the HPC-containing LSK population.
Retroviral insertional mutagenesis (RIM) in the mouse has proven to be an efficient tool in the discovery of oncogenes that play a role in hematologic tumors.32 Most entries in the Retrovirus Tagged Cancer Gene database (RTCGD; http://rtcgd.ncifcrf.gov,33 the main international repository for these types of studies) are derived from studies using Moloney murine leukemia retroviruses (MoMuLVs). These viruses mainly induce T- and B-cell lymphomas, which to a large extent is explained by the cell-type specificity of the virus long terminal repeat (LTR). Other types of MuLV retroviruses give rise to different leukemias as well as novel retroviral integration sites (RISs) as examplified by studies using the myeloid leukemia–inducing Graffi MuLV.34,35 These findings highlight a certain degree of oncogene selectivity depending on the type of MuLV used.
In the present work, we have performed a RIM screen in Cebpa+/+, Cebpa+/BRM2, and CebpaBRM2/BRM2 backgrounds with the aim to identify novel oncogenes and candidate genes that specifically collaborate with mutant C/EBPα in tumorigenesis. As C/EBPα is considered a myeloid tumor suppressor, we wanted to use a retrovirus with a broad disease spectrum. We chose the SRS19-6 MoMuLV retrovirus, which has not previously been used in RIM screens and displays a broad disease spectrum including myeloid, erythroid, T-cell and B-cell leukemias. These properties make it an attractive candidate for screening experiments.36,37
Here, we demonstrate that mice carrying either 1 or 2 copies of the CebpaBRM2 allele preferentially develop myeloid leukemia as opposed to their wild-type (WT) littermates. In addition, CebpaBRM2/BRM2 mice develop disease with significant reduced latency and fewer number of retroviral integrations than Cebpa+/+ and Cebpa+/BRM2 mice, demonstrating that abrogation of C/EBPα-mediated E2F repression is a strong tumor-suppressor function of C/EBPα. Provocatively, CebpaBRM2/BRM2 mice do not only develop myeloid leukemias with reduced latencies but also lymphoid leukemias, raising the possibility that C/EBPα could be a lymphoid tumor suppressor as well. Finally, mapping of the RISs in diseased mice allowed us to identify several putative novel oncogenes, some of which may collaborate specifically with Cebpa mutations.
Methods
Mice and retroviruses
The CebpaBRM2 allele was back-crossed to the C57B6/J and 129S6/SvEvTac backgrounds for at least 10 generations.30,31 F1 hybrid mice were obtained by intercrossing, and newborn pups were injected with 100 μL culture supernatant (105-106 infectious units) from SRS19-6–producing NIH3T3 fibroblasts (provided by Dr Hung Fan, University of California, Irvine36 ). To adjust for any variation in virus titer, we sex- and litter-matched either a Cebpa+/+ or a Cebpa+/BRM2 animal with each CebpaBRM2/BMR2 mouse analyzed. Mice were monitored by blood smear and for general fitness. When moribound, they were killed and subjected to postmortem analysis. Kaplan-Meier curves were generated using the GraphPad Prism 4 software (GraphPad Software, San Diego, CA).
Identification of RISs
The SRS19-6 RISs were identified using a splinkerette-aided 2-step polymerase chain reaction (PCR) strategy.38 Briefly, genomic tumor DNA was digested with either Sau3A, Tsp509I, or FatI. Splinkerettes was formed by annealing the corresponding “splinklong” (5′-CGAAGAGTAACCGTTGCTAGGAGAGACCGTGGCTGAATGAGACTGGTGTCGACACTAGTGG) and “splinkshort” (5′-GATCCCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA [Sau3A], 5′-AATTCCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA [TSP509I], or CATGCCACTAGTGTCGACACCAGTCTCTAATTTTTTTTTTCAAAAAAA [FatI]) oligos and ligated to enzyme-restricted genomic tumor DNA. To prevent amplification of internal retroviral fragments, the ligation reactions were subsequently digested with EarI, followed by concentration on a Microcon YM-30 column (Millipore, Copenhagen, Denmark). The resulting DNA was PCR-amplified using a hot-start protocol (94°C, 3 minutes/68°C, 30 seconds; 94°C, 20 seconds/66°C, 30 seconds/72°C, 4 seconds; 94°C, 20 seconds/66°C, 30 seconds/72°C, 6 seconds; [94°C, 15 seconds/64°C, 30 seconds/72°C, 8 seconds plus 2 seconds/cycle] 4 times; [94°C, 15 seconds/62°C, 30 seconds/72°C, 14 seconds plus 2 seconds/cycle] 10 times; [94°C, 15 seconds/62°C, 30 seconds/72°C, 45 seconds] 13 times) by Taq DNA polymerase (Invitrogen, Copenhagen, Denmark) in a reaction containing splinkerette primer-1 (5′-CGAAGAGTAACCGTTGCTAGGAGAGACC) and 5′ 32P-labeled SRS19-6-LTR-1 (5′-CC-AGGCCTTGCAAGATGGCGTTACTGTAGC). Following concentration on Microcon YM-30 columns, samples were denatured and loaded on a denaturing 4.25% polyacrylamide gel that was subjected to electrophoresis. PCR products were visualized by autoradiography, and SRS19-6 specific bands were excised and eluted. After a subsequent PCR reaction using nested primers (splinkerette primer 2, 5′-GT-GGCTGAATGAGACTGGTGTCGAC; SRS19-6-LTR-2, 5′-GATGGCGTTACTGTAGCTAGCTTGCTGAGC), the amplified bands were TOPO cloned, and the resulting plasmids were subjected to sequencing.
Computational analysis
Retroviral insertional sequences were polished for vector contribution and aligned to the Ensembl database39 (National Center for Biotechnology Information [NCBI] m36 mouse assembly). Unambigous aligning sequences were used for further analysis. Identified candidate genes were used to query the RTCGD to determine whether they have been identified in previous screens.
We performed a pathway analysis using the whole dataset, a disease-restricted dataset, and a genotype-restricted dataset. For this purpose, we used the Ingenuity software (http://www.ingenuity.com) and the National Institutes of Health (NIH) DAVID software (http://david.abcc.ncifcrf.gov/40 ). Both programs use the Fisher exact test for significance, and a description of the statistical methods can be found on their respective home pages.
Detection of chromosomal copy number aberrations by array CGH
Genomic DNA from tumor samples and matched tail reference samples was isolated using standard protocols, including RNAase treatment and desalting. Oligo comparative genome hybridization (CGH) arrays consisted of 38 467 70-mer oligonucleotides (Oligator “MEEBO” mouse genome set; Illumina, San Diego, CA) spotted onto CodeLink slides (GE Healthcare, Little Chalfont, United Kingdom) were treated according to the manufacturer's protocol. Labeling, hybridization, and scanning was performed as previously described.41 The position of all oligonucleotides were mapped to NCBI m36 Mus musculus assembly39 prior to normalization and smoothing as previously described.42 Raw and smoothed data files of all array CGH (aCGH) experiments are available via Gene Expression Omnibus (GEO) series accession number GSE8032.43
Results
SRS19-6–injected CebpaBRM2/BRM2 mice are predisposed to malignant development
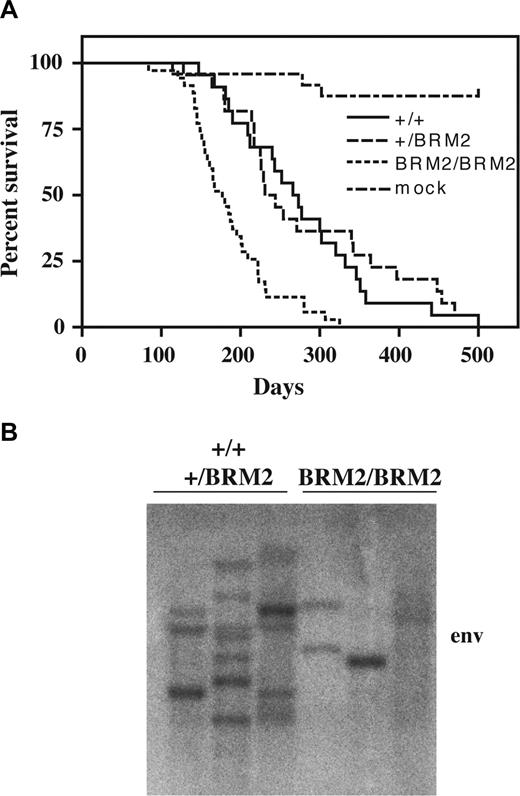
Homozygousity of the CebpaBRM2 allele results in perinatal lethality on an inbred C57B6/J background. Thus, to have our mutant allele on a defined genetic background, we intercrossed Cebpa+/BRM2 breeders backcrossed on C57B6/J and 129S6/SvEvTac backgrounds. Newborn F1 hybrid pups were injected with the SRS19-6 retrovirus (or mock), and the experimental groups were adjusted after genotyping. The SRS19-6–injected mice became moribound within the first year and displayed various symptoms of illness, including ruffled fur, hunched backs, and general inactivity. The survival curves for SRS19-6–injected Cebpa+/+ (n = 22) and Cebpa+/BRM2 (n = 21) mice were highly similar with mean latencies of 276 days and 280 days, respectively (Figure 1A). In contrast, CebpaBRM2/BRM2 (n = 35) mice developed disease much faster (P < .001), with a mean latency of 186 days. We next tested the clonality of the developing tumors by probing splenic DNA with a probe directed against the viral env gene and found that they were mainly clonal or oligoclonal (Figure 1B). Moreover, those tumors developing in CebpaBRM2/BRM2 mice contain fewer viral integrations than those developing in Cebpa+/+ and Cebpa+/BRM2 animals. Collectively, these findings demonstrate that CebpaBRM2/BRM2 mice are predisposed for malignant development and confirm that Cebpa is an important tumor suppressor gene.
Latency and clonality analysis of SRS19-6–injected mice. (A) Kaplan-Meier survival curves of SRS19-6–injected mice of the following genotypes: Cebpa+/+ (+/+; n = 22), Cebpa+/BRM2 (+/BRM2; n = 21), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 35). The CebpaBRM2/BRM2 animals have significantly shorter mean latency (P < .001; 2-tailed log-rank test) than the other genotypes. (B) Representative Southern blotting of splenic tumor DNA from SRS19-6–injected mice restricted with HindIII and probed with a probe against the SRS19-6 env open reading frame. CebpaBRM2/BRM2 animals generally have fewer integrations.
Latency and clonality analysis of SRS19-6–injected mice. (A) Kaplan-Meier survival curves of SRS19-6–injected mice of the following genotypes: Cebpa+/+ (+/+; n = 22), Cebpa+/BRM2 (+/BRM2; n = 21), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 35). The CebpaBRM2/BRM2 animals have significantly shorter mean latency (P < .001; 2-tailed log-rank test) than the other genotypes. (B) Representative Southern blotting of splenic tumor DNA from SRS19-6–injected mice restricted with HindIII and probed with a probe against the SRS19-6 env open reading frame. CebpaBRM2/BRM2 animals generally have fewer integrations.
Phenotypic characterization of SRS19-16–injected mice
Most leukemias in the SRS19-6–injected mice could be classified as either AML or T-cell acute lymphoblastic ALL (T-ALL) based on a combination of diagnostic tools. Gross necropsy of killed moribund SRS19-6–injected mice revealed splenomegaly in all mice. Enlarged thymus was observed in all mice with T-ALL and approximately one-third of mice diagnosed with AML. Enlarged lymph nodes were observed in around 60% of diseased animals, regardless of disease type or genotype. In approximately 20% of the animals, the leukemia had visibly metastasized to other organs, including liver, lung, and kidney, again regardless of the type of leukemia or genotype (data not shown).
Inspection of bone marrow (BM) and peripheral blood cells revealed massive amounts of leukemic cells in both T-ALL and AML (Figure 2A). Of the mice diagnosed with AML, 70% had more than 25% myeloblasts, and 30% had between 15% and 25% myeloblasts in the BM. Leukocyte counts of diseased mice were in the range of 10 to 170 × 109/L (10 000-170 000 cells/μL; healthy mice 2.4 × 109/L [2400 cells/μL]) and were more elevated in mice with AML (mean = 85 × 109/L cells [85 000 cells/μL]) than in mice with T-ALL (mean = 45 × 109/L cells [45 000 cells/μL]). Myeloblasts (staining positive for myeloperoxidase) were detected in the spleen and liver in some AML mice, resulting in disruption of the architecture of these organs (Figure 2B-D and data not shown).
Phenotypic characterization of SRS19-6–injected mice. (A) Morphology of bone marrow cells and peripheral blood cells derived from a nonleukemic control mouse, a mouse with AML, and a mouse with T-ALL. Cells were stained with May-Grünwald-Giemsa. (B) Splenic sections were stained for myeloperoxidase (MPO) demonstrating the accumulation of immature myeloid cells in AML mice. (C) Disruption of the splenic architechture in the AML mouse. Sections were stained with hematoxylin-eosin (HE). (D) Massive infiltration of leukemic cells in the liver of an AML mouse. Sections were stained as in panel C. Arrows mark the boundary between infiltrating myeloid cells (left) and the normal liver tissue (right). Microscopy was performed using an Olympus BX40 microscope (Olympus, Cophenhagen, Denmark) mounted with an Olympus DP10 digital camera using the following lenses: 10× Plan0.25, 40× UplanFL0.75, or 100× Plan1.25 oil. Images were processed in Adobe Photoshop CS3 v.10.0.1 and Adobe Illustrator CS3 v13.0.2 (Adobe Systems, San Diego, CA).
Phenotypic characterization of SRS19-6–injected mice. (A) Morphology of bone marrow cells and peripheral blood cells derived from a nonleukemic control mouse, a mouse with AML, and a mouse with T-ALL. Cells were stained with May-Grünwald-Giemsa. (B) Splenic sections were stained for myeloperoxidase (MPO) demonstrating the accumulation of immature myeloid cells in AML mice. (C) Disruption of the splenic architechture in the AML mouse. Sections were stained with hematoxylin-eosin (HE). (D) Massive infiltration of leukemic cells in the liver of an AML mouse. Sections were stained as in panel C. Arrows mark the boundary between infiltrating myeloid cells (left) and the normal liver tissue (right). Microscopy was performed using an Olympus BX40 microscope (Olympus, Cophenhagen, Denmark) mounted with an Olympus DP10 digital camera using the following lenses: 10× Plan0.25, 40× UplanFL0.75, or 100× Plan1.25 oil. Images were processed in Adobe Photoshop CS3 v.10.0.1 and Adobe Illustrator CS3 v13.0.2 (Adobe Systems, San Diego, CA).
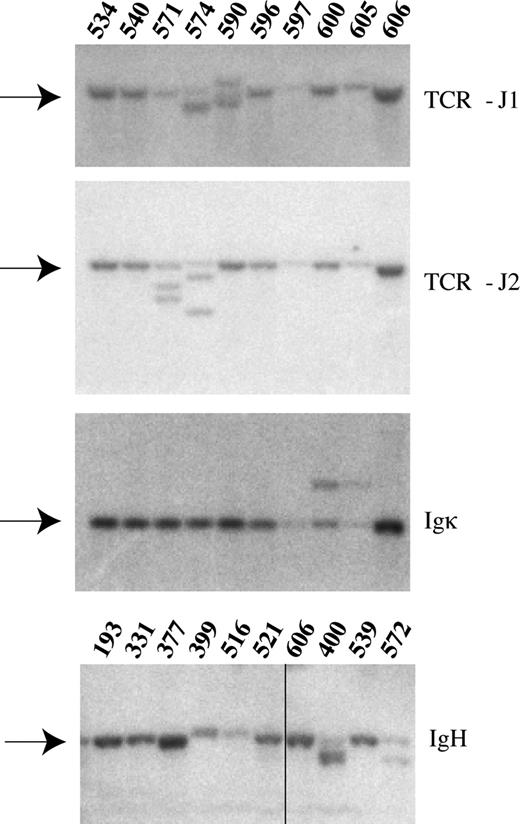
Flow cytometric analysis of bone marrow, spleen, thymus, peripheral blood, and lymph nodes using antibodies specific for various hematopoietic cellular subsets showed an enrichment of early myeloid cells (c-Kit+, Mac1+, Gr1−) and of T cells (CD4, CD8) arrested at various steps of their normal development in AML and T-ALL mice, respectively (Figure 3A-D and data not shown). As a final diagnostic tool, we used Southern blotting analysis of tumor DNA derived from moribund animals to check for the rearrangement status of the lymphoid genes encoding TCRβ, IgH, and Igκ (Figure 4). Based on this phenotypic characterization, SRS19-6–injected animals could generally be classified as suffering from either T-ALL or AML, with few occurrences of mixed leukemias and of leukemias with B-cell involvement.
Phenotypic characterization of SRS19-6–injected mice. (A) Representative flow cytometric analysis of cell derived from a nonleukemic control mouse, a mouse with AML, and a mouse with T-ALL. Cells from bone marrow (BM), spleen (Sp), and thymus (Thy) were stained with the following cocktails. (A) Progenitors: Mac1-FITC, cKit-APC. (B) Mac1-FITC, Gr1-APC. (C) CD4-FITC, CD8a-PerCP. (D) Mac1-FITC, B220-PE. Numbers indicate percentages of cells in a given gate.
Phenotypic characterization of SRS19-6–injected mice. (A) Representative flow cytometric analysis of cell derived from a nonleukemic control mouse, a mouse with AML, and a mouse with T-ALL. Cells from bone marrow (BM), spleen (Sp), and thymus (Thy) were stained with the following cocktails. (A) Progenitors: Mac1-FITC, cKit-APC. (B) Mac1-FITC, Gr1-APC. (C) CD4-FITC, CD8a-PerCP. (D) Mac1-FITC, B220-PE. Numbers indicate percentages of cells in a given gate.
Rearrangements of the TCR-β and immunoglobulin genes. Representative Southern blot analysis of 10 enzyme-restricted splenic tumor DNA samples. Radioactive probes specific for TCR-J1, TCR-J2, IgH and Igκ were used to demonstrate variable degrees of rearrangements in these genes. The germ-line bands are indicated by arrows. Mouse ID numbers are indicated above the blots. Details on the individual animals can be found in Table S1. Vertical line has been inserted to indicate a repositioned gel lane.
Rearrangements of the TCR-β and immunoglobulin genes. Representative Southern blot analysis of 10 enzyme-restricted splenic tumor DNA samples. Radioactive probes specific for TCR-J1, TCR-J2, IgH and Igκ were used to demonstrate variable degrees of rearrangements in these genes. The germ-line bands are indicated by arrows. Mouse ID numbers are indicated above the blots. Details on the individual animals can be found in Table S1. Vertical line has been inserted to indicate a repositioned gel lane.
The CebpaBRM2 allele skews disease development toward myeloid leukemias
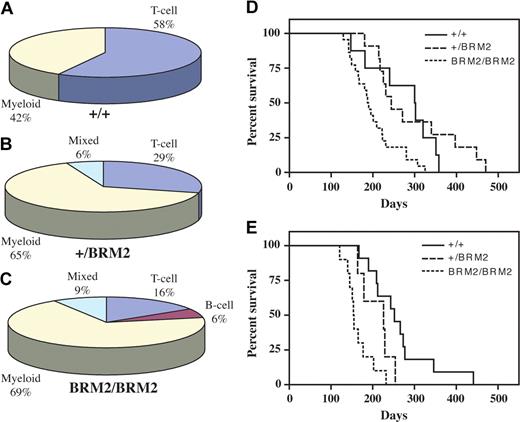
The disease distribution was dependent upon the genotype of the SRS19-6–injected mice. Development of T-cell ALL occurred in 58% of the Cebpa+/+ animals and was reduced to 29% and 16% in mice of the Cebpa+/BRM2 and CebpaBRM2/BRM2 genotypes, respectively (Figure 5A-C). Instead, 65% to 70% of these animals developed myeloid leukemias. These findings demonstrate that a single copy of the CebpaBRM2 allele is enough to skew disease development toward myeloid leukemias. To test whether development of myeloid leukemias was associated with mutations in the remaining WT Cebpa allele, we analyzed 10 tumor samples from heterozygous Cebpa+/BRM2 mice. Sequencing of 5 individual clones per tumor only revealed one recurrent mutation (L214M) in a single sample, suggesting that the preferential development of myeloid leukemias in Cebpa+/BRM2 mice was not associated with mutational inactivation of the remaining WT Cebpa allele.
Disease distribution and latency curves for SRS19-6–induced myeloid and lymphoid leukemias. Disease distributions of SRS19-6–injected (A) Cebpa+/+ (+/+; n = 19), (B) Cebpa+/BRM2 (+/BRM2; n = 17), and (C) CebpaBRM2/BRM2 (BRM2/BRM2; n = 32) mice. Mice were either diagnosed with T-cell ALL, B-cell ALL, myeloid leukemia, or mixed (T-ALL/myeloid leukemia, B-ALL/myeloid leukemia, or T-ALL/B-ALL). (D) Kaplan-Meier survival curves for myeloid leukemias of SRS19-6–injected mice of the following genotypes: Cebpa+/+ (+/+; n = 8), Cebpa+/BRM2 (+/BRM2; n = 11), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 22). (E) Kaplan-Meier survival curves for lymphoid leukemias (T-cell, B-cell) of SRS19-6–injected Cebpa+/+ (+/+; n = 11), Cebpa+/BRM2 (+/BRM2; n = 5), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 10). Statistical significance was determined as in Figure 1.
Disease distribution and latency curves for SRS19-6–induced myeloid and lymphoid leukemias. Disease distributions of SRS19-6–injected (A) Cebpa+/+ (+/+; n = 19), (B) Cebpa+/BRM2 (+/BRM2; n = 17), and (C) CebpaBRM2/BRM2 (BRM2/BRM2; n = 32) mice. Mice were either diagnosed with T-cell ALL, B-cell ALL, myeloid leukemia, or mixed (T-ALL/myeloid leukemia, B-ALL/myeloid leukemia, or T-ALL/B-ALL). (D) Kaplan-Meier survival curves for myeloid leukemias of SRS19-6–injected mice of the following genotypes: Cebpa+/+ (+/+; n = 8), Cebpa+/BRM2 (+/BRM2; n = 11), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 22). (E) Kaplan-Meier survival curves for lymphoid leukemias (T-cell, B-cell) of SRS19-6–injected Cebpa+/+ (+/+; n = 11), Cebpa+/BRM2 (+/BRM2; n = 5), and CebpaBRM2/BRM2 (BRM2/BRM2; n = 10). Statistical significance was determined as in Figure 1.
We next tested whether the latencies for the largest disease subgroups in SRS19-6–injected mice were affected by their genotype (Figure 5D,E). As expected, we found that CebpaBRM2/BRM2 mice (mean latency, 186 days) developed AML significantly faster (P < .001) than Cebpa+/+ (mean latency, 275 days) and Cebpa+/BRM2 (mean latency, 294 days) mice. Surprisingly, SRS19-6–injected CebpaBRM2/BRM2 mice also developed lymphoid leukemias (B-ALL and T-ALL) significantly faster than Cebpa+/+ animals (mean latencies of 164 days vs 261; P < .001), with Cebpa+/BRM2 mice developing disease with intermediate latency (210 days). These findings suggest a novel potential role for C/EBPα as a lymphoid tumor suppressor in addition to its well-characterized function in myeloid leukemias.
C/EBPα mRNA is detectable in the double negative (DN1-4) thymic T-cell precursor populations, and it is conceivable that the presence of the CebpaBRM2 allele may interfere with their differentiation.44 We therefore determined the relative frequencies of the DN populations in Cebpa+/+ and CebpaBRM/BRM2 mice, but found no significant differences (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
SRS19-6–induced leukemias are not generally associated with genomic instability
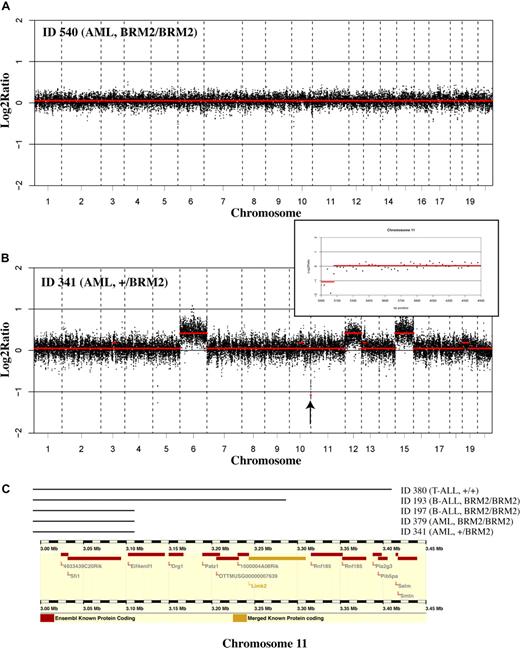
Genomic instability is often associated with cancer progression. To test for gene copy number changes during leukemic development in SRS19-6–induced leukemias, we subjected 10 samples of splenic tumor DNA to aCGH analyses using a 40K mouse Chip. The analysis of 2 Cebpa+/+ (T-ALL), 2 Cebpa+/BRM2 (AML), and 6 CebpaBRM2/BRM2 (1 T-ALL, 2 B-ALL, 3 AML) tumor samples demonstrated that the tumor genomes were essentially unperturbed (Figure 6A), and only in a single tumor did we observe large regions of chromosomal gains (Figure 6B). These findings suggest that SRS19-6 tumors are not accompanied by high degree of genomic instability. Hence, we did not observe any correlation between disease class (lymphoid or myeloid leukemias) or genotype with the level of chromosomal instability (data not shown). Interestingly, among the smaller gains and deletions that we did observe, we identified a common deletion in the acromeric region of chromosome 11 (human chromosome 22q12.2) in 5 of 10 samples (Figure 6B insert). Again, the presence of this deletion did not correlate with either genotype or disease phenotype. In 3 of 5 samples, the deletion only encompassed Sfi1 and part of the Eif4enif1 locus, strongly suggesting a role for either of these proteins in disease progression (Figure 6C). Eif4enif1 encodes a transporter of components of the translational initiation machinery, whereas Sfi1 is involved in spindle assembly.45,46
aCGH analyses of SRS19-6–induced leukemias. (A) Splenic tumor DNA from mouse 540 (AML; CebpaBRM2/BRM2) was subjected to aCGH analysis as described in “Detection of chromosomal copy number aberrations by array CGH.” The resulting profiles are displayed using a moving average of 3 (black dots) with the smoothed values superimposed (red lines). The resulting profile demonstrates an essential normal karyotype. (B) aCGH analysis of splenic tumor DNA from mouse 341 (AML; Cebpa+/BRM2) demonstrates gain of chromosomes 6, 12, and 15. The arrow indicates a small region at the tip of chromosome 11 that is deleted in 5 of 10 tumors of various genotypes and phenotypes (C). The insert highlights this small region. Here, raw normalized values are displayed without moving average, with smoothing. (C) ENSEMBL screenshot showing the genes located at the tip of chromosome 11 (due to the repetitive nature of the acromeric region, there are no probeset on the arrays upstream from 3002 kb). The lines above indicate the extent of the deletion and the identity of the mice that have a deletion in this area. The minimal deletions pinpoint Sfi1 and Eif4enif1 as candidate tumor-suppressor genes.
aCGH analyses of SRS19-6–induced leukemias. (A) Splenic tumor DNA from mouse 540 (AML; CebpaBRM2/BRM2) was subjected to aCGH analysis as described in “Detection of chromosomal copy number aberrations by array CGH.” The resulting profiles are displayed using a moving average of 3 (black dots) with the smoothed values superimposed (red lines). The resulting profile demonstrates an essential normal karyotype. (B) aCGH analysis of splenic tumor DNA from mouse 341 (AML; Cebpa+/BRM2) demonstrates gain of chromosomes 6, 12, and 15. The arrow indicates a small region at the tip of chromosome 11 that is deleted in 5 of 10 tumors of various genotypes and phenotypes (C). The insert highlights this small region. Here, raw normalized values are displayed without moving average, with smoothing. (C) ENSEMBL screenshot showing the genes located at the tip of chromosome 11 (due to the repetitive nature of the acromeric region, there are no probeset on the arrays upstream from 3002 kb). The lines above indicate the extent of the deletion and the identity of the mice that have a deletion in this area. The minimal deletions pinpoint Sfi1 and Eif4enif1 as candidate tumor-suppressor genes.
Identification of SRS19-6 retroviral insertion sites
Using a splinkerette-aided PCR strategy, we sequenced a total of 182 SRS19-6 RISs in 67 tumors from 3 genotypes38,47,48 (Table S1). The average number of identified RISs was unevenly distributed among the genotypes, ranging from 3.8 and 4.3 in Cebpa+/+ and Cebpa+/BRM2 mice, respectively, to only 2.1 in CebpaBRM2/BRM2 animals (data not shown). Again, this finding suggests that the latter mice are predisposed for malignant development. Insertions in 22 genes occurred more than once and accounted for 28% (51 of 182) of the identified insertion sites (Table 1). Of these, 17 have previously been identified as common insertion sites (CISs), including well-known oncogenes such as Myb, Myc, and Notch1. A total of 5 novel CISs (Chd4, Sesn1, Rag2, Arhgef6, and Vav3) were identified from data obtained uniquely in our study, underlining the importance of using different retroviruses for retroviral insertional mutagenesis screens. Finally, our study also converted 28 single RISs (defined as RIS 1 in Table S1) within the RTCGD to CISs by combining them with our hits. These genes, which are all potential oncogenes, include, among others Raf1, Bcl11b, and Vcl.
Multiply-tagged loci
| Genes . | No. hits . | Function . |
|---|---|---|
| Rasgrp1 | 4 | Signal transduction |
| Lmo2 | 3 | Differentiation |
| Notch1 | 3 | Differentiation |
| Hhex | 3 | Proliferation |
| Myb | 3 | Proliferation |
| Mef2c | 3 | Transcription factor |
| Gpc5 | 2 | Cell membrane protein |
| Abhd2 | 2 | Cell migration |
| Chd4* | 2 | Chromatin remodeling |
| Rag2* | 2 | DNA recombination |
| Ccnd2 | 2 | Proliferation |
| Evi1 | 2 | Proliferation |
| Itk | 2 | Proliferation |
| Myc | 2 | Proliferation |
| Sesn1* | 2 | Proliferation |
| ETS1/Fli1 | 2 | Proliferation, differentation |
| Arhgef6* | 2 | Signal transduction |
| Vav3* | 2 | Signal transduction |
| Sox4 | 2 | Transcription factor |
| Ssbp3 | 2 | Transcription factor |
| Zfpn1a1 | 2 | Transcription factor |
| AB041803 | 2 | Unknown |
| Genes . | No. hits . | Function . |
|---|---|---|
| Rasgrp1 | 4 | Signal transduction |
| Lmo2 | 3 | Differentiation |
| Notch1 | 3 | Differentiation |
| Hhex | 3 | Proliferation |
| Myb | 3 | Proliferation |
| Mef2c | 3 | Transcription factor |
| Gpc5 | 2 | Cell membrane protein |
| Abhd2 | 2 | Cell migration |
| Chd4* | 2 | Chromatin remodeling |
| Rag2* | 2 | DNA recombination |
| Ccnd2 | 2 | Proliferation |
| Evi1 | 2 | Proliferation |
| Itk | 2 | Proliferation |
| Myc | 2 | Proliferation |
| Sesn1* | 2 | Proliferation |
| ETS1/Fli1 | 2 | Proliferation, differentation |
| Arhgef6* | 2 | Signal transduction |
| Vav3* | 2 | Signal transduction |
| Sox4 | 2 | Transcription factor |
| Ssbp3 | 2 | Transcription factor |
| Zfpn1a1 | 2 | Transcription factor |
| AB041803 | 2 | Unknown |
Genes that were uniquely identified as CISs in this study.
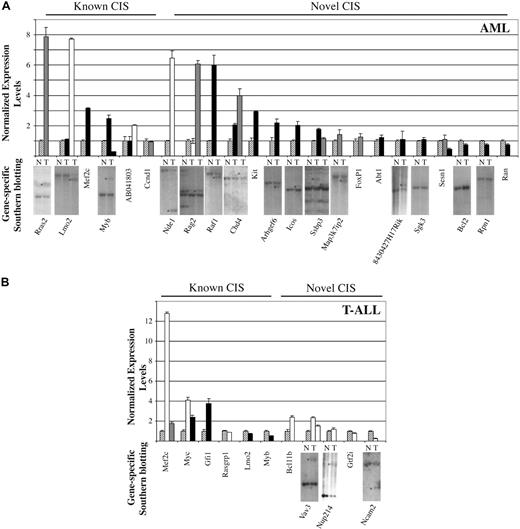
To gain more support for the potential oncogenic involvement of our RISs, we determined the expression levels of 31 genes derived from 25 tumors using quantitative reverse transcription (RT)–PCR (Figure 7A,B). As control, we used a pool of RNA derived from tumors of the same type (AML vs T-ALL), but without an integration in the gene of interest. For approximately half of the tested genes, including 4 of 5 of our novel CISs (Chd4, Rag2, Arhgef6, and Vav3), retroviral integration led to a greater than 2-fold up-regulation in at least one tagged sample, compared with the pool. The expression levels of additional novel CISs, including NdeI, Bcl11b, Raf1, Kit, and Icos, were also found to be up-regulated in samples containing retroviral integrations. The latter 3 genes are of particular interest, as they may collaborate specifically with the CebpaBRM2 allele.
Expression analysis of SRS19-6–tagged genes. Splenic tumor cDNA derived from (A) AML mice and (B) T-ALL mice was subjected to quantitative RT-PCR. Expressions levels were normalized to the level of β-actin mRNA. The expression level of each gene was further normalized to a sample consisting of pooled cDNA from 10 different tumors without retroviral integrations in the gene in question ( ; all genotypes). This sample was arbitrarily set to 1. All measurements were performed in triplicate, and standard deviations are depicted. The different genotypes are indicated by □ (Cebpa+/+),
; all genotypes). This sample was arbitrarily set to 1. All measurements were performed in triplicate, and standard deviations are depicted. The different genotypes are indicated by □ (Cebpa+/+),  (Cebpa+/BRM2), or ■ (CebpaBRM2/BRM2). To determine the contribution of the tumor clone, containing an integration in a given gene, to the total tumor mass, we performed gene-specific southern blots. Splenic genomic DNA was isolated from WT (N) and diseased (T) animals and subjected to Southern blot analysis using gene-specific probes. *Bands representing RISs. See Figure S2 for a blow-up of the Southern blots.
(Cebpa+/BRM2), or ■ (CebpaBRM2/BRM2). To determine the contribution of the tumor clone, containing an integration in a given gene, to the total tumor mass, we performed gene-specific southern blots. Splenic genomic DNA was isolated from WT (N) and diseased (T) animals and subjected to Southern blot analysis using gene-specific probes. *Bands representing RISs. See Figure S2 for a blow-up of the Southern blots.
Expression analysis of SRS19-6–tagged genes. Splenic tumor cDNA derived from (A) AML mice and (B) T-ALL mice was subjected to quantitative RT-PCR. Expressions levels were normalized to the level of β-actin mRNA. The expression level of each gene was further normalized to a sample consisting of pooled cDNA from 10 different tumors without retroviral integrations in the gene in question ( ; all genotypes). This sample was arbitrarily set to 1. All measurements were performed in triplicate, and standard deviations are depicted. The different genotypes are indicated by □ (Cebpa+/+),
; all genotypes). This sample was arbitrarily set to 1. All measurements were performed in triplicate, and standard deviations are depicted. The different genotypes are indicated by □ (Cebpa+/+),  (Cebpa+/BRM2), or ■ (CebpaBRM2/BRM2). To determine the contribution of the tumor clone, containing an integration in a given gene, to the total tumor mass, we performed gene-specific southern blots. Splenic genomic DNA was isolated from WT (N) and diseased (T) animals and subjected to Southern blot analysis using gene-specific probes. *Bands representing RISs. See Figure S2 for a blow-up of the Southern blots.
(Cebpa+/BRM2), or ■ (CebpaBRM2/BRM2). To determine the contribution of the tumor clone, containing an integration in a given gene, to the total tumor mass, we performed gene-specific southern blots. Splenic genomic DNA was isolated from WT (N) and diseased (T) animals and subjected to Southern blot analysis using gene-specific probes. *Bands representing RISs. See Figure S2 for a blow-up of the Southern blots.
For approximately 50% of the tested genes, retroviral integration did not lead to a significant up-regulation of their expression, suggesting that they may not contribute to tumor formation. Alternatively, the integrations in these genes may only lead to up-regulation during initiation of the tumor, or the cells having integrations in these genes may only constitute a minor fraction of the tumor. The latter possibility is supported by the finding that integrations into well-known CISs like Lmo2, Myb, and Myc also fail to enhance their expression in some tumors (Figure 7A,B). Furthermore, when we probe the genomic DNA from tumor tissue using gene-specific probes, most of the integrations that fail to increase transcription also fall below our detection levels in our Southern blotting analysis (Figure 7A,B).
Computational pathway analysis of SRS19-6 RISs
A recent paper describing a murine mammary tumor virus (MMTV)–based RIM screen for breast cancer–associated genes has demonstrated the power of moving from the analysis of single genes (ie, CISs) to the level of pathways.49 The concept of this approach is that since deregulation of individual genes in the same pathway is predicted to have a similar outcome, single-insertion RISs will also contain valuable information.
We first used the Ingenuity Canonical Pathway Analysis software and the NIH-DAVID software for the Kyoto Encyclopedia of Genes and Genomes50 (KEGG) pathways to search for commonly deregulated pathways in our whole RIS data set (Table 2). This analysis demonstrated a preponderance of signaling pathways, including VEGF, ERK/MAPK, B cell, T cell, GM-CSF, and PDGF signaling as the main pathways overrepresented in our dataset and thereby in SRS19-6–driven leukemogenesis. Moreover, the added value of using single-hit RISs to probe biological function using pathway analysis is validated by the multiple occurrences of genes that have not previously been identified as CISs (marked by asterisks in Table 2). The observed deregulation of multiple pathways was to a large extent driven by single RISs in Rras2 and Raf1, which further underscores the interconnectivity of signaling pathways.
Computational analysis of pathways affected by proviral insertions
| Pathways . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| VEGF signaling | Rras2, Vcl*, Bcl2*, Raf1*, Kdr, Plcg2* | < .001 |
| <G1/S checkpoint regulation | Myc2, Ccnd1, Ccnd22, Hdac6, E2f2 | < .001 |
| ERK/MAPK signaling | Rras2, Myc2, Raf1*, Prkar2b*, Mycn, Ets12, Plcg2* | < .001 |
| B-cell receptor signaling | Pou2f2, Rras2, Vav32*, Raf1*, Ets12, Plcg2* | < .001 |
| T-cell receptor signaling | Rras2, Vav32*, Raf1*, Rasgrp14, Itk2 | < .001 |
| GM-CSF signaling | Rras2, Ccnd1, Raf1*, Ets12 | < .001 |
| PDGF signaling | Rras2, Myc2, Raf1*, Plcg2* | .001 |
| Leukocyte extravasation signaling | Vav32*, Pecam1, Vcl*, Itk2, Mmp14, Plcg2* | .001 |
| Neuregulin signaling | Rras2, Myc2, Raf1*, Plcg2* | .002 |
| FcϵRI signaling | Rras2, Vav32*, Raf1*, Plcg2* | .004 |
| PTEN signaling | Rras2, Bcl2*, Ccnd1, Raf1* | .005 |
| Apoptosis signaling | Rras2, Bcl2*, Raf1*, Plcg2* | .006 |
| Natural killer cell signaling | Rras2, Vav32*, Raf1*, Plcg2* | .006 |
| JAK/Stat signaling | Socs6*, Rras2, Raf1* | .007 |
| G-protein–coupled receptor signaling | Pde1a*, Rras2, Raf1*, Prkar2b*, Rasgrp14 | .011 |
| PI3K/AKT signaling | Rras2, Bcl2*, Ccnd1, Raf1* | .013 |
| N-glycan biosynthesis | Rpn1*, Dpagt1*, Mgat4a* | .015 |
| Chemokine signaling | Rras2, Raf1*, Plcg2* | .018 |
| IGF-1 signaling | Rras2, Raf1*, Prkar2b* | .02 |
| Actin cytoskeleton signaling | Arhgef6*, Rras2, Vav32*, Vcl*, Raf1* | .021 |
| p38 MAPK signaling | Myc2, Map3k7ip2*, Mef2c3 | .025 |
| Estrogen receptor signaling | Rras2, Raf1*, Crsp2* | .035 |
| Synaptic long-term potentiation | Rras2, Raf1*, Prkar2b* | .04 |
| KEGG pathways | ||
| Dorsal-ventral axis formation | Rras2, Ets1, Notch13, Raf1* | .005 |
| Jak-STAT signaling | Ccnd1, Il12a*, Myc2, Mpl*, Ccnd22, Il21r*, Socs6* | .007 |
| Focal adhesion | Ccdn1, Vav32*, Rras2, Vcl*, Kdr, Ccnd22, Bcl2*, Raf1* | .011 |
| T-cell receptor signaling | Icos*, Vav32*, Rras2, Rasgrp14, Itk2 | .026 |
| Leukocyte transendothelial migration | Plcg2*, Vav32*, Vcl*, Pecam1, Itk2 | .049 |
| Pathways . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| VEGF signaling | Rras2, Vcl*, Bcl2*, Raf1*, Kdr, Plcg2* | < .001 |
| <G1/S checkpoint regulation | Myc2, Ccnd1, Ccnd22, Hdac6, E2f2 | < .001 |
| ERK/MAPK signaling | Rras2, Myc2, Raf1*, Prkar2b*, Mycn, Ets12, Plcg2* | < .001 |
| B-cell receptor signaling | Pou2f2, Rras2, Vav32*, Raf1*, Ets12, Plcg2* | < .001 |
| T-cell receptor signaling | Rras2, Vav32*, Raf1*, Rasgrp14, Itk2 | < .001 |
| GM-CSF signaling | Rras2, Ccnd1, Raf1*, Ets12 | < .001 |
| PDGF signaling | Rras2, Myc2, Raf1*, Plcg2* | .001 |
| Leukocyte extravasation signaling | Vav32*, Pecam1, Vcl*, Itk2, Mmp14, Plcg2* | .001 |
| Neuregulin signaling | Rras2, Myc2, Raf1*, Plcg2* | .002 |
| FcϵRI signaling | Rras2, Vav32*, Raf1*, Plcg2* | .004 |
| PTEN signaling | Rras2, Bcl2*, Ccnd1, Raf1* | .005 |
| Apoptosis signaling | Rras2, Bcl2*, Raf1*, Plcg2* | .006 |
| Natural killer cell signaling | Rras2, Vav32*, Raf1*, Plcg2* | .006 |
| JAK/Stat signaling | Socs6*, Rras2, Raf1* | .007 |
| G-protein–coupled receptor signaling | Pde1a*, Rras2, Raf1*, Prkar2b*, Rasgrp14 | .011 |
| PI3K/AKT signaling | Rras2, Bcl2*, Ccnd1, Raf1* | .013 |
| N-glycan biosynthesis | Rpn1*, Dpagt1*, Mgat4a* | .015 |
| Chemokine signaling | Rras2, Raf1*, Plcg2* | .018 |
| IGF-1 signaling | Rras2, Raf1*, Prkar2b* | .02 |
| Actin cytoskeleton signaling | Arhgef6*, Rras2, Vav32*, Vcl*, Raf1* | .021 |
| p38 MAPK signaling | Myc2, Map3k7ip2*, Mef2c3 | .025 |
| Estrogen receptor signaling | Rras2, Raf1*, Crsp2* | .035 |
| Synaptic long-term potentiation | Rras2, Raf1*, Prkar2b* | .04 |
| KEGG pathways | ||
| Dorsal-ventral axis formation | Rras2, Ets1, Notch13, Raf1* | .005 |
| Jak-STAT signaling | Ccnd1, Il12a*, Myc2, Mpl*, Ccnd22, Il21r*, Socs6* | .007 |
| Focal adhesion | Ccdn1, Vav32*, Rras2, Vcl*, Kdr, Ccnd22, Bcl2*, Raf1* | .011 |
| T-cell receptor signaling | Icos*, Vav32*, Rras2, Rasgrp14, Itk2 | .026 |
| Leukocyte transendothelial migration | Plcg2*, Vav32*, Vcl*, Pecam1, Itk2 | .049 |
The 168 RISs (out of 182) for which we have obtained accession numbers were subjected to pathway analysis using the Ingenuity and NIH-DAVID software packages. These software packages returned Ingenuity pathways and KEGG pathways, respectively. Only pathways with 3 proviral insertions in 3 or more genes are depicted. Pathways are sorted based on significance (cut-off, P < .05). A total of 32 (of 59) tumors are represented in one or more pathways. RISs affected in more than one tumor are depicted in bold, and the number of integrations is depicted in superscript. Please note that the NIH-DAVID software does not take into account multiple hits in individual genes.
Genes that are not defined as CISs in the RTCGD.
We next used pathway analysis to test whether myeloid and lymphoid SRS19-6–induced leukemias had a different propensity for deregulation of particular signaling pathways (Table 3). Indeed, leukocyte extravastion signaling (Ingenuity; similar to leukocyte transendothelial migration in KEGG), ERK/MAPK, and B-cell receptor signaling were selectively targeted in lymphoid tumors, whereas PI3K/AKT (overlapping with PTEN signaling) was selectively targeted in myeloid tumors. In addition, several pathways, including G1/S checkpoint regulation, T-cell receptor signaling, and VEGF signaling, were significantly targeted in both myeloid and lymphoid (data not shown).
Differences in affected pathways as a function of myeloid versus lymphoid leukemia
| Pathways, leukemia . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| Leukocyte extravasation signaling | ||
| Lymphoid | Vav32, Pecam1, Vcl, Itk, Plcg2 | < .001 |
| Myeloid | Itk | .5 |
| ERK/MAPK signaling | ||
| Lymphoid | Myc2, Prkar2b, Mycn, Ets12, Plcg2 | < .001 |
| Myeloid | Rras2, Raf1 | .16 |
| B-cell receptor signaling | ||
| Lymphoid | Vav32, Ets12, Plcg2 | .010 |
| Myeloid | Rras2, Raf1 | .1 |
| Actin cytoskeleton signaling | ||
| Lymphoid | Arhgef6, Vav32, Vcl | .032 |
| Myeloid | Arhgef6, Rras2, Raf1 | .052 |
| FcϵRI signaling | ||
| Lymphoid | Vav32, Plcg2 | .035 |
| Myeloid | Rras2, Raf1 | .05 |
| Natural killer cell signaling | ||
| Lymphoid | Vav32, Plcg2 | .043 |
| Myeloid | Rras2, Raf1 | .06 |
| G-protein–coupled receptor signaling | ||
| Lymphoid | Prkar2b, Rasgrp14 | .12 |
| Myeloid | Pde1a, Rras2, Raf1 | .035 |
| GM-CSF signaling | ||
| Lymphoid | Ets12 | .17 |
| Myeloid | Rras2, Ccnd1, Raf1 | .001 |
| Apoptosis signaling | ||
| Lymphoid | Plcg2 | .28 |
| Myeloid | Rras2, Bcl2, Raf1 | .007 |
| PI3K/AKT signaling | ||
| Lymphoid | ||
| Myeloid | Rras2, Bcl2, Ccnd1, Raf1 | .002 |
| PTEN signaling | ||
| Lymphoid | ||
| Myeloid | Rras2, Bcl2, Ccnd1, Raf1 | < .001 |
| KEGG pathways | ||
| Leukocyte transendothelial migration | ||
| Lymphoid | Plcg2, Vav32, Vcl, Pecam1, Itk | .003 |
| Myeloid | Itk | > .5 |
| Dorsal-ventral axis formation | ||
| Lymphoid | Ets12, Notch1 | .14 |
| Myeloid | Rras2, Raf1, Notch12 | .018 |
| Pathways, leukemia . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| Leukocyte extravasation signaling | ||
| Lymphoid | Vav32, Pecam1, Vcl, Itk, Plcg2 | < .001 |
| Myeloid | Itk | .5 |
| ERK/MAPK signaling | ||
| Lymphoid | Myc2, Prkar2b, Mycn, Ets12, Plcg2 | < .001 |
| Myeloid | Rras2, Raf1 | .16 |
| B-cell receptor signaling | ||
| Lymphoid | Vav32, Ets12, Plcg2 | .010 |
| Myeloid | Rras2, Raf1 | .1 |
| Actin cytoskeleton signaling | ||
| Lymphoid | Arhgef6, Vav32, Vcl | .032 |
| Myeloid | Arhgef6, Rras2, Raf1 | .052 |
| FcϵRI signaling | ||
| Lymphoid | Vav32, Plcg2 | .035 |
| Myeloid | Rras2, Raf1 | .05 |
| Natural killer cell signaling | ||
| Lymphoid | Vav32, Plcg2 | .043 |
| Myeloid | Rras2, Raf1 | .06 |
| G-protein–coupled receptor signaling | ||
| Lymphoid | Prkar2b, Rasgrp14 | .12 |
| Myeloid | Pde1a, Rras2, Raf1 | .035 |
| GM-CSF signaling | ||
| Lymphoid | Ets12 | .17 |
| Myeloid | Rras2, Ccnd1, Raf1 | .001 |
| Apoptosis signaling | ||
| Lymphoid | Plcg2 | .28 |
| Myeloid | Rras2, Bcl2, Raf1 | .007 |
| PI3K/AKT signaling | ||
| Lymphoid | ||
| Myeloid | Rras2, Bcl2, Ccnd1, Raf1 | .002 |
| PTEN signaling | ||
| Lymphoid | ||
| Myeloid | Rras2, Bcl2, Ccnd1, Raf1 | < .001 |
| KEGG pathways | ||
| Leukocyte transendothelial migration | ||
| Lymphoid | Plcg2, Vav32, Vcl, Pecam1, Itk | .003 |
| Myeloid | Itk | > .5 |
| Dorsal-ventral axis formation | ||
| Lymphoid | Ets12, Notch1 | .14 |
| Myeloid | Rras2, Raf1, Notch12 | .018 |
We divided our dataset (Table 2) into two datasets representing RISs identified in lymphoid (65) and myeloid (83) leukemias. A total of 23 RISs were excluded, as they were identified in animals with no clear-cut diagnosis. The 2 individual datasets were subjected to pathway analysis (Table 2). P values reaching significance (< .05) are indicated in bold. Pathways fulfilling the following two criterias are depicted: (1) must have 3 proviral insertions in 3 or more genes in a least one of the 2 groups; (2) must display selectivity (ie, only one experimental group can reach significance; P < .05). Pathways are sorted based on significance. RIS affected in more than one tumor are depicted in bold, and the number of integrations in a given disease subtype is depicted in superscript.
Finally, we used pathway analysis to test whether any of our 3 genotypes conferred selectivity for targeting of particular pathways. Here, we saw a strong correlation between RISs in genes defined as members of G1/S checkpoint regulation pathway in Cebpa+/+ mice but not in their Cebpa+/BRM2 and CebpaBRM2/BRM2 littermates (Table 4). A similar trend was observed for genes involved in leukocyte transendothelial migration/leukocyte extravasation signaling. Conversely, PI3K/AKT (PTEN) signaling was selectively targeted in Cebpa+/BRM2 and CebpaBRM2/BRM2 mice at borderline significance. However, as all the RISs in this pathway were found in myeloid leukemias, it suggests that deregulation of this pathway could be a common feature for myeloid leukemias arising in Cebpa+/BRM2 and CebpaBRM2/BRM2 mice.
Differences in affected pathways as a function of genotype
| Pathways, genotype . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| G1/S checkpoint regulation | ||
| +/+ | Myc, Ccnd22, Hdac6, E2f2 | < .001 |
| +/2 | Ccnd1 | .15 |
| 2/2 | Myc | .12 |
| Leukocyte extravasation signaling | ||
| +/+ | Vav32, Vcl, Itk, Mmp14, Plcg2 | < .001 |
| +/2 | — | — |
| 2/2 | Pecam1, Itk | .071 |
| B-cell receptor signaling | ||
| +/+ | Vav32, Ets1, Plcg2 | .007 |
| +/2 | Pou2f2, Rras2, Ets1 | .01 |
| 2/2 | Raf1 | .29 |
| ERK/MAPK signaling | ||
| +/+ | Myc, Ets1, Plcg2 | .015 |
| +/2 | Rras2, Prkar2b, Mycn, Ets1 | .003 |
| 2/2 | Myc, Raf1 | .072 |
| FcϵRI signaling | ||
| +/+ | Vav32, Plcg2 | .03 |
| +/2 | Rras2 | .26 |
| 2/2 | Raf1 | .2 |
| Natural killer cell signaling | ||
| +/+ | Vav32, Plcg2 | .035 |
| +/2 | Rras2 | .28 |
| 2/2 | Raf1 | .22 |
| GM-CSF signaling | ||
| +/+ | Ets1 | .15 |
| +/2 | Rras2, Ccnd1, Ets1 | < .001 |
| 2/2 | Raf1 | .13 |
| G-protein–coupled receptor signaling | ||
| +/+ | Rasgrp1 | .42 |
| +/2 | Pde1a, Rras2, Prkar2b, Rasgrp12 | .003 |
| 2/2 | Raf1, Rasgrp1 | .076 |
| Axonal guidance signaling | ||
| +/+ | — | — |
| +/2 | Arhgef6, Rras2, Prkar2b, Rtn4 | .025 |
| 2/2 | Arhgef6, Raf1 | .21 |
| PI3K/AKT signaling | ||
| +/+ | — | — |
| +/2 | Rras2, Ccnd1 | .062 |
| 2/2 | Bcl2, Raf1 | .037 |
| PTEN signaling | ||
| +/+ | — | — |
| +/2 | Rras2, Ccnd1 | .04 |
| 2/2 | Bcl2, Raf1 | .024 |
| KEGG pathways | ||
| Dorsal-ventral axis formation | ||
| +/+ | Ets1, Notch1 | .14 |
| +/2 | Rras2, Ets1, Notch12 | .011 |
| 2/2 | Raf1 | > .5 |
| Leukocyte transendothelial migration | ||
| +/+ | Plcg2, Vav3, Vcl, Itk | .019 |
| +/2 | — | — |
| 2/2 | Pecam1, Itk | .4 |
| JAK-STAT signaling | ||
| +/+ | Myc, Il21r, Ccnd2 | .16 |
| +/2 | Ccnd1 | > .5 |
| 2/2 | Il12a, Myc, Mpl, Socs6 | .023 |
| MAPK signaling | ||
| +/+ | Myc, Mef2c, Rasgrp1 | .36 |
| +/2 | Rras2, Map3k7ip2, Evi1, Mef2c, Rasgrp1 | .047 |
| 2/2 | Myc, Mef2c, Rasgrp1, Raf1 | .097 |
| Pathways, genotype . | Genes . | P . |
|---|---|---|
| Ingenuity pathways | ||
| G1/S checkpoint regulation | ||
| +/+ | Myc, Ccnd22, Hdac6, E2f2 | < .001 |
| +/2 | Ccnd1 | .15 |
| 2/2 | Myc | .12 |
| Leukocyte extravasation signaling | ||
| +/+ | Vav32, Vcl, Itk, Mmp14, Plcg2 | < .001 |
| +/2 | — | — |
| 2/2 | Pecam1, Itk | .071 |
| B-cell receptor signaling | ||
| +/+ | Vav32, Ets1, Plcg2 | .007 |
| +/2 | Pou2f2, Rras2, Ets1 | .01 |
| 2/2 | Raf1 | .29 |
| ERK/MAPK signaling | ||
| +/+ | Myc, Ets1, Plcg2 | .015 |
| +/2 | Rras2, Prkar2b, Mycn, Ets1 | .003 |
| 2/2 | Myc, Raf1 | .072 |
| FcϵRI signaling | ||
| +/+ | Vav32, Plcg2 | .03 |
| +/2 | Rras2 | .26 |
| 2/2 | Raf1 | .2 |
| Natural killer cell signaling | ||
| +/+ | Vav32, Plcg2 | .035 |
| +/2 | Rras2 | .28 |
| 2/2 | Raf1 | .22 |
| GM-CSF signaling | ||
| +/+ | Ets1 | .15 |
| +/2 | Rras2, Ccnd1, Ets1 | < .001 |
| 2/2 | Raf1 | .13 |
| G-protein–coupled receptor signaling | ||
| +/+ | Rasgrp1 | .42 |
| +/2 | Pde1a, Rras2, Prkar2b, Rasgrp12 | .003 |
| 2/2 | Raf1, Rasgrp1 | .076 |
| Axonal guidance signaling | ||
| +/+ | — | — |
| +/2 | Arhgef6, Rras2, Prkar2b, Rtn4 | .025 |
| 2/2 | Arhgef6, Raf1 | .21 |
| PI3K/AKT signaling | ||
| +/+ | — | — |
| +/2 | Rras2, Ccnd1 | .062 |
| 2/2 | Bcl2, Raf1 | .037 |
| PTEN signaling | ||
| +/+ | — | — |
| +/2 | Rras2, Ccnd1 | .04 |
| 2/2 | Bcl2, Raf1 | .024 |
| KEGG pathways | ||
| Dorsal-ventral axis formation | ||
| +/+ | Ets1, Notch1 | .14 |
| +/2 | Rras2, Ets1, Notch12 | .011 |
| 2/2 | Raf1 | > .5 |
| Leukocyte transendothelial migration | ||
| +/+ | Plcg2, Vav3, Vcl, Itk | .019 |
| +/2 | — | — |
| 2/2 | Pecam1, Itk | .4 |
| JAK-STAT signaling | ||
| +/+ | Myc, Il21r, Ccnd2 | .16 |
| +/2 | Ccnd1 | > .5 |
| 2/2 | Il12a, Myc, Mpl, Socs6 | .023 |
| MAPK signaling | ||
| +/+ | Myc, Mef2c, Rasgrp1 | .36 |
| +/2 | Rras2, Map3k7ip2, Evi1, Mef2c, Rasgrp1 | .047 |
| 2/2 | Myc, Mef2c, Rasgrp1, Raf1 | .097 |
Discussion
Leukemia is, like other cancers, a multistep disease that arises upon the acquisition of a series of malignant mutations. Mounting evidence has implicated C/EBPα as a myeloid tumor suppressor that is frequently targeted in human AML.4,26,28 Despite its importance as a myeloid tumor suppressor, very little is known about the additional mutations or molecular pathways that collaborate with mutated C/EBPα in disease development.
The CebpaBRM2 allele cooperates with SRS19-6–tagged genes in leukemogenesis
The CebpaBRM2/BRM2 model recapitulates several features of mutation-driven phenotypic progression.31 We took advantage of the phenotypic characteristics of these animals to screen for genes that collaborate with a mutated form of C/EBPα in the development of leukemia. SRS19-6 injection of newborn pups of the Cebpa+/+, the Cebpa+/BRM2, or the CebpaBRM2/BRM2 genotype resulted in leukemic development of T-cell or myeloid origin. Homozygous animals mainly developed myeloid leukemias, and did so with significantly reduced latency and fewer retroviral integrations compared with their WT and heterozygous littermates. These findings confirm that C/EBPα is a myeloid tumor suppressor and that interference with its functional properties predispose to disease development.
Interestingly, the skewing toward myeloid leukemia was also observed in the Cebpa+/BRM2 mice; however, in this case it was not associated with a reduction in disease latency compared with WT littermates. We have not previously observed any phenotypic differences between Cebpa+/+ and Cebpa+/BRM2 mice, but our data clearly suggest that disruption of a single Cebpa allele primes the cell for myeloid leukemias in a setting where the cells are stressed with additional oncogenic lesions. The preferential development of myeloid leukemias in heterozygous mice was not associated with mutation of the remaining WT allele. Epigenetic silencing of the Cebpa locus has recently been shown in lung cancer, squamous cell carcinoma, and in 2 patients with AML; thus, it remains a formal possibility for the phenotype of the Cebpa+/BRM2 mice.51-53 An alternative explanation for the preferential development of myeloid leukemias in these animals could be an unrecognized skewed distribution of myeloid progenitors that may serve as targets for retroviral-mediated oncogenic transformation. This hypothesis also implies that Cebpa+/BRM2 mice are not predisposed to tumor formation per se, consistent with the similar latencies of Cebpa+/+ and Cebpa+/BRM2 mice and their similar requirement for more retroviral insertions than their Cebpa/BRM2/BRM2 littermates.
A provocative result from this work is the finding that CebpaBRM2/BRM2 mice develop lymphoid tumors (mainly T-ALL) with a significantly shorter latency than Cebpa+/+ mice and with the Cebpa+/BRM2 animals displaying an intermediate latency. This is surprising, because C/EBPα has not previously been implicated in T-cell development although its mRNA is expressed in early T-cell progenitors and down-regulated during their maturation (from DN1-DN4).44 To test whether the predisposition of the CebpaBRM/BRM2 mice for T-cell leukemias was due to disturbances in thymic T-cell progenitors, we compared their distributions in Cebpa+/+ and CebpaBRM/BRM2 mice but found no significant differences. Previously, we have shown that CebpaBRM/BRM2 mice have decreased numbers of the HSC-containing LSK population, and preliminary data suggest that this is also true for the most primitive LT-HSC population (K.T.-M. and B.T.P., unpublished observations, May 2007). Hence, we speculate that the HSC population may serve as target for retroviral infection, consistent with the finding that these cells are still in cell cycle at the time of infection, and that disturbance of the HSC population may be responsible for the reduced latency of lymphoid tumors in CebpaBRM/BRM2 mice.54 Alternatively, it could be argued that the reduced latency of lymphoid tumors in CebpaBRM/BRM2 mice could be due to an increase in proliferation of myeloid progenitors in these mice, which in turn may lead to faster virus spreading even to other lineages.31 However, contrary to what we would expect in this scenario, CebpaBRM/BRM2 mice generally have fewer integrations than their control littermates.
On a final note, leukemias arising in SRS19-6–injected mice are only infrequently (1 of 10 tumors) associated with high degree of genomic instability as assayed by aCGH analysis. Interestingly, we did observe a common deletion (5 of 10 tumors) at the acromeric region of chromosome 11 minimally encompassing the Sfi1 and Eif4enif1 loci. Observations in budding yeast have demonstrated a centrin-dependent role for Sfi1 in centrosome duplication and spindle assembly.46,55 These findings and the recurrent finding of small deletion at chromosome 11 strongly suggest a tumor suppressor function of Sfi1.
Identification and analysis of SRS19-6–induced retroviral tags
The gene sequencing of RISs led to the identification of 22 CISs (5 of which were novel) and upgraded an additional 28 single RISs in RTCGD to CISs (Table S1). Hence, a relative small RIM screen as ours have yielded quite a substantial number of novel candidate oncogenes. This is likely to be due to the SRS19-6 retrovirus, which has not previously been used in RIM screens. The 5 novel CISs for which we found multiple RISs occurred in the genes encoding Chd4, Rag2, Arhgef6, Vav3, and Sesn1. All but the latter gene was demonstrated to have elevated expression levels in tumor cells bearing the integrations.
Chd4 encodes the chromodomain-helicase DNA-binding protein 4, a component of a HDAC2-containing complex, the nucleosome remodeling and deacetylating (NuRD) complex. It is found associated with Ataxia telangiectasia mutated (ATM)– and Rad3-related protein (ATR), which are implicated in DNA damage response and DNA replication checkpoint as well as in the autosomal recessive disorder Ataxia telangiectasia.56 These functional observations and the fact that its relatives Chd2, Chd3, and Chd9 have been identified as CISs in MoMuLV-based RIM screens suggest that Chd4 is indeed a likely oncogene.48,57
Rag2 encodes the recombination activating gene 2 (Rag2), which in concert with Rag1 is directly involved in V(D)J recombination in lymphoid cells. Rag2 expression is normally strictly confined to lymphoid cells, and it has been suggested that inappropriate induction of the Rag complex could induce genomic instability due to unauthorized recombination.58 Our finding that the 2 Rag2 insertions occurred in myeloid tumors is in line with this suggestion.
Arhgef6 encodes a guanine nucleotide exchange factor for Rho GTPases and has been found mutated in a screen for X-linked mental retardation.59 It has not previously been associated with cancer; however, its specific importance for Cdc42 function (which is found to be activated during phenotypic progression of MLL-AF9–induced leukemias) suggests that further studies into its functional properties are relevant.60,61
Vav3 encodes a member of the Vav family of oncoproteins and is, like the rest of the family, involved in signal transduction. Like Arhgef6, Vav3 is a guanine exchange factor for members of the Rho GTPase family, further underlining their functional relevance. Moreover, a crosstalk between Rac1/Vav and the Ras pathway in lymphocytes have been demonstrated to be mediated through PLCγ-mediated stimulation of RasGRP1.62 This finding is of particular relevance, as both Plcg2 and Rasgrp1 are targeted in our screen, thereby unfolding an interconnected network of pathways important for tumorigenesis.
Identification and analysis of SRS19-6–induced retroviral tags: the pathways
Deregulation of multiple genes in a given pathway provides increased credibility to its functional relevance in tumorigenesis. We used in silico pathway analysis on our RIS data set to detect commonly deregulated pathways in SRS19-6–induced tumors. This analysis demonstrated a profound preference for targeting of several signaling pathways, the main ones being VEGF and ERK/MAPK signaling as well as B- and T-cell receptor signaling. The finding that 16 of the 35 genes that were found in common deregulated pathways had not previously been assigned as CISs underlines the added value of this approach.
Specificity issues
Pathway analysis demonstrated specific targeting of leukocyte migration pathways in lymphoid tumors, which was mainly associated with tumors of Cebpa+/+ origin. Similarly, G1/S checkpoint regulation was mainly associated with Cebpa+/+ tumors, suggesting that the G1/S checkpoint is already deregulated in Cebpa+/BRM2 and CebpaBRM2/BRM2 premalignant cells. This is in line with the reduced ability of the BRM2 mutant of C/EBPα to repress E2F activity and implies that reduction of the gene dosage of the repressive WT Cebpa allele relieves the selective pressure for mutations that promote G1/S phase transition. In contrast, the PI3K/AKT (and PTEN) pathway(s) appears to be specifically targeted in myeloid tumors of either Cebpa+/BRM2 or CebpaBRM2/BRM2 origin, suggesting a specific cooperation between this pathway and AML in the context of the CebpaBRM2 allele.
At the gene level, we observed several single RISs in CebpaBRM2/BRM2 myeloid leukemias hinting at a specific collaboration between these loci and a mutated Cebpa. Some of these loci are targets for future studies. Moreover, we did observe a specific preference for insertions into the Myb loci in CebpaBRM2/BRM2 tumors (3 of 53 RISs in CebpaBRM2/BRM2 vs 0 of 129 RISs in Cebpa+/+ and Cebpa+/BRM2; P = .024). Myb is frequently targeted in MoMuLV screens and its exclusion from non-CebpaBRM2/BRM2 tumors could reflect a lower preference of the SRS19-6 virus for this locus coupled to a specific collaboration with the CebpaBRM2 allele. Future studies will test this hypothesis.
Conclusions
This study allows us to reach 4 main conclusions: (1) the CebpaBRM2 allele predisposes SRS19-6–injected mice to a myeloid leukemic fate in both Cebpa+/BRM2 and CebpaBRM2/BRM2 mice; (2) leukemias develop with significant reduced latency in CebpaBRM2/BRM2 mice. (3) Mapping of RISs in diseased mice led to the identification of several novel putative oncogenes, some of which may collaborate specifically with mutant Cebpa; and (4) in silico pathway analysis demonstrated differential deregulation of signaling pathways both in different leukemias and in mice of different genotypes and underlined the added value of using single RISs for pathway identification.
Finally, the identification of a total of 33 novel candidate oncogenes in a small study as ours suggest that the RTCGD is far from saturation. Larger screens with unconventional retroviruses are therefore likely to uncover new candidate oncogenes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by The Danish Medical Research Council, The Danish Cancer Society, The Association for International Cancer Research, The Danish Cancer Research Foundation, and Copenhagen University Hospital.
Authorship
Contribution: M.S.H. performed research, analyzed data, and wrote the paper; M.B.S., K.T.-M. and I.D. performed research; A.M., T.K., and B.Y. performed the CGH analysis; A.B.S., F.S.P., and C.N. designed the RIM screen; and B.T.P. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no conpeting financial interests.
Correspondence: Bo Porse, Section for Gene Therapy Research, Department of Clinical Biochemistry, Copenhagen University Hospital, Juliane Maries Vej 20-9322, DK2100 Copenhagen, Denmark; e-mail: porse@rh.dk.