Abstract
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disorder caused by mutations in the NF1 gene. Patients with NF1 have a higher risk to develop juvenile myelomonocytic leukemia (JMML) with a possible progression toward acute myeloid leukemia (AML). In an oligo array comparative genomic hybridization–based screening of 103 patients with pediatric T-cell acute lymphoblastic leukemia (T-ALL) and 71 patients with MLL-rearranged AML, a recurrent cryptic deletion, del(17)(q11.2), was identified in 3 patients with T-ALL and 2 patients with MLL-rearranged AML. This deletion has previously been described as a microdeletion of the NF1 region in patients with NF1. However, our patients lacked clinical NF1 symptoms. Mutation analysis in 4 of these del(17)(q11.2)-positive patients revealed that mutations in the remaining NF1 allele were present in 3 patients, confirming its role as a tumor-suppressor gene in cancer. In addition, NF1 inactivation was confirmed at the RNA expression level in 3 patients tested. Since the NF1 protein is a negative regulator of the RAS pathway (RAS-GTPase activating protein), homozygous NF1 inactivation represent a novel type I mutation in pediatric MLL-rearranged AML and T-ALL with a predicted frequency that is less than 10%. NF1 inactivation may provide an additional proliferative signal toward the development of leukemia.
Introduction
Neurofibromatosis type 1 (NF1) is an autosomal genetic disorder that is clinically characterized by cafe-au-lait spots and frequent fibromatous tumors of the skin and tumors of the central nervous system. The NF1 disorder is caused by genetic heterozygous mutations in the NF1 gene on chromosome 17q11.2. Most NF1 mutations are intragenic and have been found over the complete gene. They comprise a diversity of mutation types, where splicing mutations are particularly prevalent given the number of exons. This results in truncation for a large percentage of patients, thereby inactivating the encoded protein neurofibromin.1 Another genetic aberration includes microdeletions affecting the entire NF1 locus. Patients with these NF1 microdeletions display a more severe NF1 phenotype, characterized by mental retardation, facial dysmorphism, and increased risk for developing malignant tumors, including leukemias.2,3 To this end, NF1 has also been associated with juvenile myelomonocytic leukemia (JMML), with a risk of progression toward acute myeloid leukemia (AML). These malignancies are associated with loss of the wild-type allele, either through deletions or the acquisition of point mutations. In JMML, it has also frequently been reported that the wild-type allele is replaced by the mutant allele as an effect of recombinational events leading to uniparental disomy (UPD).4-6 Previously, it was shown that biallelic inactivation of NF1 are found as somatic abnormalities in patients with JMML who lack clinical evidence of NF1.7 Somatic inactivation of Nf1 in hematopoietic cells results in a progressive myeloproliferative disorder in mice,8 confirming that NF1 acts as a tumor-suppressor gene.5 The NF1 gene protein product, neurofibromin, is a GTPase-activating protein (GAP) that inhibits RAS signaling by hydrolysis of active RAS-GTP into inactive RAS-GDP.1,9 Therefore, NF1 deficiencies act as functional equivalents of activational mutations in RAS. Indeed, NF1 inactivation and RAS mutations have been found in a mutually exclusive manner in JMML.7
AML is a heterogeneous disease in which early treatment response and cytogenetic abnormalities are the most important prognostic factors. In AML, genetic aberrations can be classified as type I or type II mutations. One hypothesis about the development of AML is the coexistence of both type I and type II mutations that confer proliferative signals (type I mutations affecting the FLT3, C-KIT, NRAS, KRAS, or PTPN11 genes) in combination with type II differentiation-impairing mutations (such as PML-RARα, AML-ETO, CBFB-MYH11, or MLL-rearrangements).10
MLL-rearrangements account for 8% to 20% of all cytogenetic abnormalities in pediatric AML.11,12 HOX genes are the prime targets of MLL fusion products and regulate cellular differentiation in normal hematopoietic development. However, Eguchi et al point to another role of MLL fusion products in MLL-rearranged leukemias through the alteration of cell-cycle arrest and apoptosis.13 Most of these MLL-positive AML samples are morphologically classified as FAB-M4 and FAB-M5, and it has been suggested that MLL-rearrangements in pediatric AML are associated with a poor outcome. Interestingly, in some studies the t(9;11) subgroup has been associated with a higher sensitivity to different classes of drugs and a better prognosis.14-16 In addition, many of these patients with MLL-rearranged AML lack mutations in FLT3, C-KIT, NRAS, KRAS, and PTPN11, indicating that the type I mutations remains to be elucidated. High-resolution genomic screening of patients with MLL-rearranged AML could provide us with further insight into novel genetic aberrations with prognostic significance or new type I mutations in MLL-rearranged AML.
T-cell acute lymphoblastic leukemia (T-ALL) represents about 15% of patients with pediatric ALL and is characterized by a rapid progression of disease and a 30% relapse rate within the first 2 years after diagnosis.17 In the last decade, a large number of new genomic aberrations were identified in T-ALL, including chromosomal translocations, deletions, amplifications, and mutations.18-20 All these genetic defects target different cellular processes, including the cell cycle, T-cell differentiation, proliferation, and survival. Cooperation of these genetic events initiates leukemic transformation of thymocytes.18 RAS mutations have been found in less than 5% of patients with T-ALL, showing that proliferative hits affecting the RAS pathway remain rare.18 On the other hand, more than 50% of the patients with T-ALL are characterized by activating mutations in the NOTCH1 pathway, including the NOTCH1 gene itself21,22 or the NOTCH1-regulating U3-ubiquitin ligase FBXW7.23,24
In this study, we used oligo array–comparative genomic hybridization (array-CGH) and identified somatic NF1 microdeletions as a cryptic genetic abnormality in patients with pediatric T-ALL and patients with MLL-rearranged AML that lack symptoms of neurofibromatosis. We present further evidence for the role of NF1 inactivation as a functional equivalent to activated RAS signaling, and suggest that this can be considered as a new type I mutation in MLL rearranged AML and a proliferative hit in T-ALL.
Methods
Patients
Viably frozen diagnostic bone marrow or peripheral blood samples from 103 patients with pediatric T-ALL and 71 patients with pediatric MLL-rearranged AML were provided by the Dutch Childhood Oncology Group (DCOG), the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL), and the Berlin-Frankfurt-Münster AML Study Group (AML-BFM-SG). Informed consent was obtained according to local law and regulations and in accordance with the Declaration of Helsinki. Leukemic cells were isolated and enriched from these samples as previously described.25 All resulting samples contained 90% or more leukemic cells, as determined morphologically by May-Grünwald-Giemsa (Merck, Darmstadt, Germany)–stained cytospins. These leukemic cells were used for DNA and RNA extraction, and a minimum of 5 × 106 leukemic cells were lysed in Trizol reagent (Gibco BRL/Life Technologies, Breda, The Netherlands) and stored at −80°C. Genomic DNA and total cellular RNA were isolated as described before.25 From the patients with a deletion of NF1, remission and relapse material was only available for patient no. 2736.
Oligo array-CGH
MLPA
Multiplex ligation-dependent probe amplification (MLPA) analysis was performed using the SALSA P081/082 MLPA assay (MRC Holland, Amsterdam, the Netherlands). SALSA P081/082 consists of 2 reaction mixes containing probes for all constitutive NF1 exons except for exons 5, 7, 17, 19A, 45, and 47. The exact localization of the MLPA probes can be downloaded from the MRC Holland website (http://www.mrc-holland.com/pages/p081 p082pag.html). The 2 reactions contain 15 and 13 control probes in other regions of the genome, respectively. The patients' samples were analyzed with MLPA according to the manufacturer's protocol.27,28 Data were analyzed using GeneMarker v1.5 (Softgenetics, State College, PA).
Mutation analysis
For the detection of NF1 mutations, DNA was subjected to 40 cycles of polymerase chain reaction (PCR) of 15 minutes at 95°C and 1 minute at 60°C, using specific primers for all NF1 exons, which are being used in NF1 diagnostics (Department of Clinical Genetics, Erasmus MC, Rotterdam, The Netherlands, A.M.W.O. manuscript in preparation; primers are available on request at a.vandenouweland@erasmusmc.nl). RAS, PTPN11 and C-KIT mutation screening was performed as described in Table S1, available on the Blood website (see the Supplemental Materials link at the top of the online article). NOTCH1 and FLT3 mutational screening were done as previously described.21,29,30 PCR products were purified by standard methods and directly sequenced from both strands. The sequence data were analyzed using Seqscape V2.5 (Applied Biosystems, Foster City, CA).
NF1 expression analysis
NF1 expression was calculated based upon nonnormalized gene expression array data, performed on the human genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA) as previously described,31 which were available for 3 del(17)(q11.2)-positive and 7 del(17)(q11.2)-negative leukemia patients. For the NF1 probe sets, the expression was normalized to the median expression of GAPDH (6 probesets) for each patient sample. The difference in relative gene expression levels between patients with and without the del(17)(q11.2) was evaluated using the Mann-Whitney U test.
Results
High-resolution genomic screening of a selected subgroup of 103 patients with pediatric T-ALL and 71 patients with MLL-rearranged AML using a 44K oligo array-CGH platform led to the identification of a cryptic deletion, del(17)(q11.2). This deletion was recurrently observed in 3 patients with T-ALL and 2 patients with AML (Figure 1A,B; Table 1). These deletions were about 1.2 Mb in size and covered the NF1 gene. For all patients, the telomeric breakpoints were situated in the JJAZ1 gene, whereas the centromeric breakpoints clustered in its pseudogene JJAZ1P (Figure 1C,D). The deletion area in these samples was equivalent to those observed in patients with NF1. Genetic and clinical patient characteristics for all del(17)(q11.2)-positive leukemia patients are summarized in Table 1. One of the 3 patients with T-ALL and at least one of the 2 patients with AML relapsed.
NF1 microdeletions in pediatric acute leukemias. (A) Chromosome 17 ideogram and corresponding oligo array-CGH plot of patient DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patient no. 2736. (B) Detailed visualization of the NF1 microdeletion at chromosomal band 17q11 in T-ALL patient no. 2736. Hybridization signals around the −2X or +2X lines represent loss of the corresponding region in the patient DNA. (C) Detailed analysis of the centromeric (left panel) and telomeric (right panel) breakpoint of the NF1 microdeletion in patient no. 2736. (D) Overview of oligo array-CGH results in the chromosomal region 17q11.2 for 3 patients with T-ALL and 2 patients with AML with del(17)(q11.2). The 60-mer oligos present on the DNA array and located in this genomic area, as well as the specific genes located in this region with their transcription direction, are shown. Arrows above the indicated genes represent the direction of transcription. N indicates normal; L, loss; cen, centromere; and tel, telomere.
NF1 microdeletions in pediatric acute leukemias. (A) Chromosome 17 ideogram and corresponding oligo array-CGH plot of patient DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) for T-ALL patient no. 2736. (B) Detailed visualization of the NF1 microdeletion at chromosomal band 17q11 in T-ALL patient no. 2736. Hybridization signals around the −2X or +2X lines represent loss of the corresponding region in the patient DNA. (C) Detailed analysis of the centromeric (left panel) and telomeric (right panel) breakpoint of the NF1 microdeletion in patient no. 2736. (D) Overview of oligo array-CGH results in the chromosomal region 17q11.2 for 3 patients with T-ALL and 2 patients with AML with del(17)(q11.2). The 60-mer oligos present on the DNA array and located in this genomic area, as well as the specific genes located in this region with their transcription direction, are shown. Arrows above the indicated genes represent the direction of transcription. N indicates normal; L, loss; cen, centromere; and tel, telomere.
Patient characteristics and truncating NF1 mutations in pediatric T-ALL and AML
| ID . | Subtype . | Sex . | Age, y . | WBC, × 109/L . | Relapse or CCR, mo . | Genetic subtype . | Karyotype . | NF1 del . | NF1 mutation analysis . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon . | Mutation . | |||||||||
| 2736 | T-ALL | M | 10.1 | 13 | Relapse, 15 | CALM-AF10 | 46, XY, t(2;9)(q21;q34),?t(8,8)(?q22;q?24) | Yes | 22 | c.3734delCinsGGTTTATGGTTT |
| Remission | — | — | — | — | — | — | 46, XY | No | — | WT |
| Relapse | — | — | — | — | — | — | 46, XY, t(2;9)(q21;q34),?t(8,8)(?q22;q?24) | Yes | 22 | c.3734delCinsGGTTTATGGTTT |
| 2780 | T-ALL | F | 5 | 140 | CCR, 50+ | TLX3 | 46, XX | Yes | 4 | c.333dupA |
| 167 | T-ALL | M | 16.4 | 170.9 | CCR, 56+ | Unknown | NA | Yes | — | WT |
| 4389 | AML | F | 18.8 | 41.7 | NA | MLL-AF9 | 46, XX, t(9;11)(p22;q23)[4]/46, XX [7] | Yes | 16 | c.2849 2850insTT |
| 6421 | 1AML | F | 6.5 | NA | Relapse, 68 | MLL-AF10 | 47, XX, t(10;11)(p13;q23),+19 | Yes | — | ND |
| ID . | Subtype . | Sex . | Age, y . | WBC, × 109/L . | Relapse or CCR, mo . | Genetic subtype . | Karyotype . | NF1 del . | NF1 mutation analysis . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon . | Mutation . | |||||||||
| 2736 | T-ALL | M | 10.1 | 13 | Relapse, 15 | CALM-AF10 | 46, XY, t(2;9)(q21;q34),?t(8,8)(?q22;q?24) | Yes | 22 | c.3734delCinsGGTTTATGGTTT |
| Remission | — | — | — | — | — | — | 46, XY | No | — | WT |
| Relapse | — | — | — | — | — | — | 46, XY, t(2;9)(q21;q34),?t(8,8)(?q22;q?24) | Yes | 22 | c.3734delCinsGGTTTATGGTTT |
| 2780 | T-ALL | F | 5 | 140 | CCR, 50+ | TLX3 | 46, XX | Yes | 4 | c.333dupA |
| 167 | T-ALL | M | 16.4 | 170.9 | CCR, 56+ | Unknown | NA | Yes | — | WT |
| 4389 | AML | F | 18.8 | 41.7 | NA | MLL-AF9 | 46, XX, t(9;11)(p22;q23)[4]/46, XX [7] | Yes | 16 | c.2849 2850insTT |
| 6421 | 1AML | F | 6.5 | NA | Relapse, 68 | MLL-AF10 | 47, XX, t(10;11)(p13;q23),+19 | Yes | — | ND |
In additional mutation analysis, FLT3-ITD or D835 PM, C-KIT, RAS, and PTPN11 were all WT.
CCR indicates continued complete remission; —, not applicable; WT, wild-type; NA, not available; ND, not determined; PM, point mutation.
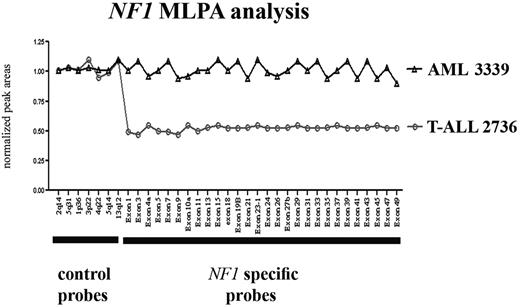
To further confirm the deletion breakpoints, 4 of 5 del(17)(q11.2)-positive leukemia patients and 15 del(17)(q11.2)-negative controls (7 with T-ALL and 8 with AML) were analyzed using an NF1 locus–specific MLPA assay.27,28 No residual material was available for patient no. 6421. These analyses confirmed that one copy of the NF1 locus was lost in all 4 of these patients (Figure 2; only T-ALL patient no. 2736 is shown), whereas all control patients retained both copies of the NF1 gene (only AML control no. 3339 is shown).
MLPA analysis of NF1 in selected cases. MLPA analysis of the NF1 locus in T-ALL patient no. 2736 and AML patient no. 3339. Normalized peak areas around 0.5 represent monoallelic loss of the corresponding genomic region.
MLPA analysis of NF1 in selected cases. MLPA analysis of the NF1 locus in T-ALL patient no. 2736 and AML patient no. 3339. Normalized peak areas around 0.5 represent monoallelic loss of the corresponding genomic region.
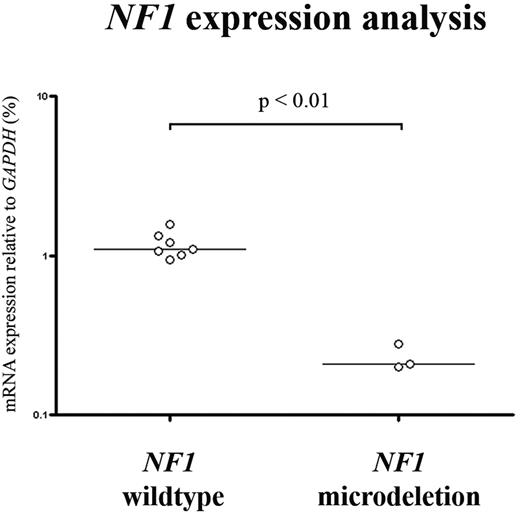
In order to investigate complete NF1 inactivation in our patients, we performed mutation analysis on all exons and exon/intron boundaries of the NF1 gene in the 4 del(17)(q11.2)-positive patients with leukemia, and in an additional group of 39 patients without a deletion involving chromosomal band 17q11.2 (including 21 patients with MLL-rearranged AML and 18 patients with T-ALL). Small frameshift mutations disrupting the NF1 coding region were only detected in 3 of 4 del(17)(q11.2)-positive patients (Table 1; Figure 3), leading to biallelic inactivation of NF1 in these patients. One patient with T-ALL and 2 patients with MLL-rearranged AML without a del(17)(q11.2) had a monoallelic mutation in nonfunctional domains, possibly reflecting rare polymorphisms. Furthermore, NF1 expression in the del(17)(q11.2)-positive patients with T-ALL and MLL-rearranged AML leukemias was significantly lower in 3 patients tested, as compared with 7 T-ALL and AML patient samples that are wild-type for NF1 (Figure 4).
Truncating NF1 mutations in pediatric T-ALL and AML. (A) Sequence analysis of patient no. 4389 (AML) showing a c.2849_2850insTT mutation in the remaining NF1 allele. (B) Sequence analysis of patient no. 2736 (T-ALL) showing a c.3734delCinsGGTTTATGGTTT mutation in the remaining NF1 allele. (C) Sequence analysis of patient no. 2780 (T-ALL) showing a c.333dupA mutation in the remaining NF1 allele.
Truncating NF1 mutations in pediatric T-ALL and AML. (A) Sequence analysis of patient no. 4389 (AML) showing a c.2849_2850insTT mutation in the remaining NF1 allele. (B) Sequence analysis of patient no. 2736 (T-ALL) showing a c.3734delCinsGGTTTATGGTTT mutation in the remaining NF1 allele. (C) Sequence analysis of patient no. 2780 (T-ALL) showing a c.333dupA mutation in the remaining NF1 allele.
NF1 expression analysis in pediatric T-ALL and MLL-rearranged AML.NF1 mRNA expression data relative to GAPDH (%), based upon gene expression array data, which were available for 3 del(17)(q11.2)-positive (patient no. 2736, 2780, and 167) and 7 del(17)(q11.2)-negative patients with leukemia. The horizontal bars represent median relative expression.
NF1 expression analysis in pediatric T-ALL and MLL-rearranged AML.NF1 mRNA expression data relative to GAPDH (%), based upon gene expression array data, which were available for 3 del(17)(q11.2)-positive (patient no. 2736, 2780, and 167) and 7 del(17)(q11.2)-negative patients with leukemia. The horizontal bars represent median relative expression.
To further verify a somatic rather than a genetic origin of NF1 inactivation, we screened relapse and remission material of T-ALL patient no. 2736, for whom material was available. At relapse, the NF1 microdeletion and NF1 mutation on the other allele were present, while in the remission sample both mutations remained undetected.
Since NF1 deficiency could act as a novel type I mutation, we screened all del(17)(q11.2)-positive patients with leukemia for activational mutations in RAS. Although NRAS or KRAS mutations have been described in MLL-rearranged AML and T-ALL, no somatic NRAS or KRAS mutations were found in our 5 del(17)(q11.2)-positive patients with leukemia. In addition, both patients with MLL-rearranged AML with a NF1 deletion lacked other type I mutations in FLT3, C-KIT, or PTPN11 in their leukemic cells. The frequency of these mutations in the 71 MLL-rearranged AML samples was low, as expected. Only 35% had one of these mutations, and all these mutations were mutually exclusive. Furthermore, the del(17)(q11.2)-positive patients with T-ALL were screened for rearrangements at the TAL1, HOX11L2, HOX11, CALM-AF10, MLL and cMYC loci or the presence of NOTCH1 mutations. One patient (no. 167) lacked rearrangements of any of the loci mentioned above, whereas a HOX11L2 translocation (no. 2780) and a CALM-AF10 fusion gene (no. 2736) were detected in 2 other patients. NOTCH1 mutations were identified in patients no. 2780 (heterodimerization domain; L1601P) and no. 167 (PEST domain; 2445insLL).
Discussion
Genetic events that lead to leukemogenesis by activating uncontrolled cell proliferation remain to be elucidated in most patients with pediatric T-ALL and MLL-rearranged AML. We used oligo array-CGH to identify new abnormalities and found somatic NF1 microdeletions as a cryptic genetic abnormality in patients lacking clinical symptoms of neurofibromatosis. This array-CGH study is currently expanded to other subtypes of leukemias. Recent single nucleotide polymorphism (SNP) array analysis of pediatric ALL by investigators from St Jude Children's Research Hospital showed that this microdeletion in NF1 may be present at low frequencies in other types of acute leukemia as well.32
NF1 microdeletions are observed in about 5% to 20% of patients with NF1.1 The majority of these patients with NF1 have a 1.4-Mb NF1 microdeletion due to interchromosomal homologous recombination between the low-copy repeats of the WI-12393 gene flanking NF1 and sequences with homology to chromosome 19 during meiosis.33 A second type of NF1 microdeletions of about 1.2 Mb in size is due to a mitotic intrachromosomal recombination between the JJAZ1 and the homologous JJAZ1P pseudogene.34,35 The NF1 microdeletions in our leukemia patients seemed identical to this 1.2 Mb microdeletion type.33 However, in contrast to patients with NF1 with similar NF1 deletions, our patients with leukemia did not meet the clinical criteria for NF1, lacking cafe-au-lait spots, mental retardation, and/or facial dysmorphism. This suggests that the NF1 deletion in our patients is somatic and leukemia specific, rather than of constitutional genetic origin, although molecular diagnostics for NF1 was not performed in these patients.
Deletion of one allele of NF1 and further inactivation of the other NF1 allele in 3 patients through the acquisition of point mutations further confirms the role of NF1 as a tumor-suppressor gene in the pathogenesis of both pediatric MLL-rearranged AML and T-ALL. This point was further strengthened by the finding of clonal stability in one of the del(17)(q11.2) patients, where the deletion of NF1 on one allele and the point mutation in the other NF1 allele were both present at diagnosis and relapse while absent in the remission sample. Therefore, the NF1 abnormalities were of somatic origin in at least patient no. 2736 and were only present in the leukemic cells. Similar findings have been described for patients with JMML,7 explaining why these patients did not have any clinical symptoms of neurofibromatosis.
Since NF1 deficiency leads to the activation of the RAS signaling pathway,9 and none of the del(17)-positive patients with leukemia had mutations in NRAS or KRAS, NF1 microdeletions presumably provide an alternative mechanism for RAS activation in both MLL-rearranged myeloid and T-lymphoid leukemias, thereby representing a novel type I abnormality. These patients with leukemia may potentially benefit from additional treatment with RAS inhibitors like farnesylthiosalicylic acid36 or downstream inhibitors.
Both del(17)-positive patients with AML were further screened for any of the other currently known type I mutations in AML. As expected, no other type I mutations were detected, indicating that NF1 microdeletions could act as a novel type I mutation which cooperate with the MLL translocation (type II mutation) in the pathogenesis of AML.
The idea of a multistep pathogenesis in T-cell leukemia is widely accepted.18-20,37 Cooperative genetic events affect cell cycle, T-cell differentiation, proliferation, and survival. We identified a number of cooperative aberrations in the del(17)(p11.2)-positive T-ALL samples. NOTCH1 mutations, generally present in about 50% of T-ALL,21 were identified in 2 of 3 del(17)(p11.2)-positive T-ALL samples. In addition, genetic aberrations that induce a T-cell differentiation arrest were identified in patient no. 2780 (HOX11L2 translocation) and patient no. 2736 (CALM-AF10 translocation). These data further suggests that loss of NF1 can be involved in the development of T-ALL, as one of the genetic hits in multistep oncogenesis.
In this study, we identified 3 patients whith a deletion of NF1 and an inactivational mutation on the remaining allele. We could not identify homozygous somatic NF1 mutations in 21 patients with MLL-rearranged AML and 18 patients with T-ALL without a microdeletion. This suggests that the frequency of biallelic inactivation, until now the only mechanism described for oncogenesis, is less than 10% in these groups.
Other mechanisms of NF1 inactivation, such as inactivation through the duplication of the mutated (UPD) NF1 allele at the expense of the remaining wild-type allele, as observed in patients with NF1 with JMML, may have been missed.6 Of interest, recent SNP array analysis of pediatric ALL, and JMML without underlying NF1, showed that there was no UPD involved in the NF1 region.6,32 In addition, in adult AML approximately 20% have large regions of UPD, but none of them involves the NF1 locus.38,39 Hence, UPD of the NF1 locus may be a rare event in leukemias of somatic origin compared with leukemias which originate from patients with clinical evidence of NF1. Therefore, the frequency of biallelic NF1 inactivation in pediatric MLL-rearranged AML and T-ALL as we reported here may be underestimated. Future studies should be extended by sequencing the NF1 locus, including the promoter region and the 3′ untranslated region (UTR), and look for abnormalities in NF1 protein expression.
In conclusion, we report the identification of NF1 microdeletions in patients with pediatric T-ALL and MLL-rearranged AML without clinical evidence of NF1. We confirmed NF1 inactivation by reduced NF1 expression levels and biallelic NF1 mutations in 3 of 5 patients, confirming the role of NF1 as a tumor-suppressor gene in cancer. NF1 inactivation is a novel type I mutation in MLL-rearranged AML and a new proliferative hit in T-ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.V.B. and P.V.V. designed and performed research and wrote the paper; G.J.L. collaborated on the MLL-AML study; E.V.W., D.R., and M.H. made this research possible by collecting patient samples and characteristics in their own study groups and providing additional information; A.M.W.O. and J.N.R.T. performed and designed NF1 analysis; and J.P.P.M, M.M.H., H.B.B, C.M.Z., and R.P. designed research and wrote the paper.
Conflict-of-interest dislosure: The authors declare no competing financial interests.
Correspondence: Jules P. P. Meijerink, Erasmus MC/Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Rm Sp 2456, Dr. Molewaterplein 60, PO Box 2060, 3000 CB Rotterdam, The Netherlands; e-mail: j.meijerink@erasmusmc.nl.
References
Author notes
*B.V.B. and P.V.V. contributed equally to this study.
†M.M.V.d.H.-E. and J.P.P.M. can be considered equally as last authors.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal