Abstract
Signaling through the transforming growth factor–β (TGF-β) pathway results in growth inhibition and induction of apoptosis in various cell types. We show that this pathway is blocked in Kaposi sarcoma herpesvirus (KSHV)–infected primary effusion lymphoma through down-regulation of the TGF-β type II receptor (TβRII) by epigenetic mechanisms. Our data also suggest that KSHV infection may result in lower expression of TβRII in Kaposi sarcoma and multicentric Castleman disease. KSHV-encoded LANA associates with the promoter of TβRII and leads to its methylation and to the deacetylation of proximal histones. Reestablishment of signaling through this pathway reduces viability of these cells, inferring that KSHV-mediated blockage of TGF-β signaling plays a role in the establishment and progression of KSHV-associated neoplasia. These data suggest a mechanism whereby KSHV evades both the antiproliferative effects of TGF-β signaling by silencing TβRII gene expression and immune recognition by suppressing TGF-β–responsive immune cells through the elevated secretion of TGF-β1.
Introduction
Kaposi sarcoma herpesvirus (KSHV) is associated with the pathogenesis of primary effusion lymphoma (PEL), Kaposi sarcoma (KS), and some forms of multicentric Castleman disease (MCD).1-3 Its genome contains an extensive number of pirated cellular homologs involved in subverting critical cellular regulatory processes.4,5 As a member of the γ-herpesvirus family, KSHV is characterized by a prolonged latency during which only a subset of its genes are expressed. These latently expressed gene products play important roles in immune evasion, cell proliferation, and inhibition of apoptosis.
One of these latently expressed proteins, latency-associated nuclear antigen (LANA), is involved in several cellular processes. It is considered an oncogenic protein because of its ability to dysregulate tumor suppressor pathways associated with p53 and pRb and to transform primary rat embryo fibroblasts in cooperation with the cellular oncogene H-ras.6,7 Its association with GSK-3β, an important modulator of the Wnt signaling pathway, leads to accumulation of β-catenin and subsequent up-regulation of Tcf/Lef-regulated genes.8 LANA inhibits expression of the reactivation transcriptional activator (RTA/Lyta), which is critical for the switch from latency to lytic reactivation.9,10 It tethers the viral episome to host chromatin during mitosis, ensuring KSHV DNA gets replicated and episomes are not lost during cellular division.11-14 LANA also regulates viral as well as cellular gene expression.6-8,15-17 Although some of the changes mediated by LANA occur indirectly via activation of β-catenin and E2F target genes, direct binding of LANA to DNA also results in transcriptional repression.18,19 Interactions with corepressors mSin3, SAP30, and CIR, the methyl CpG-binding protein MeCP2, and the histone methyltransferase SUV39H1 are consistent with a direct role for LANA in transcriptional repression.14,20,21 LANA has been shown to inhibit in vitro histone acetyltransferase activity of CREB-binding protein (CBP) and, more recently, to associate with Dnmt3a, a DNA methyltransferase involved in de novo DNA methylation, supporting a role for LANA in epigenetic gene regulation.16,22
Transforming growth factor-beta (TGF-β) is a multifunctional cytokine involved in diverse biologic processes, which include embryonic development, regulation of cell growth, differentiation, hematopoiesis, angiogenesis, immune function, and apoptosis (reviewed by Roberts and Sporn23 and Massague24 ). There are 3 isoforms of TGF-β, each of which binds to the same heterotetrameric complex of type I (TβRI) and type II (TβRII) serine/threonine kinase receptors. Initially, TGF-β binds to TβRII, which leads to the recruitment and activation of TβRI. Receptor-activated Smads (R-Smads), Smad2 and Smad3, are then phosphorylated by TβRI and translocate into the nucleus in a complex with Smad4. In the nucleus, the Smad complex binds its cognate binding site as well as several transcription factors, transcriptional activators, or transcriptional repressors (reviewed by Massague25 ). The diversity of responses that occurs under different cellular contexts is dictated by the cell specific presence/absence of these Smad complex binding partners.
TGF-β plays a role in maintaining homeostasis in many tissue types. Its antiproliferative and apoptotic effects on epithelial, endothelial, and hematopoietic lineages effectively limit their growth.26-28 The frequent loss of TGF-β responsiveness in human cancers underscores the importance of this growth regulatory role; it is considered to be a key event in the development and progression of several tumors.29-31
In our study, we looked at whether TGF-β signaling was blocked in KSHV-related diseases. We found PEL cell lines to be unresponsive to TGF-β treatment. A few KSHV gene products have previously been found to interfere with TGF-β signaling. Viral IFN regulatory factor 1 (vIRF1) interacts with both Smad3 and Smad4 and inhibits their ability to bind DNA.32 K-bZIP, on the other hand, binds CBP and consequently limits its recruitment to Smad-mediated transcription initiation complexes.33 These gene products, however, are expressed during lytic infection and therefore would not account for the defect in TGF-β signaling in PEL cells in which KSHV infection is predominantly latent. Our results indicate that the defect in TGF-β signaling in PEL is the result of epigenetic silencing of the TβRII gene by KSHV-encoded LANA.
Methods
Cell culture and treatment
PEL cell lines (BC1, BC3, BCBL1), Burkitt lymphoma cell lines (Ramos, BJAB, Namalwa), AIDS-related large B-cell lymphoma cell line (BCKN-1), and BJAB/Tet-on/LANA cell line34 were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA) and 40 μg/mL gentamicin (Sigma-Aldrich, St Louis, MO) at 37°C and 5% CO2. The telomerase-immortalized human umbilical vein endothelial cell line (TIVE) and long-term-infected TIVE cells (LTCs) were cultured as described previously.35 PEL cells were treated with recombinant human TGF-β1 (R&D Systems, Minneapolis, MN) at a concentration of 5 ng/mL, anti–human TGF-β RII (R&D Systems) at 50 μg/mL to block TβRII, MS-275 (Calbiochem, San Diego, CA) at a concentration of 0.5 μM, and 5-aza-2-deoxycytidine (5-aza-dC) at 1 μM. BJAB/Tet-on/LANA cells were induced with 2 μg/mL doxycycline (Sigma-Aldrich).
Cell growth and apoptosis assays
Cells, plated at 2.5 × 105 cells/mL in complete medium with either TGF-β1 or vehicle, were stained and counted every 24 hours over 4 days. To measure apoptosis, cells were cultured at 5 × 105 cells/mL in the presence of TGF-β1 or vehicle alone, harvested 48 hours after treatment, and stained with annexin V-fluorescein isothiocyanate as described by the manufacturer (BD Biosciences, San Jose, CA). Apoptosis was measured by annexin V inclusion with a FACSCalibur flow cytometer (BD Biosciences) and quantitated using CellQuest software (BD Biosciences). Results shown are representative of 3 independent experiments.
Plasmids, transient transfections, and reporter gene assays
The pcDNA1-TβRII expression vector (clone H2–3FF)36 was kindly provided by Dr H.Y. Lin (Massachusetts General Hospital/Harvard Medical School, Boston), and the p3TP-Lux reporter plasmid37 was a gift from Dr J. Massague (Memorial Sloan Kettering Cancer Center, New York, NY). BC1, BC3, and BCBL1 cells were plated at 6 × 105 cells/mL 24 hours before transfection. Cells were transfected with the p3TP-Lux reporter vector alone or in combination with pcDNA1-TβRII (clone H2-3FF) using Transfectin (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The pRL-RSV vector (Promega, Madison, WI) was used as an internal control. Cells were collected 48 hours after transfection and lysed in passive lysis buffer (Promega). Luciferase assays were performed using Dual Luciferase assay system (Promega) according to the manufacturer's protocol. Firefly and Renilla luciferase activities were measured with an MLX Microplate Luminometer (Dynex Technologies, Chantilly, VA). The ratio of firefly to Renilla luciferase activity was calculated to normalize for transfection efficiency. Results shown are representative of at least 3 independent experiments and are presented as means of triplicate transfections.
Immunoblotting
Whole cell lysates were prepared in radioimmunoprecipitation assay (RIPA) buffer supplemented with 5 μL/mL Protease Inhibitor Cocktail III (Calbiochem), 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM Na3VO4 (Sigma-Aldrich). Nuclear proteins were isolated as previously described.38 Proteins were quantitated using Bradford assay (Bio-Rad), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to polyvinylidene fluoride (Millipore, Billerica, MA) by semi-dry transfer. The blots were probed with primary antibodies overnight at 4°C and incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour at room temperature. Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used for signal detection. Blots were reprobed for actin after exposure to stripping buffer (100 mM β-mercaptoethanol, 2% sodium dodecyl sulfate, 62.5 mM Tris, pH 6.8) for 15 minutes at 52°C. The following antibodies were used: anti-Smad2/3 (N-19), anti–phospho-Smad2/3 (Ser 433/435), anti-TβRI (H-100), anti-TβRII (C-16), anti–cyclin A (E67.1), anti-p21 (H-164; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Smad2 (EP567Y; Abcam, Cambridge, MA), anti-HHV-8 ORF73 (Advanced Biotechnologies, Columbia, MD), and anti–actin (Sigma-Aldrich). Data shown are representative of at least 3 independent experiments.
Lentivirus production and cell transduction with lentivirus
The pHR-Sin-CSGW-TβRII lentiviral vector was made by polymerase chain reaction (PCR) amplification and cloning of TβRII into the pHR-Sin-CSGW-ΔNot1 vector39 ; 1 to 5 × 105 BC3 cells were resuspended in fresh medium to which was added 1 μL lentivirus at a multiplicity of infection of 5 genomes per cell. Both cells and virus were first incubated for 15 minutes with polybrene to neutralize surface charge and to enhance virus-cell binding. Cells were harvested for protein or cDNA 48 to 72 hours after infection.
Quantitative real-time RT-PCR
RNA was isolated with RNeasy kit (QIAGEN, Valencia, CA); 1 μg RNA was transcribed into cDNA using Reverse Transcription System (Promega) and Random Primers (Promega) in a final volume of 20 μL according to the manufacturer's instructions. Quantitative real-time PCR (RT-PCR) was performed with ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) with Sybr Green PCR Master Mix (Applied Biosystems) using 6 μL diluted cDNA in a 20-μL final reaction mixture. The following primer sets were used: TβRII (5′-GGCTCAACCACCAGGGCA-3′; 5′-CTCCCCGAGAGCCTGTCCAGA-3′), TGF-β1 (5′-TCAAGGCCGGTTTCCTGCTTCTCATG-3′; 5′-GCGGAAGTCAATGTACAGCTGCCGC-3′); glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 5′-CCACCATGGAGAAGGCTGGGGCTCA-3′, 5′-ATCACGCCACAGTTTCCCGGAGGGG-3′). Melting curve analysis was performed to verify specificity of products. Data were analyzed using the ΔCT method. Target gene expression was normalized to GAPDH by taking the difference between CT values for target genes and GAPDH (ΔCT value). These values were then calibrated to that of the control sample to give the ΔΔCT value. The fold target gene expression is given by the formula: 2−ΔΔCT. Experiments were performed 3 separate times, and results are shown as means of triplicate values.
Sodium bisulfite modification and methylation-specific PCR
Genomic DNA was extracted from BC1, BC3, BCBL1, and Ramos cell lines by GeneElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich); 1 μg of genomic DNA was bisulfite modified with EZ DNA Methylation kit (Zymo Research, Orange, CA) according to the manufacturer's protocol. Methylation-specific PCR was performed as described by Chen et al.40 Data shown are representative of 3 independent experiments.
ELISA
BC1, BC2, BC3, BC5, BCBL1, Ramos cells lines, and peripheral blood mononuclear cells (PBMCs) were plated in triplicate in 48-well plates. After 24 hours, cells were spun down and supernatants were collected and analyzed for TGF-β1 by enzyme-linked immunosorbent assay (ELISA) The level of TGF-β1 in control medium was subtracted from that of each sample to account for TGF-β1 present in FBS. Experiments were performed independently 3 times, and representative data are shown as mean plus or minus SEM.
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed as described (available at www.millipore.com/techpublications/tech1/mcproto407). Briefly, 107 cells were resuspended in sodium dodecyl sulfate lysis buffer, cross-linked with formaldehyde, sonicated, and immunoprecipitated with antiacetyl-histone H4 (Lys4), anti-Sp1 (Upstate Biotechnology, Charlottesville, VA), anti–HHV-8 ORF73 (Advanced Biotechnologies), or IgG control antibodies (Upstate Biotechnology/Millipore). Genomic DNA was isolated from the immunoprecipitated complex and used as a template for PCR with primers specific for the TβRII promoter (5′-GAGAGAGCTAGGGGCTGG-3′; 5′-CTCAACTTCAACTCAGCGCTGC-3′). ChIP assays were performed 3 times with each antibody, and representative data are shown.
Immunohistochemistry
Tissues samples of PEL, MCD, and KS were obtained after pathologic diagnosis with approval from the Weill Cornell Medical College Institutional Review Board. Double immunohistochemical staining for anti-HHV-8 ORF73 and TβRII was performed on formalin-fixed, paraffin-embedded tissue sections and fibrinogen-thrombin matrix cell clots using Bond Max Autostainer (Vision Biosystems, Waverley, Australia). Tissue sections, before immunostaining, were deparaffinized and endogenous peroxidase was inactivated. After antigen heat retrieval in Dako Cytomation Target Retrieval Solution (DakoCytomation, Glostrup, Denmark), sections were incubated sequentially with anti-HHV-8 ORF73 for 25 minutes, post primary for 15 minutes and polymer for 25 minutes (Bond Polymer Detection System; Vision Biosystems) followed by colorimetric development with diaminobenzidine (Vision Biosystems). After retrieval using Bond Epitope Retrieval solution at 99°C to 100°C for 30 minutes and blocking endogenous alkaline phosphatase using Dual Endogenous Enzyme Block (DakoCytomation), sections were incubated with the second primary antibody (anti-TβRII, C-16) twice for 25 minutes each time. Sections were then incubated with a biotinylated link and streptavidin-AP (LSAB 2 System-AP; DakoCytomation) for 25 minutes each followed by red chromagen development with permanent red (DakoCytomation). After counterstaining, slides were reviewed under an Olympus BX41 light microscope (Olympus America, Center Valley, PA) with a UPLAN F1 40×/0.75 objective lens. Images were obtained with a Q-Color 3 digital camera (Olympus America) and imported into Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) for processing.
RNA interference
siRNA duplexes were synthesized by Dharmacon RNA Technologies (Lafayette, CO). The siRNA target sequence to LANA mRNA is 5′-AAACAGGUCUCCGGAAAGAUG-3′.41 PEL cells were transfected with siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Silencing was assessed 48 hours after transfection.
Results
TGF-β signaling is defective in PEL
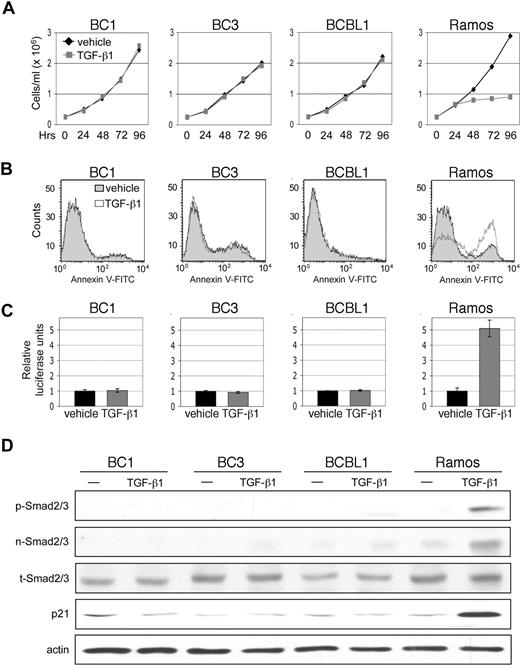
TGF-β signaling results in growth inhibition and apoptosis in lymphoid cells. We therefore examined the rate of growth and level of apoptosis in PEL cell lines in response to TGF-β1 treatment. Ramos, an EBV-negative Burkitt lymphoma cell line, was used to compare sensitivity of PELs to that of a TGF-β-responsive cell line. Treatment of PEL cell lines with TGF-β1 for 4 days resulted in no change in growth rate (Figure 1A). Examination of cell viability, in which levels of ATP production were measured, similarly showed no difference (data not shown). After 48 hours of TGF-β1 treatment, the level of apoptosis in PEL cell lines remained the same between cytokine-treated and vehicle-treated conditions (Figure 1B).
TGF-β signaling is defective in PEL. (A) Cell growth of PEL cell lines, BC1, BC3, and BCBL1, and TGF-β-sensitive, Burkitt lymphoma cell line, Ramos, over 4 days of treatment with TGF-β1. (B) Flow cytometric analysis of apoptosis 48 hours after TGF-β1 treatment. (C) TGF-β reporter, p3TP-Lux, activity in response to treatment with TGF-β1. Error bars represent SD. (D) Western blot detection of Smad2/3 phosphorylation (p-Smad2/3), Smad2/3 nuclear accumulation (n-Smad2/3), total Smad 2/3 (t-Smad2/3), and p21 expression on TGF-β1 treatment.
TGF-β signaling is defective in PEL. (A) Cell growth of PEL cell lines, BC1, BC3, and BCBL1, and TGF-β-sensitive, Burkitt lymphoma cell line, Ramos, over 4 days of treatment with TGF-β1. (B) Flow cytometric analysis of apoptosis 48 hours after TGF-β1 treatment. (C) TGF-β reporter, p3TP-Lux, activity in response to treatment with TGF-β1. Error bars represent SD. (D) Western blot detection of Smad2/3 phosphorylation (p-Smad2/3), Smad2/3 nuclear accumulation (n-Smad2/3), total Smad 2/3 (t-Smad2/3), and p21 expression on TGF-β1 treatment.
It is possible that constitutive activation of certain mitogenic pathways can override the effects of TGF-β signaling. In some instances, loss of the tumor suppressor pRb confers resistance to TGF-β.42,43 Cells that exhibit constitutive expression of c-Myc may also be refractory to TGF-β-mediated growth arrest.44,45 Overexpression of cyclin D1 has also been reported to antagonize growth inhibitory effects of TGF-β.46-48 Therefore, to determine whether a lack of TGF-β response in PEL is the result of a defect in TGF-β signaling or to dysregulation of an opposing pathway, we determined whether treatment with TGF-β1 could activate a TGF-β-responsive reporter. PEL cell lines were transfected with the p3TP-Lux plasmid, which contains 3 consecutive TPA response elements and a minimal fragment of the plasminogen activator inhibitor 1 promoter. No increase in reporter activity was observed on addition of TGF-β1 (Figure 1C).
For signal transduction to occur, activated TGF-β receptor complex must phosphorylate Smad2 and Smad3, which subsequently translocate to the nucleus. Treatment of PEL cells with TGF-β1 did not induce Smad2/3 phosphorylation or result in their nuclear accumulation. Furthermore, expression of p21, a downstream effector of TGF-β signaling, was not induced (Figure 1D).
Defective TGF-β signaling in PEL is due to lack of TβRII expression
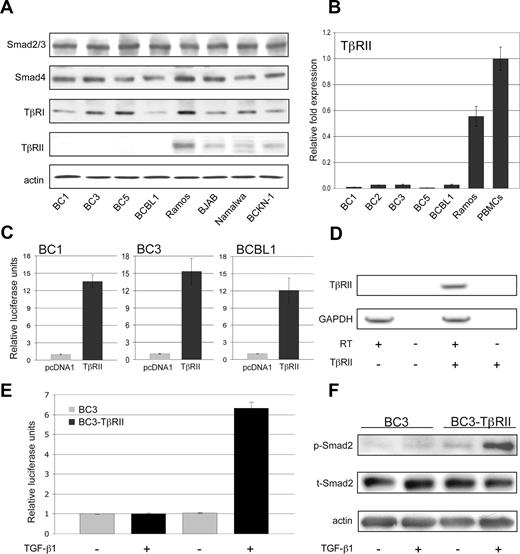
To identify the defect in TGF-β signaling, protein levels for each pathway component were analyzed in PEL cell lines (BC1, BC3, BC5, BCBL1) in relation to those of other B-cell lymphoma cell lines (Ramos, BJAB, Namalwa, BCKN-1; Figure 2A). TβRI protein expression was lower in some PEL cell lines; however, expression of TβRII was completely absent in all PEL cells. Quantitative RT-PCR analysis revealed the defect in TβRII expression to be at the mRNA level (Figure 2B).
Lack of TβRII expression is responsible for the TGF-β signaling defect in PEL. (A) Levels of TGF-β pathway components compared among PEL cells (BC1, BC3, BC5, BCBL-1) and other B-cell lymphoma cell lines (Ramos, BJAB, Namalwa, BCKN1) that express relatively normal levels of these components. (B) TβRII expression measured by real-time PCR. (C) TGF-β-responsive reporter activity on transfection of TβRII. (D) RT-PCR analysis of TβRII expression in BC3 cells transduced with pHR-Sin-CSGW-TβRII. (E) TGF-β–driven reporter activity and (F) Smad2 phosphorylation in TβRII-transduced BC3 cells. Error bars represent SD.
Lack of TβRII expression is responsible for the TGF-β signaling defect in PEL. (A) Levels of TGF-β pathway components compared among PEL cells (BC1, BC3, BC5, BCBL-1) and other B-cell lymphoma cell lines (Ramos, BJAB, Namalwa, BCKN1) that express relatively normal levels of these components. (B) TβRII expression measured by real-time PCR. (C) TGF-β-responsive reporter activity on transfection of TβRII. (D) RT-PCR analysis of TβRII expression in BC3 cells transduced with pHR-Sin-CSGW-TβRII. (E) TGF-β–driven reporter activity and (F) Smad2 phosphorylation in TβRII-transduced BC3 cells. Error bars represent SD.
We next examined whether TGF-β signaling could be rescued in PEL by restoring TβRII expression. p3TP-Lux was cotransfected with either pcDNA1-TβRII or pcDNA1 vector alone, followed by TGF-β1 treatment. Transfection of TβRII restored signaling as shown by an increase in reporter activity on addition of TGF-β1 (Figure 2C). BC3 cells were also transduced with the lentiviral construct pHR-Sin-CSGW-TβRII. TβRII expression was confirmed by RT-PCR (Figure 2D) and, in agreement with cotransfection studies, TGF-β reporter activity (Figure 2E) and Smad2 phosphorylation (Figure 2F) were stimulated on TGF-β1 treatment.
TβRII gene expression is subject to epigenetic silencing in PEL cells
Loss of TβRII expression can result from gene mutation, loss of expression or activity of transcription factors required for TβRII expression, or transcriptional silencing resulting from hypermethylation of CpG islands or promoter-proximal histone deacetylation. No mutations in the TβRII gene in PEL were found that would account for its lack of expression (data not shown).
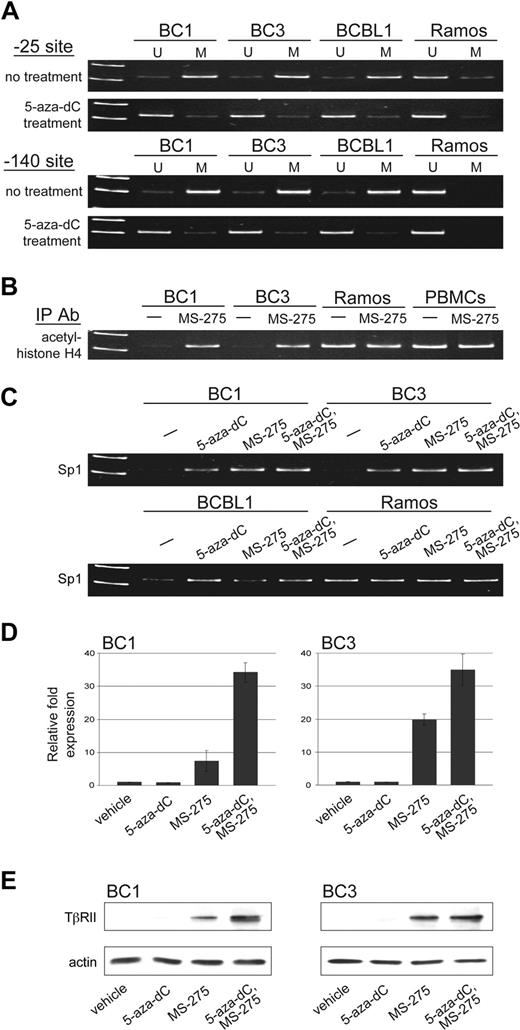
Methylation-specific PCR was performed to determine whether CpG islands found at positions −25 and −140 were methylated. Similar to other genes that lack TATA boxes in their promoters, TβRII expression relies heavily on Sp1 binding. Sp1 binding sites are located at these promoter regions and methylation at −140, and in some cases −25, has been shown to inhibit TβRII gene expression by preventing Sp1 binding.40,49-51 We observed methylation at both positions −25 and −140 in all PEL cell lines tested (Figure 3A). Methylation was observed in Ramos, which expresses TβRII, at −25, albeit to a lesser extent, but not at −140. Treatment with 5-aza-2-deoxycytidine (5-aza-dC) led to demethylation at both CpG sites.
TβRII gene expression is subject to epigenetic silencing in PEL cells. (A) Methylation status at positions −25 and −140 of the TβRII promoter in PEL cell lines, BC1, BC3, and BCBL1, and Burkitt lymphoma cell line, Ramos, before and after 5-aza-dC treatment determined by methylation-specific PCR (U indicates unmethylated; M, methylated). (B) ChIP analysis for acetylated histone H4 and (C) Sp1 under the indicated conditions for 48 hours. (D) TβRII expression analyzed by real-time PCR and (E) Western blot after indicated treatments. Error bars represent SD.
TβRII gene expression is subject to epigenetic silencing in PEL cells. (A) Methylation status at positions −25 and −140 of the TβRII promoter in PEL cell lines, BC1, BC3, and BCBL1, and Burkitt lymphoma cell line, Ramos, before and after 5-aza-dC treatment determined by methylation-specific PCR (U indicates unmethylated; M, methylated). (B) ChIP analysis for acetylated histone H4 and (C) Sp1 under the indicated conditions for 48 hours. (D) TβRII expression analyzed by real-time PCR and (E) Western blot after indicated treatments. Error bars represent SD.
ChIP assays were performed to assess the level of histone acetylation in chromatin associated with the TβRII promoter region. Acetylated histone H4, seen in Ramos and PBMCs, was not associated with the TβRII promoter in PEL cell lines (Figure 3B). Treatment with the histone deacetylase inhibitor MS-275 resulted in histone H4 acetylation in PEL cells. ChIP assays were also performed to examine association of Sp1 with the TβRII promoter. Binding of Sp1 to the TβRII promoter in PEL cells was lower compared with that in Ramos cells (Figure 3C). DNA methylation and histone deacetylation create a tightly packed DNA conformation that restricts transcription factors from binding. Treatment with agents that reverse these epigenetic changes loosens chromatin structure and allows for transcription factor binding. When we treated PEL cells with 5-aza-dC and MS-275, Sp1 binding to the TβRII promoter was restored.
Treatment with demethylating agents as well as histone deacetylase inhibitors has been shown to induce TβRII expression in various tumor types.51-57 We examined whether 5-aza-dC and MS-275 could restore expression of endogenous TβRII in PEL cell lines. Cells were treated with either vehicle alone, each drug individually, or both drugs for 48 hours and TβRII expression was analyzed by RT-PCR (Figure 3D) and immunoblot (Figure 3E). Treatment with 5-aza-dC alone had no observed effect on TβRII transcription. MS-275 treatment, on the other hand, led to induced expression of TβRII, and treatment with both drugs resulted in a synergistic increase in TβRII levels.
PEL cells express and secrete elevated levels of TGF-β1
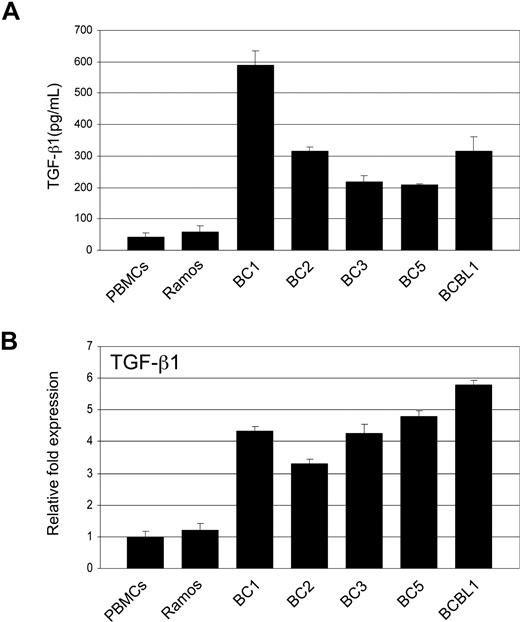
Tumor cells that have become resistant to TGF-β signaling often produce elevated levels of TGF-β. We compared TGF-β1 secretion in PEL cell lines to that of Ramos and PBMCs. ELISA analysis showed significantly higher TGF-β1 secretion in all PEL cell lines (Figure 4A). RT-PCR analysis revealed TGF-β1 gene expression to be more than 3-fold higher in PEL cells (Figure 4B).
PEL cells secrete elevated levels of TGF-β1. (A) TGF-β1 levels in supernatants taken from PEL cell lines, Ramos, and PBMCs determined by ELISA. (B) Real-time PCR analysis of TGF-β1 in PEL and Ramos cell lines and PBMCs. Error bars represent SD.
PEL cells secrete elevated levels of TGF-β1. (A) TGF-β1 levels in supernatants taken from PEL cell lines, Ramos, and PBMCs determined by ELISA. (B) Real-time PCR analysis of TGF-β1 in PEL and Ramos cell lines and PBMCs. Error bars represent SD.
KSHV-positive cells in primary cases of PEL, KS, and MCD exhibit decreased levels of TβRII
To determine whether lower TβRII expression correlates with KSHV infection in primary cases of PEL, KS, and MCD, we performed double immunohistochemical staining for TβRII and LANA on PEL cell clots and KS and MCD lesions (Figure 5). Cells in suspension were made into a clot, fixed, and embedded in paraffin to best recapitulate tissue staining (Figure 5A,B). A 1:1 mixture of BC3 and Ramos cells was stained to show the contrast in expression of TβRII and LANA between the KSHV positive PEL cell line, BC3, and the TGF-β–sensitive, KSHV negative Ramos cell line (Figure 5A). BC3 cells stained reliably for LANA only (black arrow) as indicated by brown nuclear positivity, whereas Ramos cells were positive only for TβRII (white arrow) as shown by red staining at the membrane and in the cytoplasm. Primary PEL cells consistently displayed little to no TβRII positivity, whereas some small, reactive, LANA-negative cells stained positively for TβRII (Figure 5B). Some background TβRII staining resulted from reactivity with fibrinogen used in making cell clots, but lack of TβRII could be appreciated in cells surrounded by a clear space. The correlation between LANA positivity and lower levels of TβRII staining was not absolute in MCD and KS tissue sections (Figure 5C,D). Whereas the presence of cells positive for only LANA (black arrow) or TβRII (white arrow) was frequently observed, there were also cells exhibiting positivity for both LANA and TβRII (blue arrow).
KSHV-positive cells in primary cases of PEL, KS, and MCD exhibit decreased levels of TβRII. (A) Double immunohistochemical staining for TβRII and LANA in a cell clot containing a 1:1 mixture of BC3 cells (black arrow) and Ramos cells (white arrow), (B) a primary PEL specimen cell clot containing neoplastic (black arrow) and reactive (white arrow) cells, (C) a MCD tissue section, and (D) a KS tissue section. LANA positivity appears as brown nuclear staining and TβRII appears red at the membrane and in the cytoplasm. Black arrows indicate LANA+, TβRII− cell; white arrow, LANA−, TβRII+ cell; blue arrow, LANA+, TβRII+ cell (original magnification ×132)
KSHV-positive cells in primary cases of PEL, KS, and MCD exhibit decreased levels of TβRII. (A) Double immunohistochemical staining for TβRII and LANA in a cell clot containing a 1:1 mixture of BC3 cells (black arrow) and Ramos cells (white arrow), (B) a primary PEL specimen cell clot containing neoplastic (black arrow) and reactive (white arrow) cells, (C) a MCD tissue section, and (D) a KS tissue section. LANA positivity appears as brown nuclear staining and TβRII appears red at the membrane and in the cytoplasm. Black arrows indicate LANA+, TβRII− cell; white arrow, LANA−, TβRII+ cell; blue arrow, LANA+, TβRII+ cell (original magnification ×132)
LANA inhibits TβRII expression through epigenetic silencing
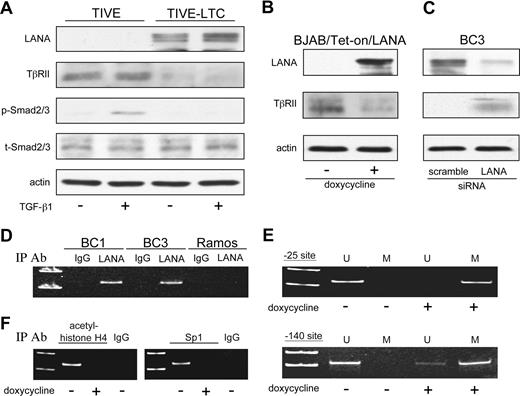
To further establish a link between down-regulation of TβRII expression and KSHV infection, TIVE that have been infected with KSHV and shown to maintain long-term infection (LTCs)35 were analyzed for expression of TβRII. LTCs exhibited lower TβRII protein expression compared with uninfected TIVE cells (Figure 6A). TGF-β1 treatment did not induce phosphorylation of Smad2/3 in these cells as it did in uninfected TIVE cells.
LANA inhibits TβRII expression through epigenetic silencing. (A) Western blot for TβRII and phosphorylated Smad2/3 in KSHV+ LTCs and their uninfected TIVE cell counterparts in response to TGF-β1 treatment. (B) Expression of TβRII on induction of LANA in BJAB/Tet-on/LANA cells and (C) on siRNA knockdown of LANA in BC3 cells. (D) ChIP analysis of LANA association with the TβRII promoter. (E) Methylation status at positions −25 and −140 of the TβRII promoter in BJAB/Tet-on/LANA cells determined by methylation-specific PCR (U indicates unmethylated; M, methylated). (F) ChIP analysis of acetylated histone H4 and Sp1 at the TβRII promoter in BJAB/Tet-on/LANA cells.
LANA inhibits TβRII expression through epigenetic silencing. (A) Western blot for TβRII and phosphorylated Smad2/3 in KSHV+ LTCs and their uninfected TIVE cell counterparts in response to TGF-β1 treatment. (B) Expression of TβRII on induction of LANA in BJAB/Tet-on/LANA cells and (C) on siRNA knockdown of LANA in BC3 cells. (D) ChIP analysis of LANA association with the TβRII promoter. (E) Methylation status at positions −25 and −140 of the TβRII promoter in BJAB/Tet-on/LANA cells determined by methylation-specific PCR (U indicates unmethylated; M, methylated). (F) ChIP analysis of acetylated histone H4 and Sp1 at the TβRII promoter in BJAB/Tet-on/LANA cells.
As mentioned previously, KSHV-encoded LANA has been linked to epigenetic gene regulation.16,22,58 Because both histone modifications and DNA methylation were found to play a role in TβRII gene transcription in PEL, we examined whether LANA was responsible for its repression. Induction of LANA in BJAB/Tet-On/LANA cells led to a decrease in TβRII (Figure 6B), whereas siRNA knockdown of LANA in BC3 cells resulted in TβRII expression (Figure 6C). ChIP assays revealed that LANA was bound to the TβRII promoter in BC1 and BC3 cells (Figure 6D). Furthermore, induction of LANA in BJAB/Tet-on/LANA cells resulted in increased methylation at the −25 and −140 sites in the TβRII promoter (Figure 6E) as well as decreased acetylation of TβRII promoter-associated histone H4 and decreased Sp1 binding to the TβRII promoter (Figure 6F).
Restoration of TβRII by 5-aza-2-deoxycytidine and MS-275 sensitizes PEL cells to apoptotic and cytostatic effects of TGF-β signaling
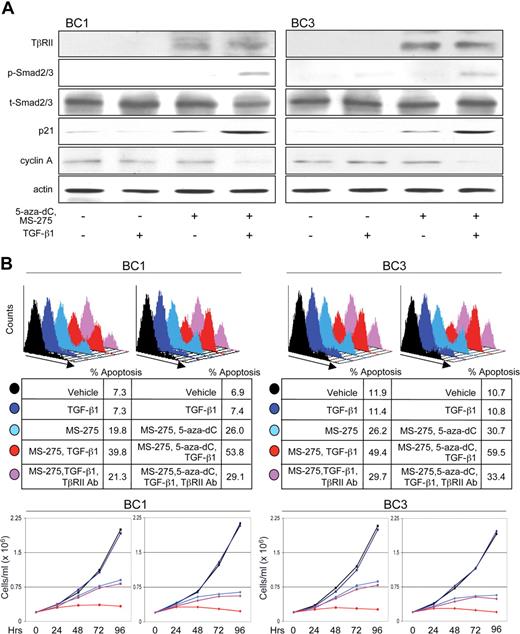
We examined whether restoring TβRII in PEL cell lines by treating with 5-aza-dC and MS-275 could rescue TGF-β signaling and lead to growth inhibition and/or apoptosis. Treatment of BC1 and BC3 cells induced expression of TβRII and, on addition of TGF-β1, resulted in Smad2/3 phosphorylation (Figure 7A). Effects on downstream targets were also observed; p21 expression was enhanced and cyclin A was down-regulated. p21 increased on treatment with MS-275 and 5-aza-dC and was further induced on addition of TGF-β1. The up-regulation in p21 expression by drug treatment alone may be due to the reversal of epigenetic silencing as this mechanism is responsible for silencing cyclin-dependent kinase inhibitors in several other malignancies.59-61
Restoration of TβRII by 5-aza-2-deoxycytidine and MS-275 sensitizes PEL cells to apoptotic and cytostatic effects of TGF-β signaling. (A) Analysis of the TGF-β signaling cascade and downstream gene targets, p21 and cyclin A, on indicated treatments. (B) Flow cytometric analysis of apoptosis (top panel) and growth curve (bottom panel) of BC1 and BC3 cells in response to indicated treatments. Line colors correspond to treatment conditions described in the top panel.
Restoration of TβRII by 5-aza-2-deoxycytidine and MS-275 sensitizes PEL cells to apoptotic and cytostatic effects of TGF-β signaling. (A) Analysis of the TGF-β signaling cascade and downstream gene targets, p21 and cyclin A, on indicated treatments. (B) Flow cytometric analysis of apoptosis (top panel) and growth curve (bottom panel) of BC1 and BC3 cells in response to indicated treatments. Line colors correspond to treatment conditions described in the top panel.
To measure PEL sensitivity to apoptotic and cytostatic effects of TGF-β signaling, cells were treated with MS-275 alone or together with 5-aza-dC in the presence or absence of TGF-β1 (Figure 7B). Treatment with drugs alone led to an increase in apoptosis and decrease in growth rate, possibly because of re-expression of other epigenetically regulated tumor suppressor genes. Addition of TGF-β1 led to a further increase in apoptosis and inhibition in growth. Treatment with a blocking antibody to TβRII before TGF-β1 lowered the level of apoptosis and restored the rate of growth essentially to that of drug treatment alone. Together, 5-aza-dC and MS-275 had an additive effect on both apoptotic and cytostatic programs, both in the presence and absence of TGF-β1.
Discussion
TGF-β signaling plays an important role in both growth and survival of several cell types, as well as in regulation of certain aspects of immune surveillance. It is not surprising, therefore, that this pathway is frequently deregulated in cancer as it creates an advantage for tumor cells in several ways. Often tumors develop a defect in a member of the signaling cascade, which makes them no longer susceptible to its adverse growth effects. In addition, they frequently secrete elevated amounts of TGF-β that inhibit proliferation of surrounding cells and preclude T cells from mounting an effective antitumor immune response.
Solid tumors frequently acquire mutations in a component of the pathway, whereas hematologic cancers often gain resistance to TGF-β through down-regulation of one of the receptors or through repression of TGF-β signaling by oncoproteins, such as Evi-1 and Tax.62 Here we show that PEL cell lines are defective in TGF-β signaling because of a lack of TβRII expression. Restoration of TβRII expression established pathway function and sensitivity to its growth inhibitory and proapoptotic effects. In addition, PEL cells secrete elevated levels of TGF-β1, which may create in vivo an immune-suppressed microenvironment favorable to their growth.
Lack of TβRII expression in PEL is the result of epigenetic alterations, rather than mutations in the TβRII gene. Silencing of the TβRII gene has been reported to result from histone deacetylation and/or promoter methylation in cancers, such as lung, breast, prostate, pancreatic, renal cell carcinoma, and B-cell lymphoma.40,50,53,54,63-66 We found that in PEL cell lines both methylation and histone deacetylation play a role in TβRII transcriptional repression.
The promoter of the TβRII gene has previously been characterized.49 It lacks a TATA box near its transcription initiation site and contains 4 regulatory elements: 2 positive regulatory elements, PRE-1 and PRE-2, located at −219 to −172 and +1 to +50, respectively; a negative regulatory element, located at −100 to −67; and a core promoter at −47 to −1. Binding of Sp1 is required for TβRII transcription. Its promoter has 4 Sp1 consensus binding sites at positions −25, −59, −102, and −143.49,67 Many transcription factor binding sites, such as those for NF-Y, AP1, CREB, and ERT, are also present. Mutations in Sp1 sites at −102 and −59 have been shown to reduce TβRII expression by 70% and 40%, respectively.67 Zhao et al reported that TβRII repression in human pancreatic cancer cells is the result of histone deacetylation at the −102 Sp1 site.64 Other studies have shown that DNA methylation at positions −140 and −25 plays a role in TβRII gene silencing.40,50,51 We show that not only is the TβRII promoter methylated at both of these sites in PEL cells but also that TβRII promoter-associated histone H4 is hypoacetylated. We further demonstrate that these interrelated epigenetic mechanisms are responsible for the decreased binding of Sp1 to the TβRII promoter and probably obstruct the binding of other transcription factors as well.
Epigenetic gene regulation is receiving increased attention as its involvement in cancer development and progression becomes more apparent. Histone deacetylase (HDAC) inhibitors and demethylating agents have shown potential as antitumor agents in many cancer cell lines, and clinical trials are being conducted. Combination drugs that affect both mechanisms have synergistic effects on apoptosis, differentiation, and growth arrest in lung, breast, thoracic, leukemia, and colon cancer cell lines. Here we show that treatment with MS-275 alone was capable of inducing TβRII expression and combined treatment with 5-aza-dC resulted in further induction in PEL cell lines. These treatments led to increased apoptosis and reduced cell growth, with addition of TGF-β1 enhancing their effects. Although cytostatic effects of drug treatment alone were probably the result of derepression of other silenced tumor suppressor genes, the increase in apoptosis and growth inhibition on TGF-β1 treatment resulted directly from signaling through the TGF-β pathway because addition of a TβRII blocking antibody negated these effects.
PEL most commonly arises in patients with underlying immunodeficiency, such as AIDS. It is generally resistant to cancer cytotoxics that are active against other lymphomas, and it carries a very poor prognosis. Our data suggest that HDAC inhibitors alone or in conjunction with demethylating agents may be a promising treatment for PEL and possibly KS and MCD. PEL cells secrete elevated levels of TGF-β1, which in vivo can suppress the antitumor immune response. Reversing epigenetic silencing of TβRII through the use of these agents would render PEL susceptible to TGF-β present in their microenvironment. In addition to turning on TGF-β signaling and other important regulatory networks, treatment with demethylating agents and HDAC inhibitors can also induce RTA expression and consequently KSHV lytic replication,68,69 suggesting that they may be used in conjunction with antivirals, such as gancyclovir, as an effective treatment for PEL and other KSHV-related diseases.
Induction of lytic replication leads to the expression of K-bZip and vIRF1, which have been reported to inhibit the TGF-β pathway.32,33 We found that treatment with 5-aza-dC and MS-275 led to KSHV lytic induction in 12% to 16% of BC1 and BC3 cells (data not shown). These cells would likely be resistant to TGF-β treatment because of the expression of K-bZip and vIRF1, suggesting that sensitivity to TGF-β signaling on treatment with 5-aza-dC and MS-275 would be even greater if they were not expressed.
Several viruses have been reported to target the TGF-β pathway.70-78 We found that KSHV targets this pathway as well. KSHV-infected TIVE cells had reduced TβRII expression and diminished signaling through this pathway. One of the KSHV latent gene products, LANA, interacts with several proteins involved in gene regulation, such as DNMT3a, SUV39HI, MeCP2, mSin3, HP1, and p300/ CBP.14,16,20-22,79 Here we show that LANA is responsible for silencing TβRII expression. A BJAB/Tet-on/LANA cell line showed lower TβRII expression on LANA induction, whereas siRNA knockdown of LANA in BC3 led to increased TβRII. We further show LANA binds the TβRII promoter in PEL cells and induction of LANA in BJAB/Tet-on/LANA leads to DNA methylation, deacetylation of histone H4, and reduced Sp1 binding at the TβRII promoter. Based on these findings, we propose a mechanism whereby LANA regulates TβRII gene expression through epigenetic silencing.
LANA represses transcription of ORF50. Lu et al found that treatment with HDAC inhibitors led to LANA acetylation and disrupted LANA association with the ORF50 promoter, Sp1, and histone H2B.10 It is possible that the effects of MS-275 on TβRII expression are not only the result of the reversal of epigenetic silencing through histone acetylation but also to LANA acetylation, which may lead to its dissociation from the TβRII promoter. Future studies will address the impact of LANA acetylation on TβRII expression.
KSHV has developed several mechanisms for blocking TGF-β signaling. In addition to LANA-mediated TβRII suppression, lytic proteins K-bZip and vIRF1 have been shown to interfere with this pathway.32,33 KSHV microRNAs were recently reported to inhibit TGF-β activity as well.80 Based on similarities to their cellular homologs, vIRFs, vIL-6, vGPCR, and microRNAs may also be involved in epigenetic regulation81 and, as such, contribute to TβRII gene silencing. The number of ways KSHV interferes with TGF-β signaling intimates an importance in blocking this pathway for virus survival. Our results suggest that the TGF-β pathway is involved in development and progression of KSHV-related malignancies and provide insight into a potentially efficacious strategy for the treatment of these diseases.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
ELISAs were performed at the Cytokine Analysis Laboratory at the Fred Hutchinson Cancer Research Center, Seattle, WA; Dr George B. McDonald, Director.
This work was supported by National Institutes of Health (NIH)/National Cancer Institute grant R01-CA68939 (E.C.), NIH/National Institute for Allergy and Infectious Diseases grant K08-AI153971 (M.C.), and Cancer Research Institute Tumor Immunology Predoctoral Training Grant (D.D.).
National Institutes of Health
Authorship
Contribution: D.L.D. designed and performed most of the research, analyzed data, and wrote the paper; M.C. performed some experiments; Y.-F.L. performed immunohistochemistry; R.R. contributed essential reagents; A.C. contributed tissues, supervised, and interpreted immunohistochemistry; C.B. supervised some experiments; E.C. designed and supervised research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ethel Cesarman, Department of Pathology and Laboratory Medicine, Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10021; e-mail: ecesarm@med.cornell.edu.